Abstract
The chicken anemia virus (CAV) protein Apoptin is a small, 13.6-kDa protein that has the intriguing activity of inducing G2/M arrest and apoptosis specifically in cancer cells by a mechanism that is independent of p53. The activity of Apoptin is regulated at the level of localization. Whereas Apoptin is cytoplasmic in primary cells and does not affect cell growth, in transformed cells it localizes to the nucleus, where it induces apoptosis. The properties of cancer cells that are responsible for activating the proapoptotic activities of Apoptin remain unclear. In the current study, we show that DNA damage response (DDR) signaling is required to induce Apoptin nuclear localization in primary cells. Induction of DNA damage in combination with Apoptin expression was able to induce apoptosis in primary cells. Conversely, chemical or RNA interference (RNAi) inhibition of DDR signaling by ATM and DNA-dependent protein kinase (DNA-PK) was sufficient to cause Apoptin to localize in the cytoplasm of transformed cells. Furthermore, the nucleocytoplasmic shuttling activity of Apoptin is required for DDR-induced changes in localization. Interestingly, nuclear localization of Apoptin in primary cells was able to inhibit the formation of DNA damage foci containing 53BP1. Apoptin has been shown to bind and inhibit the anaphase-promoting complex/cyclosome (APC/C). We observe that Apoptin is able to inhibit formation of DNA damage foci by targeting the APC/C-associated factor MDC1 for degradation. We suggest that these results may point to a novel mechanism of DDR inhibition during viral infection.
INTRODUCTION
Chicken anemia virus (CAV) is a nonenveloped, single-stranded DNA (ssDNA) virus with a small genome of approximately 2.3 kb (reviewed in reference 42). CAV causes severe anemia and immunosuppression in young chickens by replicating in thymocytes and erythroblasts, often leading to mortality. The virus encodes 3 proteins, designated VP1, -2, and -3, and of these, VP3 was shown to mediate the cytopathic properties of the virus (32). VP3 has been shown to be a potent inducer of apoptosis and was renamed “Apoptin” for this reason. Of particular interest has been the finding that Apoptin is able to selectively kill transformed cells by a mechanism that is independent of p53 (11, 52). Many types of cancer chemotherapies that function as DNA-damaging agents depend on p53 for efficacy. p53 is mutated in more than half of all human cancers, and these tumors are more resistant to many types of therapy. The study of Apoptin is therefore a unique system for identifying pathways of apoptosis that are able to kill cancer cells independent of p53.
The transformed-cell-specific killing of Apoptin is regulated at least in part at the level of subcellular localization. When expressed in transformed cells, Apoptin localizes to the nucleus, whereas in normal (primary) cells it is cytoplasmic (11). The Apoptin protein contains a C-terminal nuclear localization signal (NLS) and a central leucine-rich nuclear export sequence (NES) that cause the protein to shuttle in and out of the nucleus in both normal and transformed cells (22, 34, 46). Several studies have identified properties of the Apoptin protein that may regulate differential localization of Apoptin between primary and transformed cells, but the mechanism is poorly understood. A C-terminal phosphorylation site at threonine-108 (T108) has been reported to regulate Apoptin apoptotic activity but does not seem to be essential for transformed-cell-specific localization (27, 36). Although nuclear localization of Apoptin is required for apoptotic activity in tumor cells, additional factors seem to be necessary, since fusion of Apoptin with a strong NLS results in nuclear localization in primary cells but no killing activity (12, 22). Coexpression with SV40 large T antigen (LgT) was shown to induce translocation of Apoptin into the nucleus in primary cells, suggesting that an activity of LgT was sufficient to activate Apoptin (51). Interestingly, the domains of LgT required for binding the retinoblastoma (Rb) and p53 tumor suppressors, which are required for cellular transformation, were dispensable for activating Apoptin. Instead, Apoptin was shown to be activated by an N-terminal region of LgT implicated in causing DNA damage through an interaction with the mitotic spindle checkpoint protein Bub1 (23, 51). These observations raise the possibility that Apoptin may sense the DNA damage response (DDR) and that signaling through the DDR pathway may regulate the nuclear localization of Apoptin. Most cancer cell lines are known to have constitutive DDR activation due to intense replication stress (2, 3, 13, 18, 20), and this pathway may therefore represent a transformed-cell-specific property that triggers Apoptin nuclear localization and apoptosis.
Many viruses that replicate in the nucleus encode one or more proteins that manipulate the cellular DDR pathways. In many cases, this is because replicating viral genomes are interpreted as damaged DNA and as such can induce cell cycle arrest, DNA repair, and apoptosis, all of which can inhibit viral replication. In the current study, we present evidence that constitutive DDR signaling pathways regulate the unique transformed-cell-specific localization observed with Apoptin. Moreover, we show that Apoptin inhibits downstream effectors of the DDR and propose that these activities may be a novel mechanism utilized by CAV to evade activation of the host cell DDR.
MATERIALS AND METHODS
Cell lines and drugs.
H1299 non-small-cell lung adenocarcinoma cells, MRC5 primary lung fibroblasts, and 293T/17 cells were obtained from ATCC. All cells were maintained in Dulbecco's modified Eagle medium (Wisent Inc., QC, Canada) supplemented with 10% fetal bovine serum (HyClone; Thermo Scientific) and 0.1% gentamicin (Wisent Inc., QC, Canada). Bleomycin sulfate (BLM) was purchased from Bioshop Canada and used at concentrations from 5 to 50 μM. Puromycin was purchased from Bioshop Canada and used at concentrations of 2 μg/ml. KU55933 and NU7026 were purchased from Tocris Bioscience and used at 10 and 1 μM, respectively.
Viral infections.
The adenoviruses encoding Apoptin (Ad-Apwt) or LacZ (Ad-LacZ) were described previously (41) and used to infect H1299 cells at a multiplicity of infection (MOI) of 35 and primary cells at an MOI of 50. Adenoviruses were propagated in HEK293 cells, and titers were determined using the fluorescence-forming unit (FFU) protocol (19). The pLKO D9 Bub1 knockdown short hairpin RNA (shRNA) has been described previously (10), and the SHC002 nonsilencing control (NSC) is from Sigma. The lentiviruses were produced by the cotransfection of 3 × 106 293T/17 cells using calcium phosphate precipitation with 15 μg of the pLKO vector, 11.25 μg pCMV-dR8.2 pVPR, and 4.5 μg pCMV VSV-G (vesicular stomatitis virus G). The virus-containing supernatant was then collected, filtered through a 0.45-μm filter, and stored as 300-μl aliquots at −80°C until further use. For infections using these viruses, 5 × 105 MRC5 cells were plated on 60-mm dishes. Twenty-four hours after plating, 300 μl virus-containing medium was added in the presence of 8 μg/ml Polybrene. Twenty-four hours following the infection, the cells were trypsinized and replated in 10-cm dishes. Twenty-four hours following plating, the medium was changed for medium containing 2 μg/ml puromycin. After 48 h of selection, the cells were trypsinized, counted, and replated for experiments.
Subcellular fractionation.
