Abstract
Continuing evolution of highly pathogenic (HP) H5N1 influenza viruses in wild birds with transmission to domestic poultry and humans poses a pandemic threat. There is an urgent need for a simple and rapid serological diagnostic assay which can differentiate between antibodies to seasonal and H5N1 strains and that could provide surveillance tools not dependent on virus isolation and nucleic acid technologies. Here we describe the establishment of H5N1 SeroDetect enzyme-linked immunosorbent assay (ELISA) and rapid test assays based on three peptides in HA2 (488-516), PB1-F2 (2-75), and M2e (2-24) that are highly conserved within H5N1 strains. These peptides were identified by antibody repertoire analyses of H5N1 influenza survivors in Vietnam using whole-genome-fragment phage display libraries (GFPDLs). To date, both platforms have demonstrated high levels of sensitivity and specificity in detecting H5N1 infections (clade 1 and clade 2.3.4) in Vietnamese patients as early as 7 days and up to several years postinfection. H5N1 virus-uninfected individuals in Vietnam and the United States, including subjects vaccinated with seasonal influenza vaccines or with confirmed seasonal virus infections, did not react in the H5N1-SeroDetect assays. Moreover, sera from individuals vaccinated with H5N1 subunit vaccine with moderate anti-H5N1 neutralizing antibody titers did not react positively in the H5N1-SeroDetect ELISA or rapid test assays. The simple H5N1-SeroDetect ELISA and rapid tests could provide an important tool for large-scale surveillance for potential exposure to HP H5N1 strains in both humans and birds.
INTRODUCTION
Highly pathogenic avian influenza (HPAI) A viruses of the H5N1 subtype are still causing widespread infections and significant lethality in bird populations throughout Southeast Asia, with spread into Central Asia, Africa, and Europe (5). In unvaccinated chickens, H5N1 infectiousness develops very quickly after infection (0.25 day) and transmission among birds is very efficient (3, 18). Numerous instances of human transmission have occurred, resulting in severe disease or death (9, 23, 24, 26, 28). As of 2 August 2011, there have been 563 human cases of H5N1 infection, resulting in 330 deaths (mortality = 59%).
In the absence of preexisting immunity against these strains, it is feared that further adaptation of the HPAI viruses for human-to-human transmission will result in a global pandemic. Currently, the only assay for diagnosis of H5N1 infection is a reverse transcription-PCR (RT-PCR)-based nucleic acid detection assay that involves isolation of viral RNA from infected individuals and cannot diagnose patients more than 2 to 3 weeks post-H5N1 infection, as RNA is not detectable after the clearance of virus. Therefore, H5N1 virus is detected in individuals mainly during the period of acute illness. Hence, the number of human H5N1 influenza cases is likely underreported, missing nonsymptomatic H5N1 virus exposure cases (household and contact cases and also people exposed to poultry and environmental sources). Currently available serologic tests include hemagglutination inhibition (HI), microneutralization (MN) assay, enzyme-linked immunosorbent assay (ELISA), and the agar gel precipitation test. However, these assays suffer from low sensitivity, are labor-intensive, and often require specialized biocontainment facilities, which prevents the usage of these assays for rapid screening outside a central laboratory setting. The development of a simple and rapid serodiagnostic method that can detect antibodies soon after infection as well as antibodies lasting long (months to years) after H5N1 infection is urgently required for large-scale surveillance of humans and domestic poultry and could provide an early warning of an impending pandemic with HP H5N1 viruses. Moreover, it is very important to differentiate between vaccine-induced antibody responses and natural infection in poultry and humans. Most of the currently available influenza virus rapid diagnostic tests are capable of detecting seasonal influenza virus infection but have demonstrated low sensitivity for detection of H5N1 infections and do not discriminate between infection with seasonal virus and that with avian influenza virus (21, 25). Previously developed ELISAs for H5N1 infection serodiagnosis suffered from false-positive results due to cross-reacting antibodies against seasonal influenza viruses (7, 8, 20).
In an earlier publication, we reported the construction of whole-genome-fragment phage display libraries (GFPDLs) expressing fragments of 15 to 350 amino acids (aa) covering all the open reading frames of the H5N1 A/Vietnam/1203/2004 strain. These GFPDLs were used to analyze sera of five individuals who had recovered from H5N1 infection. Among the epitopes recognized by the H5N1 convalescent-phase sera, we have identified several potentially protective targets in HA1, the NA catalytic site, and the M2 ectodomain. In addition, for the first time in humans, we identified strong reactivity against PB1-F2, a putative virulence factor, following H5N1 infection. Importantly, a few novel epitopes were identified, which were recognized by H5N1 convalescent-phase sera but did not react with sera from control individuals (H1N1 or H3N2 seropositive but H5N1 negative) (15). Three of these immunodominant epitopes, mapping to conserved regions in H5N1 HA2, M2e, and PB1-F2, were further evaluated as the basis for development of a serodiagnostic assay in ELISA as well as a rapid test platform. The current study demonstrates the specificity and sensitivity of the new H5N1 serodiagnostic assays termed H5N1-SeroDetect using samples from both acute-phase H5N1 infections (from individuals who died or survived) and long-term convalescent individuals in Vietnam. Importantly, the H5N1-SeroDetect tests did not react with sera containing antibodies against seasonal influenza virus following either vaccination or infection, nor with samples from individuals vaccinated with inactivated H5N1 vaccines.
MATERIALS AND METHODS
Plasma and serum samples.
