Abstract
Background
Cytoplasmic class XI myosins are the fastest processive motors known. This class functions in high-velocity cytoplasmic streaming in various plant cells from algae to angiosperms. The velocities at which they process are ten times faster than its closest class V homologues.
Results
To provide sequence determinants and structural rationale for the molecular mechanism of this fast pace myosin, we have compared the sequences from myosin class V and XI through Evolutionary Trace (ET) analysis. The current study identifies class-specific residues of myosin XI spread over the actin binding site, ATP binding site and light chain binding neck region. Sequences for ET analysis were accumulated from six plant genomes, using literature based text search and sequence searches, followed by triple validation viz. CDD search, string-based searches and phylogenetic clustering. We have identified nine myosin XI genes in sorghum and seven in grape by sequence searches. Both the plants possess one gene product each belonging to myosin type VIII as well. During this process, we have re-defined the gene boundaries for three sorghum myosin XI genes using fgenesh program.
Conclusion
Molecular modelling and subsequent analysis of putative interactions involving these class-specific residues suggest a structural basis for the molecular mechanism behind high velocity of plant myosin XI. We propose a model of a more flexible switch I region that contributes to faster ADP release leading to high velocity movement of the algal myosin XI.
Background
Myosin is an actin based motor protein that generates motion using chemical energy released through ATP hydrolysis. Myosins play many important roles within plant cells such as organelle trafficking [1,2], remodelling [3,4], and inheritance [5]. They are also known to be involved in development of various plant parts like root hairs, pollen etc [6,7]. Though there are around 24 classes of myosins reported in eukaryota [8], only three classes - class VIII, XI and XIII are seen in plants. The similarity of plant myosin sequences with animal and fungal class V myosins [9], derived from phylogenetic analysis, suggests a common ancestor from which plants and Opisthokonts [10] might have evolved.
Myosins, in general, function through an ATP hydrolysis cycle by converting the hydrolysis energy to allosteric conformational changes within the motor head as well as the neck region leading to motion. Swinging cross-bridge hypothesis, proposed by H. E. Huxley, has been the most popular model to explain the molecular mechanism of energy transduction in myosins. Numerous biochemical and biophysical experiments thereafter helped to improve it to the present day swinging lever arm model [11], according to which, immediately after the release of ADP from the previous cycle, ATP binds to myosin head that is in an actin-bound post-stroke conformation. Upon hydrolysis of ATP, myosin head gets transformed to pre-stroke conformation which is actually actin unbound form. Upon rebinding to the next actin molecule, Pi is released first and the ADP bound myosin head changes conformation from weak to strong actin binding state. This will be followed by ADP release and conformational changes at the head domain back to post-stroke state where myosin strongly interacts with actin.
In plants, from algae to angiosperms, the high velocity cytoplasmic streaming (of the range 40-60 μm/s) is driven by myosins [12]. Myosin XI, found in plants, is the fastest known motor and it moves processively along actin filaments towards the plus-end, performing cellular functions like cytoplasmic streaming and vesicle transport [1,2]. Myosin XI is architecturally similar to class V myosin in animals, with a motor head followed by six IQ motifs, a coiled-coil and a globular cargo-binding domain called DIL [10]. Myosin XI, just like myosin V, functions as a dimer formed through coiled-coil interaction between the α-helical tail regions of the monomers. Both the classes have comparable step size (an average of 35nm) and same directionality towards the plus end of actin filament.
Due to the difficulties in crystallization of actin-myosin complex, the actual actin binding residues are not known even in well-characterized myosins like myosin II and V. However, docking studies have revealed the actomyosin interface residues (see ref. [13] for a review). Actomyosin interface is extensive in the rigour state because of the interaction of a single head with regions on two adjacent actin molecules. Rigour state contact between actin and myosin head can be divided into four regions: a large primary binding site on the face of actin, which on three sides, is flanked by three additional sites from surface loops [14]. In this study, using the Evolutionary Trace method, we have identified crucial residues at the actin binding site, at the ATP binding domain and at the beginning of neck region that could contribute to the fast release of ADP and the high velocity. During the process of accumulation of myosin XI sequences from plants, we have recognised nine Myosin XI sequences from sorghum and seven from grape through genome-wide survey and gene prediction.