MRC5 cells (6 × 105) were plated on 10-cm dishes. They were then infected with Ad-Apwt or Ad-LacZ and left for 48 h. They were then treated for 18 h overnight with BLM and harvested. Fractionation was performed by scraping the cells and washing them twice with phosphate-buffered saline (PBS). The cells were then resuspended in 100 μl buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 5 mM dithiothreitol [DTT], 0.5% NP-40) per 106 cells on ice for 10 min. The lysate was then centrifuged for 3 min at 3,000 × g at 4°C. The supernatant containing the cytoplasmic fraction was then set aside, and 4× Laemmli buffer was added to achieve a concentration of 1×. The pelleted nuclei were then resuspended in 50 μl buffer C (20 mM HEPES, 25% glycerol, 0.42 M NaCl, 0.4 mM EDTA, 1 mM DTT) per 106 cells on ice for 30 min. The lysate was then centrifuged at maximum speed for 10 min at 4°C. The supernatant containing the nuclear fraction was then set aside, and 4× Laemmli buffer was added to achieve a concentration of 1×. The final insoluble pellet was then resuspended in 50 μl 1× Laemmli buffer using a 23-gauge needle and a 1-ml syringe. All lysates were then boiled for 5 min and stored at −20°C until further use.
Immunoprecipitation.
Cells were treated as for the fractionation assay and then lysed in 1 ml buffer X (50 mM Tris [pH 8.5], 250 mM NaCl, 1% NP-40, 1 mM EDTA) per 106 cells on ice for 20 min. Cell debris was pelleted by centrifugation at maximum speed for 15 min at 4°C. The supernatant was then incubated with 30 μl anti-FLAG affinity gel (Sigma) for 2 h at 4°C. The beads were then pelleted and washed 4 times with buffer X. Following the washes, the beads were pelleted and resuspended in 1× sample buffer, boiled for 5 min, and stored at −20°C until further use. Buffers A, C, and X were supplemented with 1 protease inhibitor cocktail tablet (Roche) per 10 ml.
Antibodies and Western blotting.
The rabbit polyclonal and mouse monoclonal anti-FLAG antibodies and the mouse monoclonal antibody against TATA box binding protein were obtained from Sigma. The rabbit polyclonal anti-ATM antibody was obtained from EMD Biochemicals. The anti-DNA-dependent protein kinase (anti-DNA-PK) antibody was purchased from Cell Signaling Technology. The goat anti-MDC1, mouse monoclonal anti-CDC27, and mouse monoclonal anti-PML were all obtained from Santa Cruz. The anti-53BP1 mouse monoclonal antibody was obtained from BD Biosciences. The rabbit polyclonal against γH2AX was obtained from Millipore. The rabbit polyclonal antibodies against Bub1 and APC1 have been described previously (16, 41). The goat anti-rabbit Alexa Fluor 594-conjugated and goat anti-mouse and donkey anti-goat Alexa Fluor 488-conjugated antibodies were from Invitrogen-Life Technologies.
Western blotting was performed using standard protocols for SDS-PAGE and wet transfer onto nitrocellulose membranes (Bio-Rad). Conditions for Western blots were the use of 5% nonfat dry milk in TBS-T (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween 20). The bands were visualized by enhanced chemiluminescence (Western Lightning [PerkinElmer]; Super Signal West Pico or Femto [Pierce-Thermo Scientific]) and exposure on film. Quantitation Western blot signals were measured using ImageJ software.
Immunofluorescence.
Cells were seeded at low confluence on glass coverslips in 6-well plates and, 24 h later, infected as described above. The cells were fixed by immersing the coverslips in ice-cold methanol for 10 min. Staining was performed by washing the coverslips twice in PBS and then blocking for 15 min in 10% ADB (10% horse serum, 0.05% Triton X-100 in PBS). The coverslips were then incubated with the primary rabbit anti-FLAG and anti-53BP1 antibodies at 1:1,000 and 1:10,000, respectively, in 10% ADB for 1 h at room temperature. The coverslips were then washed 3 times each for 5 min in PBS, followed by incubation with the fluorescence-conjugated secondary antibodies at 1:500 in 10% ADB for 1 h at room temperature. The coverslips were then washed again, counterstained with DAPI (4′,6-diamidino-2-phenylindole), and mounted on slides using DABCO mounting medium (Sigma). The cells were then viewed using a Zeiss Axiovert fluorescence microscope and processed with Zeiss Axiovision software.
For analysis of green fluorescent protein (GFP)-fused apoptin mutants, 1.0 × 105 cells were plated onto 6-well plates. The cells were allowed to recover for 24 h and were then transfected using 1 μg of plasmid DNA and 2.5 μl Lipofectamine 2000 in opti-MEM medium. Four hours following transfection, the medium was changed and caffeine added. Twenty-four hours following transfection, the cells were fixed in cold methanol, stained with DAPI, and examined at ×400 magnification. The GFP-fused apoptin constructs used have been described previously (22).
Apoptosis assays.
MRC5 cells (7 × 104) were plated on 35-mm dishes and then infected with Ad-Apwt or Ad-LacZ as described above. The cells were then treated with bleomycin as indicated in the figure legends. The cells were stained by washing them once with PBS and once with AnnexinV binding buffer (2.5 mM CaCl2, 140 mM NaCl, 7.75 mM HEPES [pH 7.4]) and then incubated with 200 μl AnnexinV binding buffer containing 5 μl AnnexinV (BD Biosciences) and 0.25 μg 7-amino-actinomycin D (7AAD) (A.G. Scientific) per sample at room temperature in the dark on a rocking platform at slow speed for 15 min. The cells were then washed twice with AnnexinV binding buffer, collected by scraping, and analyzed on a Cell Lab Quanta SC flow cytometer (Beckman Coulter) in conjunction with Quanta Collection and Quanta Analysis software (Beckman Coulter). For examination of nuclear morphology, 25,000 cells per well were plated on 12-well plates, infected, and treated as indicated in Fig. 3A and B. Following treatment, the cells were fixed in cold methanol, stained with DAPI, and analyzed under the microscope at ×400 magnification.
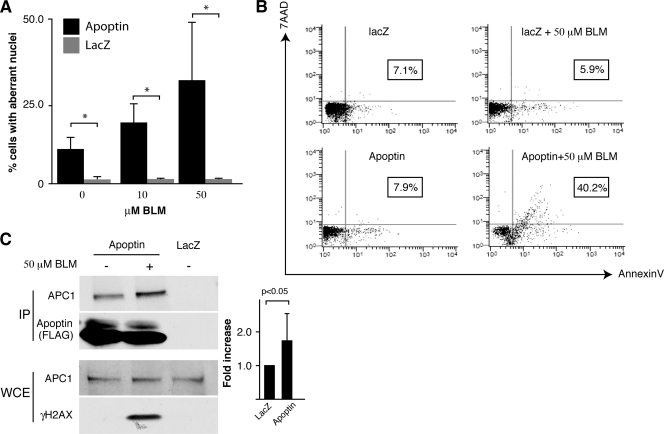
Fig. 3.
Treatment of cells with apoptin and BLM induces apoptosis in primary cells. (A) MRC5 fibroblasts were infected with Ad-Apwt or Ad-LacZ and then treated with BLM or vehicle control. Cells were fixed and stained with anti-FLAG antibody to detect Apoptin and DAPI to visualize nuclei. The morphology of the nuclei was examined by microscopy, and the percentage of cells with aberrant nuclei was determined. *, P < 0.05. (B) MRC5 fibroblasts were treated as described for panel A, stained with AnnexinV and 7AAD, and analyzed by flow cytometry. The percentage of Annexin-positive cells is indicated for each treatment (boxed). (C) MRC5 fibroblasts were infected with Ad-Apwt or Ad-LacZ and then treated with BLM. Apoptin was immunoprecipitated (IP) from cell extracts using an anti-FLAG antibody. IPs were resolved by SDS-PAGE, and immunoblotting was performed for APC1 and FLAG (Apoptin). Whole-cell extracts (WCE) used for IP were analyzed by Western blotting to confirm equal amounts of APC1 and to confirm activation of DDR (γH2AX). Band intensities of APC1 IPs were quantitated from 4 separate experiments and plotted as fold induction (right). Error bars indicate standard deviations.
siRNA transfection.