Serum samples from 40 H5N1 virus-exposed individuals who died or survived following H5N1 infection in Vietnam were obtained at multiple time points within 4 to 1,449 days following onset of symptoms from 2004 to 2010. Plasma samples from 82 Vietnamese community influenza cohort members (aged 5 to 80 years, 31 [38%] male) resident in Ha Nam province, where H5N1 poultry outbreaks and human cases have occurred, but with no known history of H5N1 virus exposure, were used as controls in antibody binding experiments. The samples were collected in December 2007 from 31 subjects, in December 2008 from 45 subjects, and at both times from 6 subjects. Of the 51 subjects assessed in December 2008, 32 had a seasonal H1N1 infection during 2008 and 4 had a seasonal H3N2 infection. Additional plasma samples from 25 Vietnamese adults (all females, aged 20 to 39 years) resident in Vietnam but with no history of H5N1 virus exposure were also used as controls.
Plasma samples from 50 U.S. residents with culture-confirmed human seasonal influenza virus infections and high HI titers against H1N1 and H3N2 strains circulating during the 2004 to 2007 seasons were used as additional negative controls (no possible H5N1 virus exposure). Pre- and postvaccination samples were obtained from subjects enrolled in U.S. clinical trials with either the seasonal trivalent influenza vaccine (TIV) or with the H5N1 vaccine. For the H5N1 vaccine study, inactivated unadjuvanted subunit H5N1 vaccine for the clade −1 H5N1 strain (rgA/Vietnam/1203/04 × A/PR/8/34) that contained 90 or 120 μg/ml hemagglutinin (HA) was used as a vaccine (Sanofi Pasteur) (1). For these vaccinated individuals, ∼50% seroconversion was observed following two vaccinations with HI titers ranging from 10 to 640.
All study protocols were in accordance with the Helsinki Declaration and with Good Clinical Practice principles and were approved by the local ethical committees. All samples were deidentified. All protocols were evaluated by the CBER/NIH Research Involving Human Subjects Committee (RIHSC) and were conducted under RIHSC exemption 03-118B.
H5N1-SeroDetect ELISA.
The inserts of the phage clones demonstrating high reactivity with H5N1-seropositive sera were sequenced and mapped to different H5N1 proteins (Fig. 1). Three immunodominant sequences that did not show cross-reactivity with normal (H5N1 virus-uninfected) serum samples from Vietnam were selected for the development of a new H5N1 serodiagnostic assay, termed H5N1-SeroDetect. Based on preliminary screening of H5N1-seronegative and -seropositive sera (as determined by HI or microneutralization [MN] assays), the optimal conditions for the H5N1 peptide PB1-F2, M2e, and HA2 ELISA were determined. Biotinylated peptides (1 μg/well) were captured onto wells coated with 200 ng of streptavidin. Following three washes with phosphate-buffered saline (PBS) containing Tween 20 (PBST) (20 mM PBS, 0.1% Tween 20), plates were blocked with PBST containing 2% whole milk (WMPBST). For testing, all specimens were diluted 1:100 in WMPBST and were added to peptide-coated wells for 1 h at room temperature (RT). After three washes with PBST, the wells were reacted with horseradish peroxidase (HRP)-conjugated goat anti-human IgG Fc-specific antibody (diluted 1:5,000) (Jackson ImmunoResearch, West Grove, PA) at RT for 1 h, followed by addition of O-phenylenediamine (OPD) substrate.
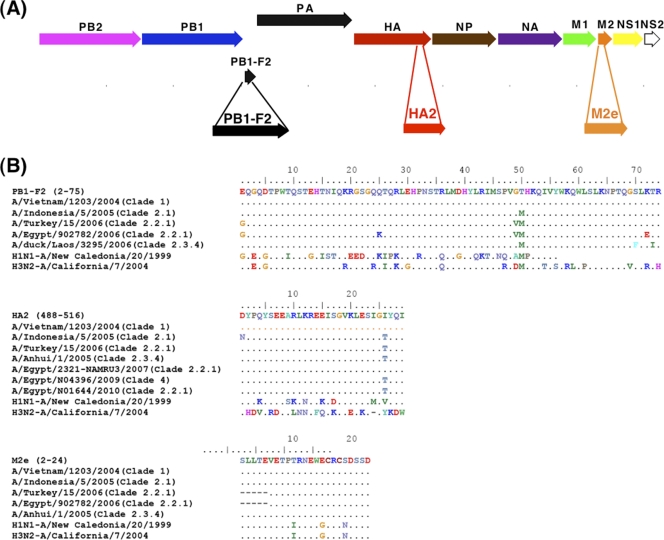
Fig. 1.
Identification of H5N1 peptides for serodiagnosis of H5N1 infection using a gene fragment phage display library. (A) The 11 proteins encoded by eight RNA segments in the H5N1 virus are depicted. Alignment of H5N1 sequences displayed on the affinity-selected phage clones after the second round of panning with the H5N1 genome sequence (H5N1 A/Vietnam/1203/2004) led to identification of the immunodominant regions recognized by antibodies generated after H5N1 infection in PB1-F2, HA2, and M2e. (B) Sequence alignment of selected peptides in PB1-F2, HA2, and M2e that are being used in H5N1-SeroDetect with other H5N1 strains from different clades and seasonal influenza virus strains is shown. The numbers in each epitopic site indicate the positions of the amino acid residues in the corresponding H5N1-encoded proteins for the A/Vietnam/1203/2004 strain.
The cutoff values used are the average absorbance of negative sera (at 1:100 dilution) + 5 standard deviations (for each peptide).
Peptides used in new H5N1-SeroDetect.