Algal myosin XI, isolated from Chara corallina [15], slides F-actin in vitro at a speed equivalent to cytoplasmic streaming speed of 40- 60 μm/s, which is 10 times the speed generated by myosin V [16,17]. Studies with tobacco myosin XI heavy isoform, by Tominaga and coworkers, revealed that single myosin XI molecules move at velocity 7 μm/s along the actin and generate relatively smaller force, in the order of 0.5 pN [8] much smaller than the force generated by muscle myosin II [18] and by myosin V [19-21]. In an attempt to elucidate the mechanism of this fast movement of Chara myosin, Ito and co-workers measured its kinetic properties [22]. The rate constant of ADP dissociation from actin-motor domain complex of Chara myosin XI was estimated to be more than 2800 s-1 and the rate constant of ATP-induced dissociation of motor domain from the actin was 2200 s-1 at a physiological concentration of ATP. The estimated time spent on actin, in strongly bound state, was estimated to be <0.82ms. This value is the shortest among known values for various myosins and it has a duty ratio of <0.3 and a Vmax of actin-activated ATPase activity of 390 s-1 [22]. ADP release, which is the rate limiting step in all other myosin types, is dramatically accelerated in this plant myosin. Most of the myosins possess positively charged residues on loop 2 where as Chara myosin has no net positive charge on loop2. Instead, positively charged residues are harboured on loop 3. Ito and coworkers investigated the effect of positively charged residues on the loops at the actin binding region and provided evidence for its partial role on the high velocity movement through mutation studies [23]. Still, the sequence signatures that lead to the specialization of these myosins as the fastest motors and the actual molecular mechanism of such a rapid process are not known completely. In this study, based on sequence analysis and molecular modelling, we propose that the sequence signatures at the switch I region, the ATP-binding site and the neck region as partly responsible for the observed high rate of ADP release, which in turn lead to the specialization of these myosins as the fastest ones.
Methods
Sequence search and validation
A database of 143 myosin sequences belonging to 19 different classes was developed through text search in NCBI and by consulting a database of myosins - The myosin home page (http://www.mrc-lmb.cam.ac.uk/myosin/myosin.html). The members of this database were used as queries in PSI-BLAST against sorghum and grape genomes. A relatively relaxed E-value cut-off of 10-5 and a query length coverage filter of 25% were used in PSI-BLAST in order to retrieve even distantly related myosins. Hits were filtered for myosins through a three-fold validation procedure. In the first level, string-based scripts were used to gather completely or partly annotated myosin hits (Category 1) from respective GENEPEPT flat files. Mis-annotated or unannotated hits (Category 2) were gathered by using perl scripts based on the percentage identity of the hit with the query sequence. An identity cut-off of 35% was used here. The two categories of myosin hits obtained from sorghum and grape were checked for their domain architecture using PFAM [24], CDD [25], SMART [26] and COILS2 [27] servers. The consensus domain architecture, after consulting the domain repositories, was obtained for each of the validated hits. This forms the second level of filtering where false positive hits that deviate from the general domain architecture of myosins are eliminated. Such myosins were next subjected to gene prediction using fgenesh program [28]. Wherever the NCBI annotations were partial, full-length genes were obtained through fgenesh program and the gene boundaries are re-defined. The final sequence set obtained after these filters were annotated based on the co-clustering of motor domain (on the basis of sequence similarities) with representative sequences from 19 known myosin classes using neighbour-joining method of clustering in Phylip package [29]. This representative sequence dataset, obtained from 19 classes of myosins, were also seeded with representative sequences from three non-myosins - kinesin, ATPase and helicases - as outgroups to ensure the phylogenetic relationship of putative myosins with known myosins. This method of genome scan and validation is pictorially represented in a flowchart [Figure 1].
Figure 1.
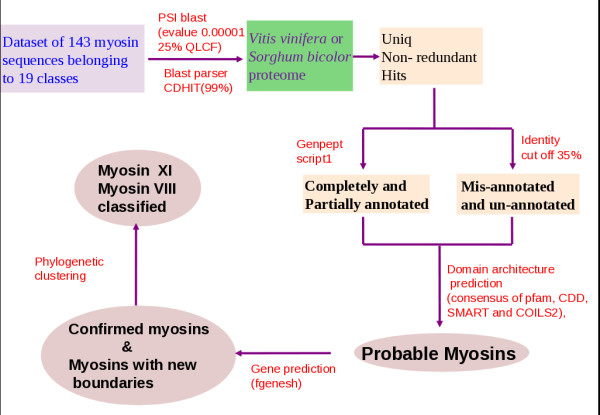
Schema of extensive sequence search in two plant genomes: sorghum and grape.