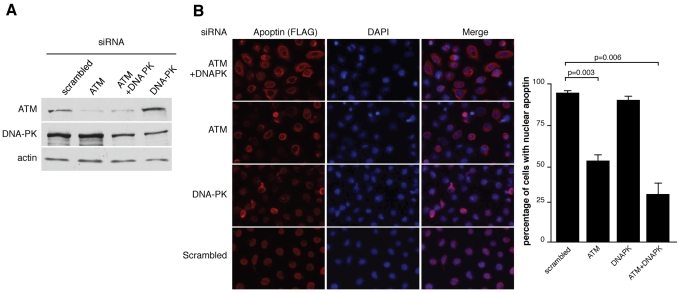
H1299 cells (5 × 104) were seeded in 24-well plates and then transfected overnight using 2 μl Lipofectamine 2000 (Invitrogen-Life Technologies) per well and 50 nmol ATM (5′-AAU UCA GAA AGC AAC AUU CUU dTdT-3′), DNA-PK (5′-GAU CGC ACC UUA CUC UGU UdTdT-3′), or scrambled small interfering RNA (siRNA) (5′-AAT TCT CCG AAC GTG TCA CGT dTdT-3′) in Opti-MEM I reduced-serum medium (1×) (Invitrogen-Life Technologies) as indicated in Fig. 5. All siRNAs were purchased from Sigma. The cells were allowed to recover for 24 h and then were infected with Ad-Apwt for 24 h, at which point they were either fixed with cold methanol and stained with anti-FLAG and anti-mouse Alexa Fluor 594-conjugated antibodies as described above or harvested for SDS-PAGE and Western blotting. The lower quality of the DNA-PK antibody required that nuclear extracts be prepared in order to analyze the levels of DNA-PK protein.
Fig. 5.
Inhibition of DNA damage signaling by RNAi induces cytoplasmic localization of Apoptin in cancer cells. (A) H1299 cells were transfected with siRNAs targeting ATM, DNA-PK, or a scrambled control siRNA. Forty-eight hours following transfection, nuclear extracts were generated and separated by SDS-PAGE, and the levels of ATM and DNA-PK were monitored by immunoblotting. Actin was used as a loading control. (B) H1299 cells were transfected with siRNAs as described for panel A and then infected with Ad-Apwt. The cells were then fixed and stained with anti-FLAG antibody to detect apoptin and DAPI to visualize nuclei. The percentages of cells displaying nuclear localization of apoptin were determined by microscopy and are graphed on the right. Error bars indicate standard deviations.
MDC1 time course experiments.
For experiments involving immunoblots of MDC1, 5 × 105 H1299 cells were plated on 60-mm dishes and infected 24 h later with Ad-Apwt or Ad-LacZ. Cells were treated 40 h postinfection (hpi) with cycloheximide (CHX) (Sigma) at 20 μg/ml for the amounts of time indicated in the figure legends. In order to test for proteasome dependency, cells were treated with various concentrations of MG132 (Sigma) from 42 to 48 hpi and then harvested. Nuclear extracts were prepared, and the protein concentration in each sample was measured by the Bradford assay (Bio-Rad).
Statistical tests.
Where indicated, statistical significance was calculated using Student's two-tailed t test, with data from at least 3 independent biological repeats.
RESULTS
Knockdown of bub1 induces DNA damage and causes Apoptin to localize to the nucleus in primary fibroblasts.
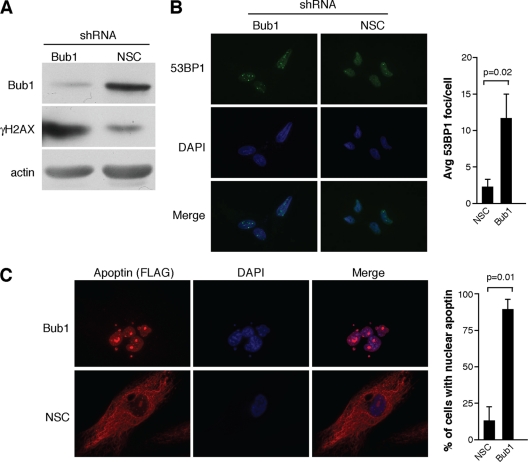
Simian virus 40 (SV40) LgT was previously shown to induce DNA damage signaling through a region in the N-terminal region that interacts with and inhibits the mitotic checkpoint protein bub1 (23). This same region of LgT was also demonstrated to induce nuclear localization of Apoptin (51). We therefore hypothesized that Bub1 inhibition and the resulting activation of DNA damage signaling may regulate Apoptin localization. In order to test this hypothesis, we knocked down bub1 expression in normal MRC5 fibroblasts by using lentiviral shRNA constructs. Figure 1A shows that the expression levels of bub1 could be efficiently knocked down using the shRNA construct and that these conditions resulted in activation of DDR, as measured by levels of γH2AX, as was previously reported (23). Figure 1B shows that Bub1 knockdown cells also displayed increased numbers of DNA damage foci, as measured by immunofluorescence against 53BP1. In order to determine if Bub1 knockdown could affect localization of Apoptin, MRC5 fibroblasts were stably transduced with the lentiviral shRNA construct targeting Bub1 or a nonsilencing control (NSC). Knockdown cells were then infected with an adenoviral vector expressing an epitope-tagged wild-type Apoptin (Ad-Apwt) or a control virus expressing LacZ (Ad-LacZ). Figure 1C shows that Bub1 knockdown resulted in a dramatic localization of Apoptin from the cytoplasm to the nucleus that was not observed in the NSC. These data show that bub1 knockdown and possibly the resulting DNA damage signaling are able to trigger nuclear localization of Apoptin.
Fig. 1.
Bub1 knockdown in primary cells induces DNA damage and causes Apoptin to localize to the nucleus. (A) Immunoblot for γH2AX and bub1 from MRC5 normal fibroblasts infected with lentiviral vectors encoding an shRNA targeting bub1 or a nonsilencing control. An actin blot is included as a loading control. (B) Immunofluorescence detecting DNA damage foci in MRC5 fibroblasts treated as described for panel A. Foci were detected using an antibody against 53BP1, and cell nuclei were visualized with DAPI. Average numbers of DNA damage foci counted in knockdown and control cells are shown in the graph on the right. (C) Immunofluorescence detecting FLAG-tagged Apoptin in bub1 knockdown and control cells infected with Ad-Apwt. Cells were fixed and stained with anti-FLAG antibody, and cell nuclei were visualized with DAPI. The percentages of cells displaying nuclear localization of Apoptin are shown in the bar graph on the right. Error bars indicate standard deviations.
Treatment with the radiomimetic drug bleomycin causes a dose-dependent relocalization of Apoptin to the nucleus of MRC5 cells.