Biotinylated peptide sequences from H5N1-PB1-F2 (2-EQGQDTPWTQSTEHTNIQKRGSGQQTQRLEHPNSTRLMDHYLRIMSPVGTHKQIVYWKQWLSLKNPTQGSLKTR-75), HA2 (488-DYPQYSEEARLKREEISGVKLESIGIYQI-516), and M2e (2-SLLTEVETPTRNEWECRCSDSSD-24) were chemically synthesized (amino acid residues are numbered based on the positions of the amino acid residues in the corresponding H5N1-encoded proteins for the A/Vietnam/1203/2004 strain). All peptides were synthesized at the Facility for Biotechnology Resources, CBER, FDA, on Applied Biosystems peptide synthesizer models 431 and 433 (Foster City, CA) by standard 9-fluorenylmethoxy carbonyl (Fmoc) chemistry. Peptides were purified by reverse-phase high-performance liquid chromatography (RP-HPLC) and characterized by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS).
H5N1-SeroDetect rapid test.
Three test zone membrane regions containing H5N1 peptides PB1-F2 (2–75), M2e (2–24), and HA2 (488–516), were striped by capture onto a streptavidin-coated nitrocellulose membrane (Whatman; 5-μm nitrocellulose) at 1 mg/ml of peptide, with approximately 1 μl of antigen per cm of membrane along with a positive control. The control line was goat anti-human IgG (H&L) (Jackson ImmunoResearch), which functioned as an internal control for the addition of specimen and the proper functioning of the test. At one end of the strip (distal to the control line), a sample pad (containing fibrous mesh material and protein A colloidal gold conjugate) was present. The membrane capture reagent (protein A colloidal gold conjugate) reacts primarily with human immunoglobulins via binding to immunoreactive domains on the Fc portion of IgG. To run the test, the sample pad at the bottom of the strip was immersed in a 50-fold dilution of a serum/plasma sample to absorb the sample and allow it to migrate through the membrane by capillary lateral flow. H5N1 peptide PB1-F2, peptide M2e, and peptide HA2 on the membrane surface react with antigen-specific antibodies present in the sample, which are themselves bound to the protein A-coated gold conjugate, resulting in a colored line in the corresponding test zone region of the membrane. A positive reaction in any of the test antigen lines was termed H5N1 seropositive.
RESULTS
Identification of peptides for serodiagnosis of H5N1 infection using gene fragment phage display library.
The genome localization of the three immunodominant epitopes identified by complete epitope repertoire analysis using GFPDL screening of five convalescent-phase plasma samples from H5N1 (A/Vietnam/1203/2004) influenza survivors (13) is shown in Fig. 1A. Sequence alignment of the selected peptides in PB1-F2, HA2, and M2e with other H5N1 human isolates revealed a very high degree of sequence conservation among multiple H5N1 isolates from diverse clades and subtypes, including recent isolates (Fig. 1B; see also Fig. S1 in the supplemental material). In contrast, seasonal type A influenza virus as well as the recently circulating swine-like H1N1pdm09 strain shows multiple amino acid differences in these regions (Fig. 1B).
Development of H5N1-SeroDetect ELISA: specificity and sensitivity.
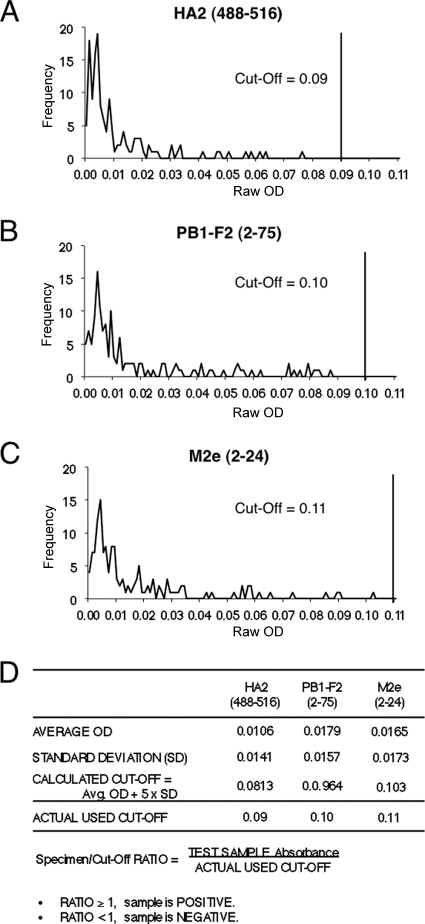
Biotinylated H5N1 peptides PB1-F2 (aa 2 to 75), M2e (aa 2 to 24), and HA2 (aa 488 to 516) were chemically synthesized and captured onto wells coated with streptavidin. Figure 2 shows the reactivity of each individual peptide with 139 serum samples from H5N1-negative individuals in the United States and Vietnam. Based on the results with seronegative samples, cutoff values were determined for the three peptides individually. The cutoff values ranged between 0.09 and 0.11 for the three peptides used in the H5N1-SeroDetect ELISA (Fig. 2D).
Fig. 2.
Seroreactivity of H5N1-negative samples with H5N1-SeroDetect peptides and determination of cutoff values. ELISA conditions were described in Materials and Methods. (A to C) Reactivities of 139 H5N1-seronegative serum/plasma samples from the United States and Vietnam at a 1:100 dilution with the peptides HA2 (488–516) (A), PB1-F2 (2–75) (B), and M2e (2–24) (C) peptides are shown. Values on the x axis are the test specimen ODs with the H5N1 peptide in specific ELISA. The y axis represents the number of serum/plasma samples that exhibited a specific ELISA absorbance reading as shown on the x axis. (D) The cutoff value for each peptide was determined as the mean absorbance + 5 standard deviations obtained with H5N1-seronegative samples.