Evolutionary Trace (ET) analysis
Class-specific residues were identified using ET method for myosin V and XI members. Multiple sequence alignment for 30 sequences from 10 genomes was done using CLUSTALW [30]. ET analysis was initially guided by Trace Suite II [31]. Sequences were divided into three parts based on domain architecture: 1) N-terminal SH3-like domain 2) motor domain and 3) light chain binding neck region. Class-specific residues for the motor domain and light chain binding neck domains were mapped on to myosin V crystal structure (PDB ID: 2DFS) and clustering of conserved residues were analysed.
Molecular modelling of algal myosin XI
Homology model of the algal myosin XI (Chara corallina) was obtained using MODELLER [32]. Myosin V from chicken (PDB code: 1W7J) was used as a template [33]. Sequence identity between algal and chicken myosin head domains was 41%. Further, the predicted secondary structures of algal myosin head were comparable with the secondary structures identified in the template (chicken myosin head), except for fine-tuned boundaries. The best energy model from a set of 20 models, generated by MODELLER, was passed through various validation processes viz. WHATIF [34] and VERIFY-3D [35]. The model was energy minimized to relieve any inherent strain in the model and to obtain the most favourable conformation including side chains. The energy minimized structures were used to visualize the class-specific residues using PYMOL [36].
Results and discussion
Sequence search and validation identify putative myosins in soghum and grape
Proteomes of a monocot and a dicot (Sorghum bicolor and Vitis vinifera ) were scanned for myosin types using PSI-BLAST preceding a three-fold validation as mentioned in Methods. Additional myosin class V and XI sequences were obtained from three different plant genomes: Arabidopsis thaliana, Oryza sativa and Zea mays. Motor domains of validated hits, including those with new predicted boundaries (Table 1) were subjected to clustering (Figure 2) so as to annotate their subfamily types and classified them as belonging to myosin VIII or XI subfamily. Sorghum genome harbours 10 myosin genes distributed in seven chromosomes, whereas grape genome contains only eight myosins out of which seven are myosin XI isoforms (Figure 2) spread over eight chromosomes (Table 1). Sorghum has one type VIII and nine type XI myosin genes. For three of the predicted myosin XI in sorghum, we have re-defined the gene boundaries using fgenesh gene prediction program. We assigned new boundaries to XP_002458397 in Chromosome 3 and merged two partial genes (XP_002440148 & XP_002440149) into a single gene with complete domain architecture of type XI myosin.
Table 1.
Details of confirmed myosins from sorghum and grape after multi-fold validation
| Organism | Id/Acc number | Chromosome region | Gene ID | Protein length | %ID* | Annotation | Motor domain |
|---|---|---|---|---|---|---|---|
| VvMXI1 | XP_002281615 | Chr4 1597682 - 1630481 |
100254166 | 1517 | 55 | XI | 54-730 |
| VvMXI2 | XP_002285579 | Chr5 10420215 - 10531891 |
100259730 | 1513 | 61 | XI | 57-734 |
| VvMXI3 | XP_002268099 | Chr7 8436820 - 8456586. |
100256590 | 1518 | 62 | XI | 63-739 |
| VvMXI4 | XP_002274978 | Chr9 1674607 - 1696216. |
100266890 | 1535 | 77 | XI | 66-743 |
| VvMXI5 | XP_002279028 | Chr11 1332352 - 1348606 |
100243893 | 1586 | 75 | XI | 113-790 |
| VvMXI6 | XP_002263591 | Chr14 360176 - 377881 |
100243373 | 1587 | 70 | XI | 113-790 |
| VvMXI7 | XP_002263354 | Chr7 6130176 - 6182090 |
100265237 | 1204 | 59 | XI(DILminus) | 126-803 |
| VvMVIII | XP_002281748 | Chr19 3770898 - 3797194 |
100241905 | 1229 | 30 | VIII | 210-888 |
| SbMXI1 | XP_002463461 | Chr1 186074 - 205207 |
8061371 | 1557 | 56 | XI | 85-810 |
| SbMXI2 | XP_002466464 | Chr1 7057755 - 7066246 |
8059553 | 1464 | 60 | XI | 57-733 |
| SbMXI3 | XP_002461898 | Chr2 14576408 - 14597301 |
8057935 | 1497 | 59 | XI | 56-732 |
| SbMXI4 | XP_002458046 | Chr3 52499134 - 52510404 |
8078219 | 1529 | 70 | XI | 108-783 |
| SbMXI5 | XP_002458397 | 61226792 - 61232762 new:61219493-61232763 |
8069786 | 499 1498 |
49 | XI | 56-952 |
| SbMXI6 | XP_002452906 | Chr4 64687149 - 64704955 |
8082670 | 1520 | 53 | XI | 51-731 |
| SbMXI7 | XP_002453038 | Chr4 66880218 - 66891041 |
8056444 | 1347 | 60 | XI | 1 to 566 |
| SbMXI8 | XP_002440148 & XP_002440149 | Chr9 56017722 - 56023175 & 56024467-56037464 New:56017040-56037361 |
8061068 & 8061069 | 610 & 1399 2176 |
51 | XI | 113-796 |
| SbMXI9 | XP_002436758 | Chr10 8229550 - 8241900. |
8057883 | 1539 | 66 | XI | 67-743 |
| SbMVIII | XP_002467046 | Chr1 19620879 - 19630164 |
8067384 | 1196 | 32 | VIII | 190-868 |
* Full-length percentage identity with Arabidopsis myosin XI
Figure 2.