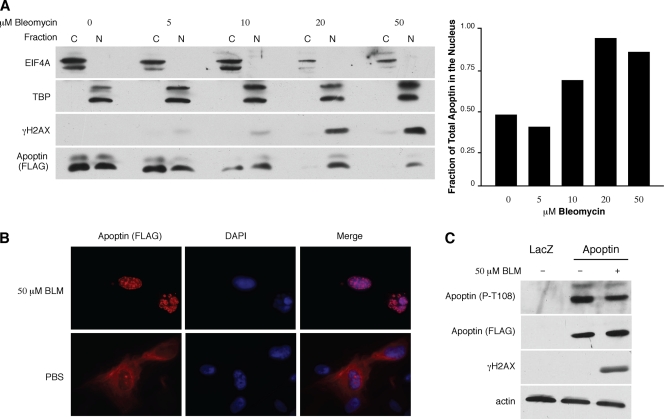
Since Bub1 knockdown is able to induce in primary cells a nuclear localization of Apoptin that correlates with the activation of the DDR, we asked whether activation of DDR signaling alone would be sufficient to activate Apoptin. To test this hypothesis, we chose to use the anticancer drug bleomycin (BLM). BLM induces nonspecific double-stranded DNA (dsDNA) and ssDNA breaks through the generation of superoxide and hydroxide free radicals. To examine the effects of BLM treatment on Apoptin localization, MRC5 cells were infected with Ad-Apwt and subsequently treated with BLM at various concentrations. Biochemical fractionation was then performed to isolate nuclear and cytoplasmic fractions, which were analyzed by Western blotting for the presence of Apoptin. Figure 2A shows that treatment with BLM caused Apoptin to localize in the nuclear fractions in a dose-dependent manner. Nuclear fractions in which Apoptin appeared correlated with the appearance of γH2AX, one of the major markers for DNA damage, in the nucleus. Quantitation of the Apoptin signal in each fraction shows that Apoptin localizes almost exclusively to the nucleus at higher BLM concentrations (Fig. 2A, right). To confirm this result, we also performed immunofluorescence microscopy and found that in cells treated with BLM, Apoptin localized predominantly to the nucleus (Fig. 2B).
Fig. 2.
DNA damage induces nuclear localization of Apoptin in primary cells. (A) MRC5 fibroblasts were infected with Ad-Apwt and subsequently treated with the indicated concentrations of BLM for 18 h. The cells were then fractionated into cytoplasmic and nuclear fractions and separated by SDS-PAGE, and the levels of Apoptin present in each fraction were examined by immunoblotting with anti-FLAG antibody. EIF4A (eukaryotic translational initiation factor 4) and TATA box binding protein (TBP) are shown as markers for the cytoplasmic (C) and nuclear (N) fractions, respectively. Band intensities of Apoptin were quantitated from blots and graphed as the fraction of total apoptin present in the nucleus under each condition (right). (B) MRC5 fibroblasts were infected with Ad-Apwt and then treated with 50 μM bleomycin or PBS for 18 h. The cells were then fixed and stained with anti-FLAG antibody to detect Apoptin and DAPI to visualize nuclei. (C) MRC5 fibroblasts were infected and treated as described for panel A. Whole-cell lysates were separated by SDS-PAGE, and levels of phosphorylated Apoptin on threonine 108 (P-T108) were determined by immunoblot analysis using a phosphospecific antibody. Levels of total Apoptin were detected using anti-FLAG antibody; activation of DDR was confirmed with γH2AX, and an actin immunoblot was used as a loading control.
Since the C-terminal phosphorylation site of Apoptin (T108) was previously reported to regulate Apoptin activity, we determined whether activation of the DDR results in differential phosphorylation at this site. To address this question, we performed immunoblot analysis with a phosphospecific antibody previously shown to recognize the phosphorylated form of T108 (kindly provided by M. Tavasolli) (14). We show in Fig. 2C that there is no significant change in T108 phosphorylation following induction of DNA damage by BLM. Taken together, these data indicate that induction of DNA damage is itself sufficient to trigger the translocation of Apoptin to the nucleus in primary cells. Furthermore, phosphorylation on T108 does not appear to mediate these effects.
Apoptin and bleomycin synergize to kill normal cells.
Forced nuclear targeting of Apoptin in primary cells does not induce apoptosis, indicating that additional events are required to fully activate the protein (12, 22). Since we observed that DNA damage can induce localization of Apoptin to the nucleus of primary cells, we hypothesized that Apoptin activated by DNA damage may induce apoptosis. To address this question, MRC5 fibroblasts were infected with Ad-Apwt or Ad-LacZ control adenovirus and then treated with sublethal doses of BLM. When expressed in transformed cells, Apoptin induces changes in nuclear morphology typical of apoptosis (11, 41). Figure 3A shows that primary cells treated with BLM in combination with Apoptin expression displayed aberrant nuclear morphology typical of apoptosis. To further confirm this result, we repeated the experiment and analyzed cells using the annexinV/7AAD apoptosis assay. Figure 3B shows that following treatment, a 5-fold increase in apoptosis was observed in cells treated with Ad-Apwt and BLM relative to that in cells expressing Apoptin alone.
We previously demonstrated that in tumor cells Apoptin interacts with and inhibits the anaphase-promoting complex/cyclosome (APC/C) and that this activity contributes to the apoptotic effect (22, 41). We therefore determined whether treatment with BLM in normal fibroblasts enhanced interaction of Apoptin with the APC/C. Figure 3C shows that there is a significant, 1.7-fold increase in association of Apoptin with APC1 upon treatment with BLM. Although the increased binding with the APC/C in the presence of DNA damage was modest, it was highly reproducible and was observed in four separate biological repeats (Fig. 3C, right). Therefore, disruption of APC/C activity may contribute to the increased apoptosis observed upon treatment with Apoptin and BLM.
Inhibition of DDR signaling prevents Apoptin activation in tumor cells.
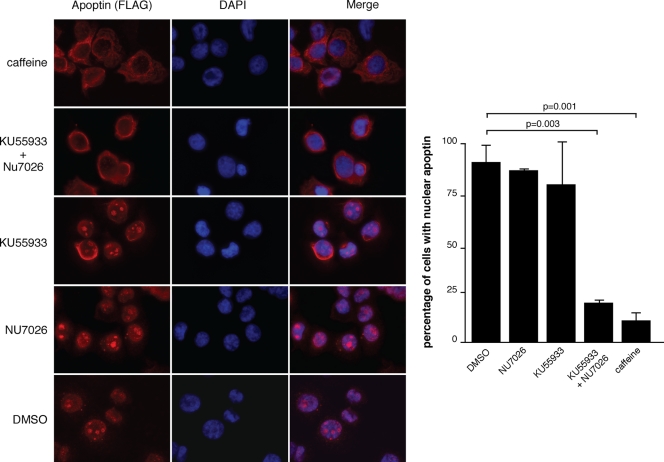
Recent studies from several groups show that a common property of cancer cells is constitutive DDR signaling due to rapid cell division and resulting replication stress (2, 3, 13, 18, 20). We therefore asked whether DDR signaling is required for nuclear Apoptin localization in tumor cells. In order to address this question, we used three chemical inhibitors of DDR signaling. We first used caffeine, which is a well-characterized broad-spectrum inhibitor of the phosphatidylinositol 3-kinase (PI-3K)-like kinases (PIKK). Caffeine inhibits MTOR, PI-3K, ATM, ATR, DNA-PK, and chk1 to various degrees depending on the concentration used (37). Figure 4 shows that treatment of H1299 cells expressing Apoptin with 5 mM caffeine was sufficient to relocalize Apoptin to the cytoplasm in 95% of cells, whereas in control cells, Apoptin remained almost exclusively nuclear. Since caffeine is a pan-inhibitor of PIKK, we further examined the effects of KU55933 and NU7026, which specifically inhibit ATM and DNA-PK, respectively. Figure 4 shows that when used individually, neither inhibitor had significant effects on the overall localization of Apoptin. However, Apoptin relocalized to the cytoplasm in almost all cells when KU55933 and NU7026 were combined. Therefore, pharmacological inhibition of both ATM and DNA-PK, two key kinases in DDR signaling, is able to prevent nuclear localization of Apoptin in tumor cells.