Table 1 summarizes the results obtained with serum samples from five patients who survived H5N1 infection in Vietnam in 2004. The ELISA absorbance values at 490 nm for an average of triplicates are reported. These samples were obtained within 2 to 6 months postinfection and were previously described (15, 22). All five samples reacted with the HA2 and PB1-F2 sequences, and 4/5 samples reacted with M2e peptide in ELISA. Five control plasma samples from H5N1 virus-uninfected Vietnamese individuals were negative (Table 1).
Table 1.
H5N1-SeroDetect reactivities of control and post-H5N1 infection human sera
| Peptide | ELISA OD reading for serum sample reactivity with individually coated peptide |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Post-H5N1 infection human sera |
Control human sera |
|||||||||
| Viet-1 | Viet-2 | Viet-3 | Viet-4 | Viet-5 | C-1 | C-2 | C-3 | C-4 | C-5 | |
| HA2 (488–516) | 1.857 | 2.842 | 3.217 | 2.981 | 3.458 | 0.049 | 0.045 | 0.047 | 0.043 | 0.051 |
| PB1-F2 (2–75) | 0.541 | 0.538 | 0.497 | 0.61 | 0.109 | 0.044 | 0.043 | 0.074 | 0.075 | 0.071 |
| M2e (2–24) | 3.658 | 1.076 | 3.492 | 0.819 | 0.082 | 0.043 | 0.044 | 0.043 | 0.045 | 0.043 |
The specificity of the assay was further established using multiple panels of plasma samples from residents of Vietnam and the United States with no history of H5N1 virus exposure. These included panels from individuals with confirmed infections with seasonal influenza virus or vaccination with seasonal trivalent influenza vaccine (TIV) (Table 2). All the specimens from H5N1-negative individuals scored negative in the H5N1-SeroDetect ELISA (Table 2). In addition, we evaluated the reactivities of individuals who were vaccinated with inactivated subunit H5N1 vaccine (1, 17). Since this vaccine virus was from reverse genetics-derived vaccine seed where the internal genes are derived from PR8 (H1N1) and lack significant M2e and PB1-F2 in the subunit vaccine, we did not observe any reactivity of these sera against H5N1-M2e or PB1-F2 peptides in H5N1-SeroDetect ELISA. To our surprise, these postvaccination sera (with confirmed HI titers ranging from 10 to 640 against A/Vietnam/1203/04) gave negative results with the peptides in the H5N1-SeroDetect ELISA, implying that they did not bind to the HA2 epitope, which was included in the vaccines as part of intact hemagglutinin (Table 2). Thus, H5N1-SeroDetect ELISA exhibited 100% specificity (no false positives) with normal (H5N1 virus-uninfected) serum samples.
Table 2.
Summary of H5N1-SeroDetect specificity: no reactivity with serum samples from individuals vaccinated with H5N1 vaccine or individuals with seasonal influenza virus vaccination and infections
| Sample set | Total no. of samples | Country of origin | H5N1-SeroDetect ELISAa |
|||
|---|---|---|---|---|---|---|
| No. of samples reacting positively |
Combined specificity (%) | |||||
| HA2 (488–516) | PB1-F2 (2–75) | M2e (2–24) | ||||
| Samples from seasonal influenza virus vaccination and infection | ||||||
| Prevaccination | 50 | USA | 0 | 0 | 0 | 100 |
| Postvaccination | 50 | USA | 0 | 0 | 0 | 100 |
| Postinfection (seasonal influenza virus) | 50 | USA | 0 | 0 | 0 | 100 |
| Normal human serum samples | 117 | Vietnam | 0 | 0 | 0 | 100 |
| Samples from H5N1 vaccination | ||||||
| Prevaccination | 15 | USA | 0 | 0 | 0 | 100 |
| Postvaccination (H5N1 vaccine) | 128 | USA | 0 | 0 | 0 | 100 |
H5N1-SeroDetect reactivity is considered negative if a cutoff value of <1 is obtained in the peptide ELISA. The cutoff value (specimen ELISA OD/control ELISA OD readings) for each peptide was determined as the mean absorbance + 5 standard deviations obtained with H5N1-seronegative samples (Fig. 2) and was 0.09 to 0.11 for the three peptides.
H5N1-SeroDetect reacts with H5N1 serum panels at early time points after H5N1 infection from nonsurvivors.
Reactivity in the H5N1-SeroDetect of sequential plasma samples obtained soon after acute infection from two representative individuals who succumbed to H5N1 (clade 2.3.4) infection in Vietnam is shown in Fig. 3. The 30-year-old male died within 12 days. All the bleeds reacted positively with the HA2 and M2e peptides (optical density [OD] >, 0.1), but only the last bleed contained antibodies reactive with the PB1-F2 peptides (Fig. 3, blue lines). A female patient who died on day 20 developed antibodies reactive with all three peptides between days 7 and 20 (Fig. 3, red lines). No gender differences in reactivity patterns were observed with any one of the three peptides for the 19 fatal case samples (11 females and 8 males) tested.
Fig. 3.
H5N1-SeroDetect ELISA reacts with H5N1 serum panels at early time points postinfection from nonsurvivors. Reactivity in the H5N1-SeroDetect ELISA of sequential plasma samples obtained soon after acute infection from individuals who succumbed to H5N1 (clade 2.3.4) infection in Vietnam. Reactivities of two representative sequential samples, one female (aged 20 years) and one male (aged 30 years), with HA2 (488–516) (A), PB1-F2 (2–75) (B), and M2e (2–24) (C) peptides are shown. In these panels, all plasma samples were tested at a 1:100 dilution. All data are represented as test specimen ODs on the y axis. Day 0 represents the date of onset of symptoms. The upper and lower limits for the average cutoff values + 5 standard deviations of the plasma sample upon repeated testing, representing the 95% confidence intervals for the given sample, ranged from 0.09 to 0.11 for each of the three peptides. The cutoff value for each peptide is shown as a dashed line in each graph.