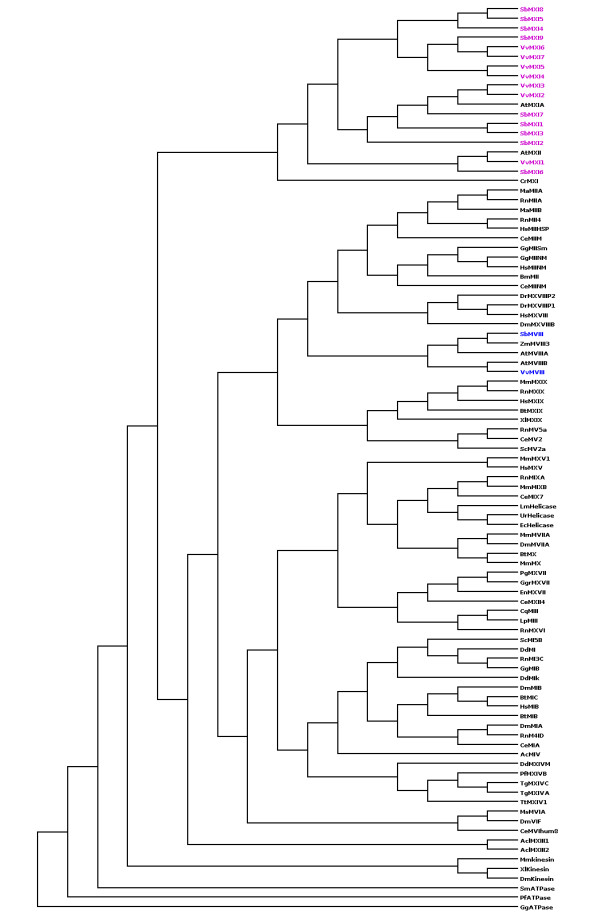
Unrooted phylogenetic tree showing the evolutionary relationship of putative myosins from sorghum and grape. Myosin XI types are in pink colour and Myosin VIII types are in blue.
Both sorghum and grape genomes contain only one copy of myosin VIII type in chromosome 1 and 19, respectively (Table 1).
The domain architecture predicted for these new myosins from sorghum and grape are shown in Figures 3 and Figure 4, respectively. All the myosin XI gene products from sorghum and grape followed the common domain architecture of motor head-coiled coil-DIL except one, namely VvMXI7 (Table 1). Myosin VIII from both the genomes had the two-domain architecture of motor head followed by coiled coil. Myosin VIII sequences do not possess features of a compact domain at the N-terminal region before the head domain, where as many of the myosin XI types have an N-terminal SH3-like domain that is involved in interaction with actin [37]. The effects of presence or absence of the co-existing domains on their motility and cellular behaviour are yet to be understood.
Figure 3.