Fig. 4.
Chemical inhibitors of DNA damage signaling induce cytoplasmic localization of Apoptin in cancer cells. H1299 non-small-cell lung carcinoma cells were infected with Ad-Apwt and then treated with the indicated inhibitors of DNA damage signaling for 18 h. The cells were fixed and stained with anti-FLAG to detect apoptin and DAPI to visualize nuclei. The percentages of cells displaying nuclear localization of Apoptin were determined by microscopy and are graphed on the right. DMSO, dimethyl sulfoxide. Error bars indicate standard deviations.
To further confirm our result with small-molecule inhibitors of DDR signaling, we proceeded to inhibit ATM and DNA-PK using RNA interference (RNAi). Two very effective siRNA sequences were previously reported to inhibit expression of these kinases (26, 49). DNA-PK and ATM levels were significantly reduced when cells were treated with these siRNAs either individually or in combination (Fig. 5A). Figure 5B shows that ATM knockdown resulted in cytoplasmic localization in approximately 50% of cells, compared to a control siRNA sequence, which displayed greater than 90% nuclear Apoptin. Cotransfection of siRNAs targeting ATM and DNA-PK resulted in an even greater effect, with approximately 70% of cells with cytoplasmic Apoptin. Knockdown of DNA-PK by itself, however, did not have a significant effect. Therefore, DDR-induced nuclear targeting of Apoptin requires signaling through the ATM kinase and, to a lesser extent, DNA-PK.
Nucleocytoplasmic shuttling activity is essential for Apoptin response to DDR.
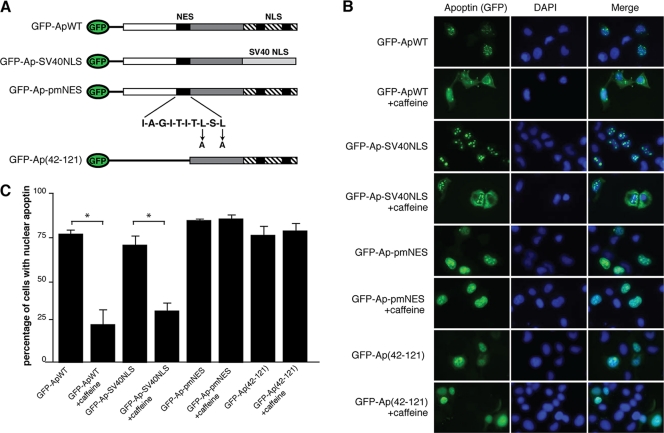
Studies from our lab and others have shown that Apoptin contains both NLS and NES motifs that confer nucleocytoplasmic shuttling activity to the protein (22, 34, 46). Intriguingly, regulation of NES activity appears to be a major factor that determines transformed-cell-specific localization of Apoptin (22, 34). We therefore asked whether the NES and the NLS sequence of Apoptin were also required to respond to DDR signaling. We have previously generated a GFP-fused Apoptin construct as well as a series of mutants with mutations in the NES and the NLS sequence (Fig. 6A) (22). Figures 6B and C show that wild-type Apoptin fused to GFP (GFP-Apwt) localizes primarily to the nucleus of H1299 cells but upon treatment with caffeine relocalizes to the cytoplasm. However, an N-terminal deletion of Apoptin that truncates the NES, GFP-Ap(42–121), as well as a point mutation of the Apoptin NES (GFP-Ap-pmNES), failed to relocalize to the cytoplasm when DDR signaling was inhibited with caffeine. It has previously been reported that the C-terminal T108 phosphorylation site of Apoptin can regulate localization of the protein. The T108 site lies within the NLS sequence of the protein and could potentially regulate its activity. In order to address the role of the Apoptin NLS in response to the DDR, we utilized an Apoptin mutant that replaces the Apoptin NLS, which includes T108, with the SV40 LgT NLS (GFP-Ap-SV40NLS). Figures 6B and C show that GFP-Ap-SV40NLS responds like the wild type to caffeine and, therefore, that regulation by T108 is not required for response to DDR signaling. This result is in agreement with the data in Fig. 2C showing no change in T108 phosphorylation upon induction of DNA damage. Taken together, these data indicate that the nucleocytoplasmic shuttling activity of Apoptin is essential for DDR-induced changes in localization but does not require T108 phosphorylation.
Fig. 6.
Apoptin nucleocytoplasmic shuttling activity is required to respond to DNA damage. (A) Schematic diagram of N-terminal GFP-tagged Apoptin mutants. (B) H1299 cells were transfected with the indicated GFP-Apoptin constructs and monitored by fluorescence microscopy for GFP (left) and DAPI (center). Merged GFP/DAPI images are shown on the right. (C) Percentages of cells displaying nuclear Apoptin under each of the experimental conditions shown in panel B were determined by microscopy. Error bars indicate standard deviations. *, P < 0.05. WT, wild type.
Apoptin inhibits formation of 53BP1-containing DNA damage foci.
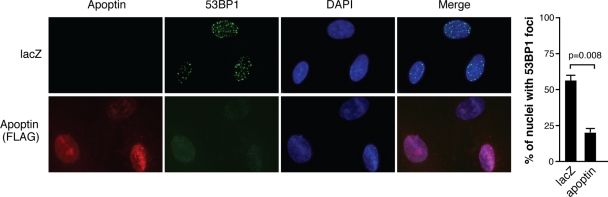
Figures 1C and 2B show that nuclear Apoptin is concentrated into discrete foci. Given that DNA damage is accompanied by the formation of nuclear foci, we asked whether Apoptin foci colocalize with markers of DNA damage. To address this question, control and Apoptin-expressing MRC5 cells were treated with BLM to induce DNA damage and analyzed by immunofluorescence with an antibody recognizing 53BP1, which is a marker of DNA damage. Figure 7 shows that cells infected with a control adenovirus expressing LacZ and treated with BLM displayed intense DNA damage foci in the nucleus. Surprisingly, when BLM-treated cells were infected with Ad-Apwt, DNA damage foci were less intense and much fewer in number than those in controls. Therefore, although DDR induces nuclear localization of Apoptin, once in the nucleus Apoptin may attenuate the response by preventing formation of DNA damage foci.
Fig. 7.
Apoptin reduces numbers of DNA damage foci. MRC5 fibroblasts were infected with Ad-Apwt or Ad-LacZ for 48 h and then treated with 10 μM BLM. Following 18 h of treatment, the cells were fixed and stained with anti-FLAG to detect Apoptin and anti-53BP1 to detect DNA damage foci. Nuclei were visualized using DAPI. Percentages of cells displaying DNA damage foci were determined by microscopy and are graphed on the right. Error bars indicate standard deviations.
Apoptin targets MDC1 through the APC/C.