H5N1-SeroDetect reactivity with seroconversion panels from H5N1 influenza survivors.
Sequential plasma samples were obtained soon after acute infection and up to 2 months later from individuals who survived H5N1 (clade 2.3.4) infection in Vietnam and were tested in the H5N1-SeroDetect. In a 21-year-old male (Fig. 4, black triangles), reactivity against PB1-F2 and M2e peaked early (<14 days postinfection) and declined gradually thereafter. In contrast, reactivity against the HA2 peptide peaked after 14 days and was maintained through day 60. In the second patient (red circles), reactivity against PB1-F2 was very low, but all bleeds (including day 60) reacted positively with M2e and HA2. Again, reactivity against the HA2 peptide peaked after 3 weeks and high titer was maintained until day 60 postinfection. Reactivity patterns with additional convalescent-phase serum panels from H5N1 influenza survivors demonstrated that seroconversions against the three peptides in the H5N1-SeroDetect test are not linked. H5N1 influenza survivors responded to at least two of the three peptides as early as 1 week post-H5N1 infection. Importantly, reactivity against the HA2 peptide remained strong (OD > 1) for at least 2 months. These observations were made with survivors of either clade 2.3.4 (Fig. 4) or clade 1 (Table 3) H5N1 infections.
Fig. 4.
Seroreactivity of H5N1 seroconversion panels in H5N1-SeroDetect ELISA soon after infection in H5N1 survivors. Sequential plasma samples obtained soon after acute infection up to 2 months from individuals who survived H5N1 (clade 2.3.4) infection in Vietnam were tested in the H5N1-SeroDetect. Reactivities of two representative sequential samples from males (aged 21 and 30 years) with HA2 (488–516) (A), PB1-F2 (2–75) (B), and M2e (2–24) (C) peptides. All plasma samples were tested at a 1:100 dilution. All data are represented as test specimen ODs on the y axis. The cutoff value for each peptide was determined as the mean absorbance + 5 standard deviations obtained with H5N1-seronegative samples (Fig. 2) and was <0.11 for each of the three peptides. The cutoff value for each peptide is shown as a dashed line in each graph.
Table 3.
Summary of H5N1-SeroDetect ELISA with H5N1 virus-infected serum samples
| Time since onset | Clade | Outcome | Total no. of samples | Rapid H5N1-SeroDetect |
|||
|---|---|---|---|---|---|---|---|
| No. of samples reacting positively |
Combined (% positive)a | ||||||
| HA2 (488–516) | PB1-F2 (2–75) | M2e (2–24) | |||||
| <21 days | 2.3.4 | Died | 21 | 11 | 7 | 12 | 95.3 |
| Survived | 18 | 13 | 7 | 13 | 100 | ||
| >21 days to 1 year | 1 | Survived | 7 | 7 | 4 | 6 | 100 |
| 2.3.4 | Survived | 7 | 7 | 3 | 6 | 100 | |
| >1 year | 1 | Survived | 25 | 18 | 9 | 15 | 100 |
| 2.3.4 | Survived | 2 | 2 | 0 | 2 | 100 | |
Combined reactivity depicts positivity to any one or more antigens in the H5N1-SeroDetect.
H5N1-SeroDetect ELISA detects long-lasting H5N1-specific antibodies following recovery from H5N1 infection.
For surveillance for H5N1 exposure in areas of endemicity, it is important to establish for how long antibodies against key epitopes are maintained and how they could be detected using a serodiagnostic assay for estimating the true incidence of H5N1 infection. To that end, we were able to obtain additional long-term post-H5N1-infection convalescent-phase samples from three of the original five H5N1 influenza survivors (Table 1). As can be seen in Fig. 5, reactivity against the HA2 peptide was maintained at high levels in all three individuals. Reactivities against M2e and PB1-F2 were much lower but remained above the cutoff values in two of the three survivors of H5N1 A/Vietnam (clade 1) infection. Testing of additional large numbers of samples ranging from 1 month to 4 years after H5N1 infection (either clade 1 or clade 2.3.4) showed frequent and strong positive reactivity with HA2 and M2e, with gradually less frequent and moderate to weak reactivity against PB1-F2 peptide (Table 3). At any time point, all the H5N1 influenza survivors reacted positively with at least two peptides in the H5N1-SeroDetect ELISA. The one false-seronegative sample (in the <21-day group) that did not react in H5N1-SeroDetect was from a fatal case and was collected 5 days after the onset of symptoms.
Fig. 5.