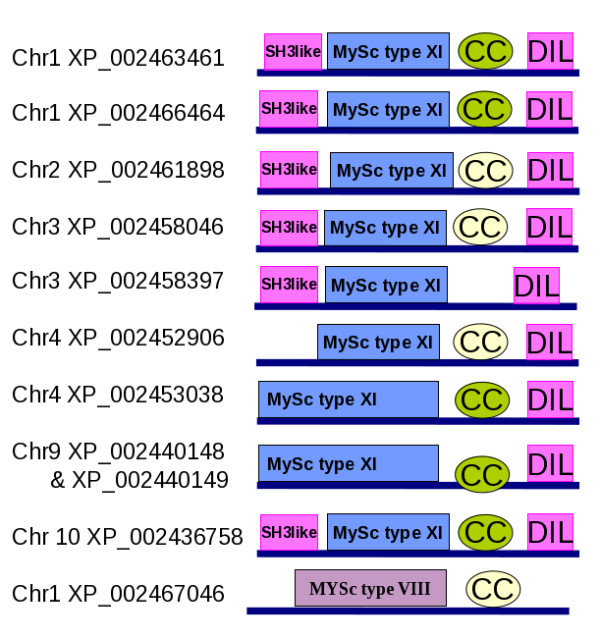
Domain architectures of putative myosins from Sorghum. XP_002458397, on Chromosome3, was a hypothetical partial gene and assigned a new gene boundary according to fgenesh. The domain architecture corresponds to the new gene boundary. XP_002440148 & XP_002440149 of chromosome 9 are two adjacent incomplete genes, which are merged to a single gene with new gene boundaries and the corresponding domain architecture is shown here. 'CC' denotes the coiled coil domain. Green colour indicates that it is detected by domain prediction servers and off-white colour indicates that it was predicted using COILS program. 'Chr' indicates the chromosome on which the gene is located.
Figure 4.
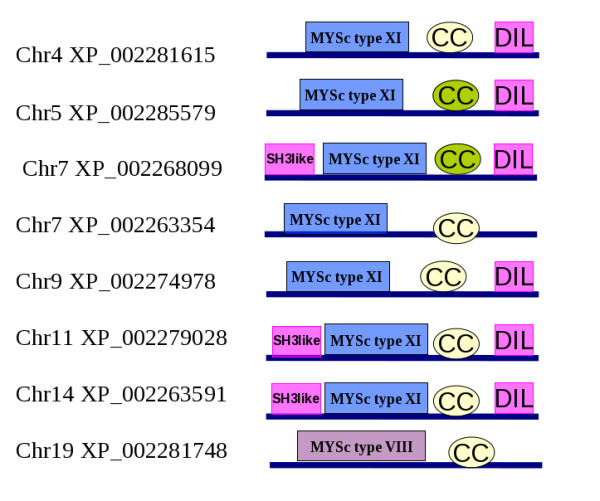
Domain architectures of putative myosins from grape, their chromosome and the domain architecture.
Class-specific residues in myosin XI lines actin binding site
The full-length of myosin XI shares 31% overall identity with myosin V and the motor head domains alone share a high sequence identity (above 40%). There is a high level of sequence conservation between the two classes at the active site residues. Nevertheless, the two classes differ considerably at functionally important sites and mechanistic details, according to the literature (as mentioned in Introduction). Myosin XI head contains 21 class-specific residues, out of which six appear on the actin binding sites. Known actin binding sites on myosin II fall into three regions: A primary binding site comprising of a helix-loop-helix and an adjacent helix, a secondary site which is a charged loop and a tertiary site lying at the front or nose of the myosin molecule [13]. The corresponding regions on myosin V are marked on the 2DFS structure in red color (Figure 5). All the three sites contain class-specific residues. This implicates the possible involvement of these residues in imparting the differential actin-binding strength of the two types of myosins.
Figure 5.
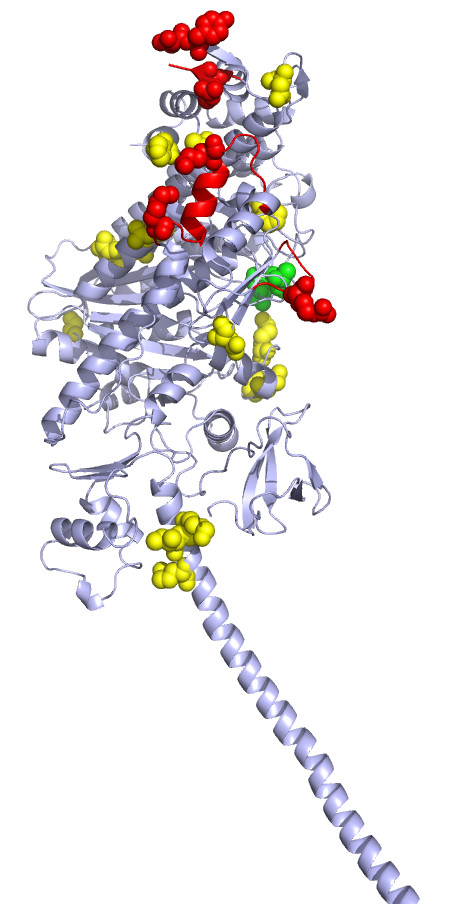
Class-specific residues (shown as spheres) mapped on to the structure of myosin V (PDB Id: 2DFS). Red spheres are the class-specific residues at the putative actin binding site. Few class-specific residues observed near the actin binding site and neck region are shown in yellow. The green spheres are the class-specific residues at the ATP binding site.