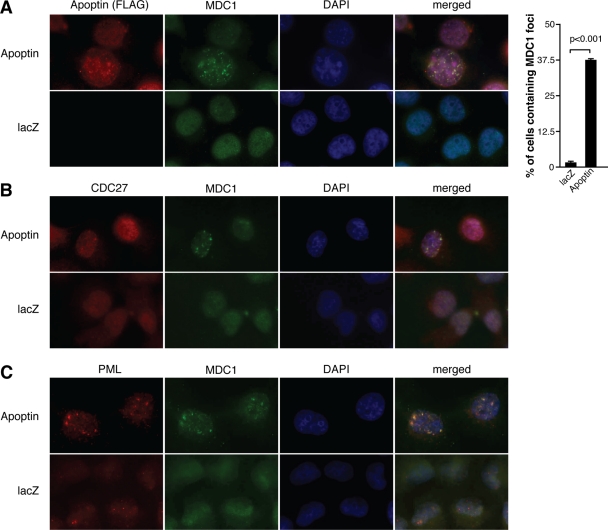
We previously showed that interaction of Apoptin with the APC/C causes the complex to relocalize to PML bodies, where it becomes inactivated through degradation (22, 41). Recent findings also established a link between the APC/C and a key adaptor in DDR signaling called the mediator of DNA damage checkpoint 1 (MDC1). These studies show that MDC1 is constitutively associated with several subunits of the APC/C (9, 43). MDC1 is an essential component of DDR signaling and is required to form DNA damage foci by recruiting several critical proteins, including 53BP1 (17, 29, 40). We therefore investigated the possibility that the Apoptin-mediated inhibition of DNA damage foci may be due to targeting the MDC1 protein through the APC/C. We first characterized the localization of MDC1 in Apoptin-expressing cells by immunofluorescence (Fig. 8A). As expected, MDC1 displayed a diffuse nuclear localization in control cells. In sharp contrast, approximately 40% of cells displayed MDC1 aggregated in foci in cells expressing Apoptin. Furthermore, Apoptin and MDC1 were often observed to colocalize within foci. As we previously found that Apoptin recruits the APC/C into foci that colocalize with PML, we examined whether MDC1 also colocalizes with these structures. Figures 8B and C show that MDC1 colocalizes in foci with an APC/C subunit (CDC27) in PML bodies specifically when Apoptin is expressed.
Fig. 8.
Apoptin induces MDC1 localization to PML bodies with the APC/C. H1299 cells were infected with Ad-Apwt or Ad-LacZ for 48 h and then fixed, and immunofluorescence was performed with the following antibodies: anti-FLAG (Apoptin) and anti-MDC1 antibodies (A), anti-MDC1 and anti-CDC27 antibodies (B), and anti-MDC1 and anti-PML antibodies (C). In all experiments, cell nuclei were visualized using DAPI. The percentages of cells containing MDC1 foci were determined by microscopy and are graphed on the right. Error bars indicate standard deviations.
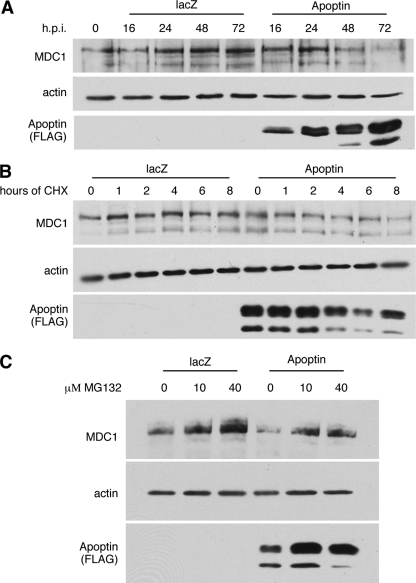
Since we previously demonstrated that localization of APC/C to PML bodies resulted in the degradation of APC/C subunits, including CDC27, we determined the protein levels of MDC1 in Apoptin-expressing cells. Immunoblot analysis of MDC1 from cells infected with Ad-Apwt or Ad-LacZ revealed a dramatic reduction in MDC1 steady-state levels specifically in Apoptin-expressing cells (Fig. 9A). In order to determine if the reduction in MDC1 protein results from a decrease in protein stability, we measured the half-life of MDC1 with a cycloheximide (CHX) experiment. Figure 9B shows that MDC1 protein levels decreased more rapidly over time in the presence of Apoptin than in LacZ control cells. To determine if the observed degradation of MDC1 is proteasome dependent, the experiment was repeated in the presence of MG132, a specific inhibitor of the 26S proteasome. Figure 9C shows that addition of MG132 resulted in a dose-dependent inhibition of Apoptin-induced MDC1 degradation. Taken together, these results demonstrate that, as with the APC/C, MDC1 in the presence of Apoptin is sequestered into PML bodies, where it is targeted for degradation in a proteasome-dependent manner.
Fig. 9.
Apoptin induces the degradation of MDC1 in a proteasome-dependent manner. (A) H1299 cells were infected with Ad-Apwt or Ad-LacZ and harvested at the indicated time points. Equal quantities of nuclear extract were loaded in each lane and separated by SDS-PAGE. The levels of MDC1 protein were then monitored by immunoblotting. Apoptin expression was detected using anti-FLAG, and actin was included as a loading control. (B) H1299 cells were infected with Ad-Apwt or Ad-LacZ for 40 h. Cells were then treated with cycloheximide for the indicated amounts of time, and nuclear extracts were prepared. Equal quantities of protein were separated by SDS-PAGE, and the levels of MDC1 protein were then monitored by immunoblotting. Apoptin expression was detected using anti-FLAG, and actin was included as a loading control. (C) H1299 cells were infected with Ad-Apwt or Ad LacZ for 42 h. Cells were then treated with MG132 at the indicated concentrations for 6 h, and nuclear extracts were prepared. Equal quantities of protein were separated by SDS-PAGE, and the levels of MDC1 protein were then monitored by immunoblotting. Apoptin expression was detected using anti-FLAG, and actin was included as a loading control.
DISCUSSION
Apoptin is thought to function by sensing a specific aspect of transformed-cell physiology, but a precise mechanism has yet to be elucidated. We show here for the first time that the localization of Apoptin responds to DNA damage signaling within cells and that the constitutive DDR signaling present in transformed cells may account for the unique localization characteristics of the protein. Our observations explain the curious result that the N-terminal domain of SV40 LgT is able to induce nuclear localization of Apoptin in primary cells, since this region also binds Bub1 and induces DNA damage (10). Apoptin expressed in primary cells is generally cytoplasmic, but we observe that upon induction of DNA damage, the protein translocates to the nucleus. Conversely, Apoptin expressed in transformed cells is almost entirely nuclear, but inhibition of DDR signaling either chemically or by RNAi results in cytoplasmic localization of the protein. Interestingly, although DDR signaling seems to be required for Apoptin activation, the result of Apoptin nuclear localization is the elimination of MDC1 and inhibition of DNA damage focus formation.