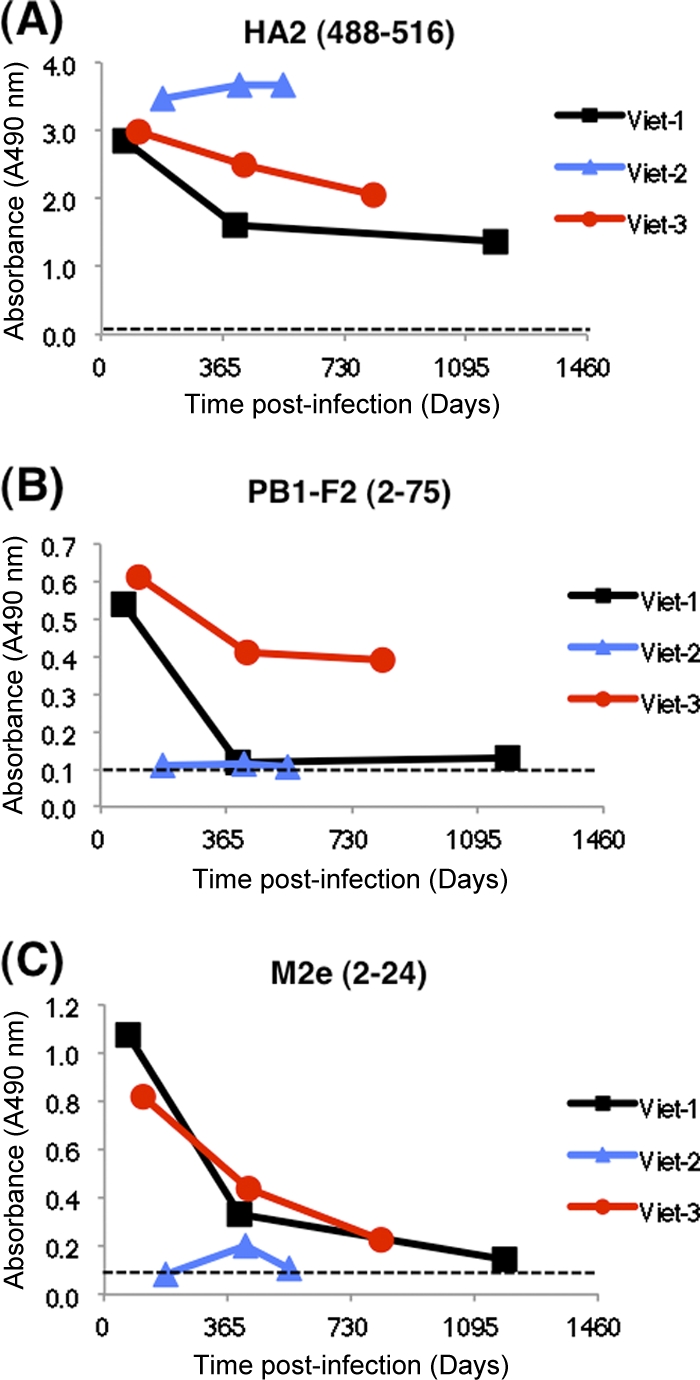
H5N1-SeroDetect detects long-lasting H5N1-specific antibodies up to 4 years following recovery from H5N1 infection. Long-term follow-up of sequential plasma samples obtained after 2 months and up to 4 years post-H5N1 infection from individuals who survived H5N1 (clade 1) infection in Vietnam tested in the H5N1-SeroDetect. Reactivities of three representative sequential samples (at a 1:100 dilution) with HA2 (488–516) (A), PB1-F2 (2–75) (B), and M2e (2–24) (C) peptides are shown. All data are represented as test specimen ODs on the y axis. In all three H5N1-infected individuals, the H5N1-SeroDetect was positive (OD > respective cutoff value) with at least one of the three peptides from the earliest bleed. The cutoff value for each peptide is shown as a dashed line in each graph.
Development of H5N1-SeroDetect rapid test.
Large-scale surveillance will be greatly enhanced by developing a high-throughput point-of-care rapid test that captures antibodies against all three H5N1 peptides on one platform. To that end, three test zone membrane regions containing H5N1 peptides PB1-F2 (2–75), M2e (2–24), and HA2 (488–516) were striped by capture onto a streptavidin-coated nitrocellulose membrane at 1 mg/ml of peptide, with approximately 1 μl of antigen per cm of membrane. The control line was goat anti-human IgG (H&L), which functions as an internal control for the addition of a plasma specimen. As can be seen in Fig. 6A (bottom three strips), three sera from H5N1 influenza survivors (clade 1 and clade 2.3.4) reacted positively with all three peptides in the SeroDetect rapid test. In contrast, H5N1 virus-uninfected individuals, including a subject who received H5N1 vaccine, did not react with the H5N1 SeroDetect rapid test (Fig. 6A, top three strips). Samples from multiple bleeds from 10 clade 1 and 10 clade 2.3.4 culture-confirmed H5N1 virus-infected individuals were tested in the rapid test. Most H5N1 clade 1-infected samples represent samples that were >1 year post-H5N1 infection, while clade 2.3.4 samples were from >7 days after onset of symptoms. A summary of all the panels tested to date is shown in Fig. 6B. All the H5N1-infected samples tested seropositive in the H5N1-SeroDetect rapid assay. In most cases, binding to the HA2 and M2e peptide bands was positive, while PB1-F2 provided an additional confirmation in 25 to 30% of cases, especially in samples soon after acute H5N1 infection. Importantly, all panels tested individually from H5N1 virus-uninfected individuals did not react positively with any of the peptides in the H5N1 SeroDetect rapid test and confirmed high specificity (Fig. 6B).
Fig. 6.
H5N1-SeroDetect rapid test for detection of H5N1 infections. (A) The rapid test was developed by striping H5N1 peptides [HA2 (488–516), PB1-F2 (2–75), and M2e (2–24)] and control protein (goat anti-human IgG), and data were assessed using a panel of H5N1 patient and control plasma samples at a 1:50 dilution. H5N1 virus-infected samples were from the survivors (clade 1 and clade 2.3.4) and individuals who died from infection (clade 2.3.4) in Vietnam. (B) Summary of reactivities of H5N1-negative and H5N1-positive samples in rapid assay format of H5N1-SeroDetect. Combined reactivity depicts positivity to any one or more antigens in the H5N1-SeroDetect. In all the H5N1 virus-infected individuals, the H5N1-SeroDetect rapid test was positive with at least one of the peptides.