It was recently shown that the ATP-induced motor head dissociation in myosin XI occurs at a very fast rate of 2200s-1, resulting in stunningly fast movement of myosin XI [22]. In our analysis, we identified two myosin XI-specific residues in the primary binding site (helix-loop-helix) that corresponds to K502 and K514 of chicken myosin V. In myosin XI, K502 and K514 are mutated in a class-specific manner to proline and methionine, respectively. This may introduce a structural kink and local rigidity to the loop and disrupt the charged interaction of the primary binding site to actin. This can help the myosin XI motor head to get unbound sooner from the actin upon ATP binding after the release of Pi and ADP in the previous ATP-hydrolysis cycle.
Apart from the three key actin binding sites, surrounding residues may also get involved in actin binding. Few of the class-specific residues observed near the actin binding site may also contribute to binding strength (yellow spheres at the actin binding -region in Figure 5). The second actin binding site on myosin XI also harbours a class-specific residue: Lysine, but the mutation R542K seems to be too subtle to introduce a difference in the interaction with actin.
Putative involvement of two myosin XI specific residues in switch I flexibility
The switch I region, which drives an open-close conformational change in the myosin head during ATP hydrolysis [38], has two class-specific residues (shown in green in Figure 5). One of these is Asn 214 (algal myosin XI numbering), corresponding to Asp215 of myosin V. In myosin V, Asp215 has a favourable charged interaction with Lys668, whereas in myosin XI, due to changes of Asp215 to Asn214 and Lys688 to Leu640, this stabilizing interaction is likely abolished which could lead to a more flexible switch I region. These class-specific residues are mapped on the three-dimensional model of algal myosin head (Figure 6A). Since the movement at the switch I region is intimately associated with γ-phosphate binding, coordination of Mg2+ and there by ADP binding and release, we propose a more flexible switch I region in myosin XI that might facilitate an early or easy release of ADP.
Figure 6.
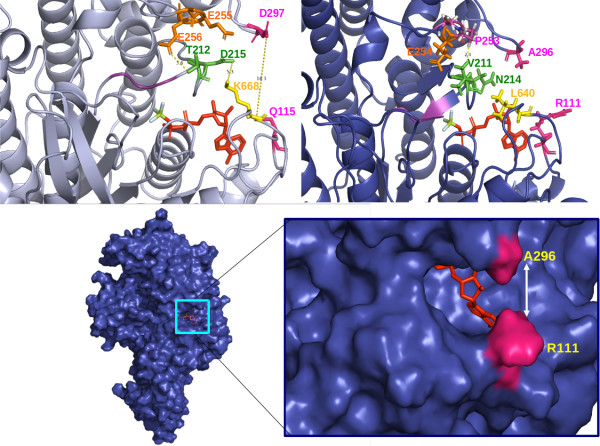
Residues lining the ATP binding site of A) chicken myosin V and B) Chara myosin XI model are shown in stick representation C) Structural model of Chara myosin XI showing the ATP binding site and the shutter residues coloured in pink and D) a close-up of shutter region.
In Myosin V, Thr 212 is located close to a Glutamate triad (Glu254, Glu255 and Glu256) and it stabilizes the switch I through a favourable interaction with Glu256. In plant myosin XI, there is an interesting co-evolution at this location, where a spatially proximate Thr212 is replaced by Val and Glu255 by Pro (Figure 6A &6B). As a result of this, stabilizing polar interaction of Thr212 with Glu255 observed in myosin type V appears obsolete in myosin XI. A new possible interaction between Valine and Proline seems to be stabilizing the switch I region of myosin XI (Figure 6A, 6B).