The observation that Apoptin senses and ultimately inhibits DDR focus formation provides insights toward understanding the role of the protein during CAV replication. Replicating viral genomes are interpreted as damaged DNA. Triggering the DDR during DNA replication could induce cell cycle arrest, DNA repair, and apoptosis, all of which can be deleterious to viral replication. For this reason, many viruses that replicate in the nucleus encode one or more proteins that manipulate the cellular DNA damage response of host cells (reviewed in reference 8). Adenovirus E1B-55K/E4orf6 (6), human T-cell leukemia virus tax (7), and SV40 T antigen (47) are good examples of proteins that exhibit this type of activity. The CAV genome is comprised of single-stranded DNA that replicates by rolling-circle replication and would be predicted to elicit strong DDR activation. Alternatively, some types of viruses have been shown to highjack the DNA repair machinery to enhance viral genome replication. ATM signaling during infection with human papillomavirus, for example, has been shown to increase viral DNA replication (31). Similarly, DDR signaling during parvovirus infection has also been shown to increase replication (30). Inhibiting the formation of DNA damage foci could potentially redirect DNA repair factors, such as DNA polymerases, from sites of cellular DNA damage to viral replication centers.
An intriguing possibility is that activation of Apoptin by the DDR may be a sequential step during viral replication. The production of viral DNA would trigger the host DDR, which would then induce the translocation of Apoptin into the nucleus, where it would trigger apoptosis. CAV replication in vivo has been reported to induce potent apoptosis in thymocytes, and virus particles are observed to be contained in apoptotic bodies that are absorbed by surrounding cells (24). The process of viral DNA replication may therefore trigger apoptosis in order to facilitate viral egress from infected cells and to promote viral spreading to surrounding cells.
In its natural setting, CAV kills thymocytes and hematopoietic precursor cells, resulting in catastrophic anemia, immunodeficiency, and often death (1). Both of these types of cells divide rapidly, which is a trait they share with cancer cells. A consequence of the rapid division is replication stress. This is defined as the occurrence of DNA damage as a result of DNA replication irregularities during the S phase of the cell cycle. DNA damage can result from the collision of replication forks, which then stall and degenerate into single- or double-stranded breaks. A common characteristic of cancer cells is therefore constitutive DNA damage signaling (reviewed in reference 20).
Nuclear localization of Apoptin was shown to be essential for apoptotic activity (12, 22); however, it is equally clear that other factors are required. For example, it has been shown that forced nuclear localization of Apoptin by fusion with the SV40 nuclear localization signal does not induce cell death in primary cells (12, 22). Our observations suggest that it is the combination of nuclear localization and DDR activation that can induce apoptosis in primary cells. Although it would be interesting to determine if inhibition of DDR signaling could protect cells from Apoptin-mediated death, we observed that inhibition of the ATM and ATR kinases either by RNAi or by small molecules resulted in high toxicity after extended treatments, and so it was not possible to conduct these experiments (data not shown). A previous study has suggested a potential relationship between DNA damage responses and Apoptin-induced apoptosis. Zhang et al. showed that low doses of UV-C or ionizing radiation could cause Apoptin to migrate to the nucleus and induce apoptosis of normal cells that originated from individuals carrying inherited mutations causing xeroderma pigmentosum (XP) (50). Cells from XP individuals are unable to repair some types of DNA damage and would therefore accumulate these lesions, which could potentially enhance Apoptin nuclear localization.
Although our results indicate that DDR signaling regulates Apoptin localization, the molecular mechanisms that mediate these effects remain unclear. One possibility is that Apoptin may itself bind damaged DNA. It was previously shown that Apoptin binds to DNA, preferentially to single-stranded DNA and to double-stranded DNA ends (28), but the significance of this observation has not been investigated further. Previous studies have reported that a phosphorylation site at the C terminus of Apoptin on T108 is differentially phosphorylated between primary and transformed cells and that this regulates localization and apoptosis (27, 36). However, a follow-up study has shown that although T108 phosphorylation does seem to enhance the apoptotic activity of the protein, it is not required for transformed-cell-specific localization (27). Our own published data have also shown that replacement of the C-terminal NLS of Apoptin, which includes the T108 site, with the SV40 NLS results in localization like that of the wild-type protein, also suggesting that this site is not absolutely required (22). In addition, as shown in Fig. 2C, a significant amount of T108 phosphorylation is observed even under conditions where Apoptin is highly cytoplasmic. In the current study, we show that although DDR signaling can dramatically affect Apoptin localization, it does not significantly change phosphorylation on T108, suggesting that other phosphorylation sites or possibly other types of posttranslational modification may play a role.
Although we observe that ATM and, to a lesser extent, DNA-PK are required for Apoptin activation, direct phosphorylation by these kinases appears unlikely since the Apoptin sequence does not contain any consensus motifs for ATM or DNA-PK phosphorylation [(S/T)QE or (S/T)Q]. Recently, protein kinase C (PKC) was shown to associate with Apoptin and induce its phosphorylation (25). Intriguingly, PKCδ displays characteristics similar to those of Apoptin in that it is also regulated by localization during apoptosis. Various types of apoptotic stimuli, including DNA damage, were shown to induce caspase-dependent cleavage of PKCδ (4, 5, 33, 35, 38). The cleaved PKCδ then translocates to the nucleus, where it phosphorylates a range of substrates and potentiates cell death. Interestingly, activation of PKCδ in response to DNA damage was shown to be downstream of ATM (48). Thus, it is possible that PKCδ may phosphorylate Apoptin in response to DNA damage signaling as well. In support of this notion, the amino acid sequence of Apoptin has seven potential PKC phosphorylation sites (S/TXK/R or S/TXXK/R).
Apoptin appears to potently inhibit the accumulation of 53BP1 in repair foci. These observations may be explained at least partly by the decrease in levels of MDC1, as shown in Fig. 6. Two recent publications (9, 43) have shown an association between MDC1 and the APC/C that appears to be important for regular mitotic progression as well as DNA damage. Apoptin sequesters and destabilizes MDC1 along with the APC/C, which would eliminate a critical factor required to form DNA damage foci. Apoptin interaction with the APC/C therefore seems to disrupt a central node controlling DNA repair mechanisms as well as mitotic checkpoint functions. Several types of animal viruses have been reported to bind and deregulate APC/C function (reviewed in references 21 and 39). In particular, cytomegalovirus (CMV) has been shown to inhibit the APC/C by inducing degradation of key subunits of the complex (44, 45). Interestingly, CMV has also been shown to inhibit the DDR by inducing the mislocalization of checkpoint proteins (15). It will be interesting to determine if APC/C targeting during CMV infection is also responsible for attenuation of the DDR.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institute of Health Research (CIHR) (MOP-179122) and the Natural Science and Engineering Research Council (NSERC) of Canada to J.G.T. J.G.T. is a CIHR new investigator and is a Chercheur-Boursier of the Fonds de la Recherche en Santé du Québec (FRSQ). T.J.K. is supported by a studentship from the FRSQ.
We thank Anthony Yu for technical assistance and Josée Dostie for reviewing the manuscript.