DISCUSSION
Recent spread of H5N1 infections into pig populations along with H1N1 coinfection has raised alarms for potential adaptation of this highly pathogenic H5N1 avian virus into efficient human-to-human transmission. Thus, there is an urgent need for a rapid, simple, sensitive, and specific serological diagnostic assay for detection of antibodies to H5N1 and it is of prime importance for large-scale surveillance in areas of endemicity (10, 12, 20). Virus isolation and detection of viral RNA by RT-PCR from infected individuals can be done only within the first 2 to 3 weeks post-H5N1 infection in the symptomatic cases who report to a clinic/hospital, but asymptomatic cases could be easily missed, resulting in underestimation of exposure rates (16).
Peptide-based enzyme immunoassays have been widely used for the serodiagnosis of bacterial and viral infections. They provide the advantage of enhanced specificity and can be easily implemented in a simple, rapid, sensitive, and relatively cheap diagnostic kit. In the current study, we describe the development of simple serodiagnostic tests based on reactivity with three peptides from highly conserved H5N1 regions in HA2, M2e, and PB-1F2. These peptides were initially identified using a large GFPDL to screen plasma of H5N1 (clade 1) influenza survivors in Vietnam (15). The ELISA and rapid test platforms of H5N1-SeroDetect demonstrated a high degree of specificity, discriminating between H5N1 virus-infected individuals in Vietnam and uninfected individuals in Vietnam and the United States. Importantly, individuals who were either vaccinated against or infected with (culture-confirmed) seasonal influenza virus (with HI titers against H3N2 and H1N1) did not react with the H5N1-SeroDetect test. This is one of the primary properties required from an H5N1-specific surveillance tool in geographical areas where both seasonal and HP H5N1 viruses are circulating.
PB1-F2 was originally identified as an alternate reading frame in the PB1 gene of A/PR/8/34 PB1, which was found to contribute to the pathogenicity of the 1918 H1N1 Spanish influenza pandemic and H5N1 HP virus infections (4, 6, 29). This protein is not incorporated into viral particles, and expression is short lived in influenza virus-infected cells. However, the GFPDL analyses with serum antibodies from H5N1 influenza survivors provided the first evidence that this protein is expressed during H5N1 infection and is immunogenic in humans (15). The current study extended these findings and showed that antibodies against PB1-F2 appear relatively soon after infection and usually return to background levels by 2 to 3 months. The short duration of antibody response to PB1-F2 could be quite important and relevant in differentiating between recent and past H5N1 infections.
The role of M2e in the immune response to influenza virus during natural infections with seasonal influenza virus is still debated (2, 11). The current study confirms our previous finding in H5N1 influenza survivors demonstrating a fairly broad reactivity of postinfection sera against the M2e peptide, including some individuals who died from infection. Among long-term survivors, antibody reactivity against M2e can be detected for several years in some individuals, although at low levels. The overall percentage of M2e-positive sera in the current study was similar to anti-HA2 reactivity. The HA2 peptide used in the H5N1-SeroDetect tests is one of 9 HA2 sequences that were reactive with H5N1 convalescent-phase sera (15). It was selected for the tests because of its high degree of conservation among H5N1 strains and sufficiently large differences from human seasonal virus HA2 sequences (27). Furthermore, our data demonstrated that such antibodies are particularly long lived, up to many years post-H5N1 infection.
An epitope-blocking ELISA for detection of human antibodies against H5N1 viruses has been described elsewhere (19). However, this assay is based on a single monoclonal antibody (5F8) targeting a short sequence in HA2 (aa 274 to 281) and has low H5N1 specificity, as some samples from seasonal influenza convalescent patients reacted positively in the H5N1 ELISA. Our H5N1-SeroDetect ELISA and rapid test have demonstrated >99% specificity to date, including seronegative results with sera from individuals vaccinated against licensed prepandemic subunit H5N1 vaccine without adjuvant. Since this vaccine virus was from reverse genetics-derived vaccine seed where the internal genes are derived from PR8 (H1N1) and lack significant M2e and PB1-F2 in the subunit vaccine, we did not observe any reactivity of these post-H5N1 vaccination sera against H5N1-M2e or PB1-F2 peptides in H5N1-SeroDetect ELISA. However, lack of reactivity to HA2 peptides was unexpected, since the HA2 epitope (488–516) is expected to be part of the HA subunit vaccines. It is possible that this hydrophobic peptide sequence in the membrane-proximal domain of HA2 is somewhat sequestered in the inactivated subunit H5N1 vaccine formulations and therefore not exposed for immune response but is recognized by the immune system for replicating influenza virus during H5N1 infections. This phenomenon is similar to the observation made for the corresponding sequences in the cytoplasmic domain sequence of HIV-gp41 envelope, which were not recognized by serum antibodies from individuals vaccinated with HIV envelope-based vaccines but were strongly reactive to serum antibodies from HIV-infected individuals. These findings led to development of a novel serodiagnostic assay termed HIV-Selectest for differential diagnosis of true HIV infections in the face of HIV vaccine-induced antibodies that is currently being developed for feasibility studies (13, 14).
Alignment of the PB1-F2, HA2, and M2e peptide sequences used in H5N1-SeroDetect with other H5N1 human isolates revealed a very high degree of sequence conservation in H5N1 isolates from diverse clades and subtypes, including recent isolates (Fig. 1B; also see Fig. S1 in the supplemental material). The few amino acid variations observed in the recent isolates from Egypt in the HA2 immunodominant peptide sequence (Fig. S1) did not affect the reactivity of the HA2 peptide with human sera derived from clade 2.2.1 H5N1 infections from Egypt (unpublished data). Together, the three-peptide-based ELISA and rapid test may provide the required sensitivity to detect H5N1-specific antibodies starting within 6 to 7 days postinfection and persisting for >3 years. Both the ELISA and rapid test require very small volumes of test sample. It will be important to also evaluate the reactivity of infected or vaccinated poultry samples as previously described by Cui et al. (7, 8). Our data support the use of peptide-based H5N1-SeroDetect ELISA and rapid tests for a serodiagnostic assay of H5N1 exposure in humans that can be easily adapted for surveillance of H5N1 infection in birds.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vladimir Lugovtsev and Zhiping Ye for a thorough evaluation of the manuscript. We thank Nguyen Thi Thu Yen and Tran Nhu Duong for their assistance with sample collection.
All authors declare that they have no competing interests.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Beigel J. H., Voell J., Huang C. Y., Burbelo P. D., Lane H. C. 2009. Safety and immunogenicity of multiple and higher doses of an inactivated influenza A/H5N1 vaccine. J. Infect. Dis. 200:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Black R. A., Rota P. A., Gorodkova N., Klenk H. D., Kendal A. P. 1993. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J. Gen. Virol. 74:143–146 [DOI] [PubMed] [Google Scholar]
- 3. Bouma A., et al. 2009. Estimation of transmission parameters of H5N1 avian influenza virus in chickens. PLoS Pathog. 5:e1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chanturiya A. N., et al. 2004. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J. Virol. 78:6304–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen H., et al. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. U. S. A. 103:2845–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conenello G. M., Zamarin D., Perrone L. A., Tumpey T., Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui S., Chen C., Tong G. 2008. A simple and rapid immunochromatographic strip test for monitoring antibodies to H5 subtype avian influenza virus. J. Virol. Methods 152:102–105 [DOI] [PubMed] [Google Scholar]
- 8. Cui S., Tong G. 2008. A chromatographic strip test for rapid detection of one lineage of the H5 subtype of highly pathogenic avian influenza. J. Vet. Diagn. Invest. 20:567–571 [DOI] [PubMed] [Google Scholar]
- 9. de Jong M. D., et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gambotto A., Barratt-Boyes S. M., de Jong M. D., Neumann G., Kawaoka Y. 2008. Human infection with highly pathogenic H5N1 influenza virus. Lancet 371:1464–1475 [DOI] [PubMed] [Google Scholar]
- 11. Gerhard W., Mozdzanowska K., Zharikova D. 2006. Prospects for universal influenza virus vaccine. Emerg. Infect. Dis. 12:569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katz J. M., et al. 1999. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. J. Infect. Dis. 180:1763–1770 [DOI] [PubMed] [Google Scholar]
- 13. Khurana S., et al. 2006. Human immunodeficiency virus (HIV) vaccine trials: a novel assay for differential diagnosis of HIV infections in the face of vaccine-generated antibodies. J. Virol. 80:2092–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khurana S., et al. 2010. HIV-Selectest enzyme immunoassay and rapid test: ability to detect seroconversion following HIV-1 infection. J. Clin. Microbiol. 48:281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khurana S., et al. 2009. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 6:e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li F. C., Choi B. C., Sly T., Pak A. W. 2008. Finding the real case-fatality rate of H5N1 avian influenza. J. Epidemiol. Community Health 62:555–559 [DOI] [PubMed] [Google Scholar]
- 17. Lu H., et al. 2011. A rapid Flp-In system for expression of secreted H5N1 influenza hemagglutinin vaccine immunogen in mammalian cells. PLoS One 6:e17297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poetri O. N., et al. 2009. An inactivated H5N2 vaccine reduces transmission of highly pathogenic H5N1 avian influenza virus among native chickens. Vaccine 27:2864–2869 [DOI] [PubMed] [Google Scholar]
- 19. Prabakaran M., et al. 2009. Development of epitope-blocking ELISA for universal detection of antibodies to human H5N1 influenza viruses. PLoS One 4:e4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rowe T., et al. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakai-Tagawa Y., et al. 2010. Sensitivity of influenza rapid diagnostic tests to H5N1 and 2009 pandemic H1N1 viruses. J. Clin. Microbiol. 48:2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simonsen L., Taylor R. J., Viboud C., Miller M. A., Jackson L. A. 2007. Mortality benefits of influenza vaccination in elderly people: an ongoing controversy. Lancet Infect. Dis. 7:658–666 [DOI] [PubMed] [Google Scholar]
- 23. Smith G. J., et al. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U. S. A. 103:16936–16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith G. J., et al. 2006. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology 350:258–268 [DOI] [PubMed] [Google Scholar]
- 25. Stelzer-Braid S., et al. 2008. A commercial ELISA detects high levels of human H5 antibody but cross-reacts with influenza A antibodies. J. Clin. Virol. 43:241–243 [DOI] [PubMed] [Google Scholar]
- 26. Tang X. C., et al. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vareckova E., Mucha V., Kostolansky F., Gubareva L. V., Klimov A. 2008. HA2-specific monoclonal antibodies as tools for differential recognition of influenza A virus antigenic subtypes. Virus Res. 132:181–186 [DOI] [PubMed] [Google Scholar]
- 28. Xu K. M., et al. 2007. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J. Virol. 81:2635–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zamarin D., Ortigoza M. B., Palese P. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.