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Adair B. M. 2000. Immunopathogenesis of chicken anemia virus infection. Dev. Comp. Immunol. 24:247–255 [DOI] [PubMed] [Google Scholar]
- 2. Bartkova J., et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870 [DOI] [PubMed] [Google Scholar]
- 3. Bartkova J., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633–637 [DOI] [PubMed] [Google Scholar]
- 4. Bertho N., et al. 2002. MHC class II-mediated apoptosis of mature dendritic cells proceeds by activation of the protein kinase C-delta isoenzyme. Int. Immunol. 14:935–942 [DOI] [PubMed] [Google Scholar]
- 5. Blass M., Kronfeld I., Kazimirsky G., Blumberg P. M., Brodie C. 2002. Tyrosine phosphorylation of protein kinase Cdelta is essential for its apoptotic effect in response to etoposide. Mol. Cell. Biol. 22:182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carson C. T., et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandhasin C., Ducu R. I., Berkovich E., Kastan M. B., Marriott S. J. 2008. Human T-cell leukemia virus type 1 tax attenuates the ATM-mediated cellular DNA damage response. J. Virol. 82:6952–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaurushiya M. S., Weitzman M. D. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coster G., et al. 2007. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 282:32053–32064 [DOI] [PubMed] [Google Scholar]
- 10. Cotsiki M., et al. 2004. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc. Natl. Acad. Sci. U. S. A. 101:947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danen-Van Oorschot A. A., et al. 1997. Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc. Natl. Acad. Sci. U. S. A. 94:5843–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Danen-Van Oorschot A. A., et al. 2003. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J. Biol. Chem. 278:27729–27736 [DOI] [PubMed] [Google Scholar]
- 13. DiTullio R. A., Jr., et al. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998–1002 [DOI] [PubMed] [Google Scholar]
- 14. Flinterman M., et al. 2009. Delivery of therapeutic proteins as secretable TAT fusion products. Mol. Ther. 17:334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaspar M., Shenk T. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. U. S. A. 103:2821–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gjoerup O. V., et al. 2007. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc. Natl. Acad. Sci. U. S. A. 104:8334–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldberg M., et al. 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952–956 [DOI] [PubMed] [Google Scholar]
- 18. Gorgoulis V. G., et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907–913 [DOI] [PubMed] [Google Scholar]
- 19. Groitl P., Dobner T. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 130:29–39 [DOI] [PubMed] [Google Scholar]
- 20. Halazonetis T. D., Gorgoulis V. G., Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science (New York) 319:1352–1355 [DOI] [PubMed] [Google Scholar]
- 21. Heilman D. W., Green M. R., Teodoro J. G. 2005. The anaphase promoting complex: a critical target for viral proteins and anti-cancer drugs. Cell Cycle 4:560–563 [PubMed] [Google Scholar]
- 22. Heilman D. W., Teodoro J. G., Green M. R. 2006. Apoptin nucleocytoplasmic shuttling is required for cell type-specific localization, apoptosis, and recruitment of the anaphase-promoting complex/cyclosome to PML bodies. J. Virol. 80:7535–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hein J., et al. 2009. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 83:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeurissen S. H., Wagenaar F., Pol J. M., van der Eb A. J., Noteborn M. H. 1992. Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J. Virol. 66:7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang J., et al. 2010. Crucial roles for protein kinase C isoforms in tumor-specific killing by apoptin. Cancer Res. 70:7242–7252 [DOI] [PubMed] [Google Scholar]
- 26. Kulkarni A., Das K. C. 2008. Differential roles of ATR and ATM in p53, Chk1, and histone H2AX phosphorylation in response to hyperoxia: ATR-dependent ATM activation. Am. J. Physiol. 294:L998–L1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y. H., Cheng C. M., Chang Y. F., Wang T. Y., Yuo C. Y. 2007. Apoptin T108 phosphorylation is not required for its tumor-specific nuclear localization but partially affects its apoptotic activity. Biochem. Biophys. Res. Commun. 354:391–395 [DOI] [PubMed] [Google Scholar]
- 28. Leliveld S. R., Dame R. T., Rohn J. L., Noteborn M. H., Abrahams J. P. 2004. Apoptin's functional N- and C-termini independently bind DNA. FEBS Lett. 557:155–158 [DOI] [PubMed] [Google Scholar]
- 29. Lou Z., Minter-Dykhouse K., Wu X., Chen J. 2003. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421:957–961 [DOI] [PubMed] [Google Scholar]
- 30. Luo Y., et al. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J. Virol. 85:8046–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moody C. A., Laimins L. A. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noteborn M. H., et al. 1994. A single chicken anemia virus protein induces apoptosis. J. Virol. 68:346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pongracz J., et al. 1999. Spontaneous neutrophil apoptosis involves caspase 3-mediated activation of protein kinase C-delta. J. Biol. Chem. 274:37329–37334 [DOI] [PubMed] [Google Scholar]
- 34. Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A. 2005. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 65:7059–7064 [DOI] [PubMed] [Google Scholar]
- 35. Reyland M. E., Anderson S. M., Matassa A. A., Barzen K. A., Quissell D. O. 1999. Protein kinase C delta is essential for etoposide-induced apoptosis in salivary gland acinar cells. J. Biol. Chem. 274:19115–19123 [DOI] [PubMed] [Google Scholar]
- 36. Rohn J. L., et al. 2002. A tumor-specific kinase activity regulates the viral death protein Apoptin. J. Biol. Chem. 277:50820–50827 [DOI] [PubMed] [Google Scholar]
- 37. Sarkaria J. N., et al. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382 [PubMed] [Google Scholar]
- 38. Shizukuda Y., Reyland M. E., Buttrick P. M. 2002. Protein kinase C-delta modulates apoptosis induced by hyperglycemia in adult ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol. 282:H1625–H1634 [DOI] [PubMed] [Google Scholar]
- 39. Smolders L., Teodoro J. G. 2011. Targeting the anaphase promoting complex: common pathways for viral infection and cancer therapy. Expert Opin. Ther. Targets 15:767–780 [DOI] [PubMed] [Google Scholar]
- 40. Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966 [DOI] [PubMed] [Google Scholar]
- 41. Teodoro J. G., Heilman D. W., Parker A. E., Green M. R. 2004. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 18:1952–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Todd D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29:373–394 [DOI] [PubMed] [Google Scholar]
- 43. Townsend K., et al. 2009. Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J. Biol. Chem. 284:33939–33948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tran K., Kamil J. P., Coen D. M., Spector D. H. 2010. Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. J. Virol. 84:10832–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tran K., et al. 2008. Accumulation of substrates of the anaphase-promoting complex (APC) during human cytomegalovirus infection is associated with the phosphorylation of Cdh1 and the dissociation and relocalization of APC subunits. J. Virol. 82:529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Q. M., Fan G. C., Chen J. Z., Chen H. P., He F. C. 2004. A putative NES mediates cytoplasmic localization of Apoptin in normal cells. Acta Biochim. Biophys. Sin. (Shanghai) 36:817–823 [DOI] [PubMed] [Google Scholar]
- 47. Wu X., et al. 2004. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 18:1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoshida K., Wang H. G., Miki Y., Kufe D. 2003. Protein kinase Cdelta is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J. 22:1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., et al. 2007. Partial deficiency of DNA-PKcs increases ionizing radiation-induced mutagenesis and telomere instability in human cells. Cancer Lett. 250:63–73 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y. H., Abrahams P. J., van der Eb A. J., Noteborn M. H. 1999. The viral protein Apoptin induces apoptosis in UV-C-irradiated cells from individuals with various hereditary cancer-prone syndromes. Cancer Res. 59:3010–3015 [PubMed] [Google Scholar]
- 51. Zhang Y. H., Kooistra K., Pietersen A., Rohn J. L., Noteborn M. H. 2004. Activation of the tumor-specific death effector apoptin and its kinase by an N-terminal determinant of simian virus 40 large T antigen. J. Virol. 78:9965–9976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhuang S. M., et al. 1995. Apoptin, a protein derived from chicken anemia virus, induces p53-independent apoptosis in human osteosarcoma cells. Cancer Res. 55:486–489 [PubMed] [Google Scholar]