Shutter residues might make the ATP binding cavity closure inefficient in Chara coralina
A closer investigation of the ATP binding site (Figure 6C &6D) of the myosin V crystal structure, reveal two mutations in plant myosins that would probably decide the opening and closing of the small cleft at the ATP binding site. Asp297 is located in an upper loop at the face of the cleft and has a potential polar interaction partner Gln115 located in a lower loop of myosin V. In Figure 6A, this interaction is highlighted through a dotted line in the near-rigour-open conformation, as seen in the crystal structure of myosin V. This interaction can form a vertical shutter for the ATP binding cavity. These 'shutter' residues are Ala296 and Arg111 in myosin XI from Chara coralina, which are stabilised by weak dispersion forces, and hence the closure of the cleft may become inefficient in Chara myosin.
Weakened interaction between the neck and the converter domain affects lever arm tilting
We have also examined the neck region (also termed as 'lever arm') for class-specific residues and the conservation of amino acid types. We identified a cluster of three class-specific residues, two of which (Ile762 and Lys766, chicken myosin V numbering) have potential interactions with the converter domain in the case of myosin V (coloured yellow in Figure 5, Additional files 1 and 2).
(a) Ile762 (in chicken myosin V, please see Additional file 2) at the beginning of the neck region, is embedded between two hydrophobic residues - a highly conserved Leu714 and a conserved hydrophobic residue at position 735 - of the converter domain forming a "hydrophobic triad". The corresponding residue of Ile762 in plant myosin XI neck region is a charged arginine that possibly weaken the hydrophobic interaction of the N-terminal neck with the converter domain.
(b) Spatially close to this "hydrophobic triad" in myosin V (please see Additional files), the side chain of Lys766, the second class specific residue, interacts with the backbone oxygen of Val713 located at the converter domain by forming a hydrogen bond. In plant myosin XI, Lys766 is replaced by a Valine in a class specific manner. As a result of this change, a weak hydrophobic interaction alone could prevail between the neck and the converter domains of myosin XI (Additional file 1). Such amino acid differences could provide higher conformational flexibility to plant myosins in tilting the lever arm (neck region) with ease, which in turn is associated with a faster ADP release and actin dissociation. The third class-specific residue (not shown in Additional file 1) is likely to interact with SH3 domain.
Conclusion
For a processive myosin to move at high speed on the actin filament, the best way is to strike an optimal duty ratio so that the dwell time is just sufficient to locate the next binding site on actin. ADP release is the rate limiting step of ATP hydrolysis cycle and the associated movement of myosin along the actin filament. One of the possible ways to attain speed is to hasten the ADP release process. We propose that altered sequence conservation patterns could redefine structural interactions and impart differences in speed. A poorly closed ATP binding cavity, together with a more flexible switch I region, is compelling to think of a model for faster release of ADP in plant myosins. From our sequence analysis, the switch I region bears unique sequence signatures in the fast-acting myosin. As shown in Figure 7, this region may be pulled away from the ADP binding site resulting in the loss of co-ordination of Mg2+, which in turn would make the ADP release easier. As the gating loops of the ATP binding cavity in Myosin XI are not directly interacting, due to class-specific amino acid differences, the closing of the cavity may be for a very short period and this may facilitate the release of ADP smoothly without any further delay.
Figure 7.
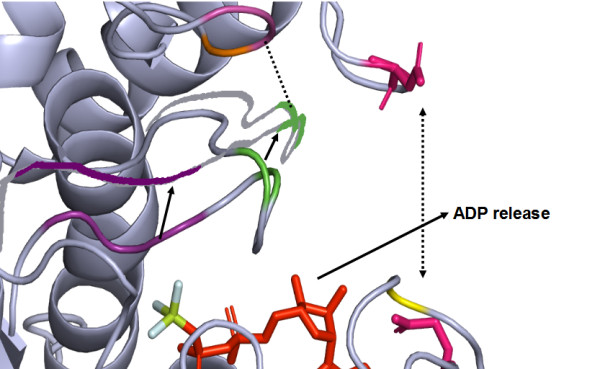
A model depicting the possible mechanism of ADP exit in plant myosin XI and the associated conformational changes.
From this comparative sequence analysis, we could recognise most crucial residue changes that may be responsible for decreased/altered affinity of myosin XI to the actin and rapid ADP release in comparison with myosin V. A molecular mechanism of fast movement of myosin XI could be proposed: As long as ADP is bound to the myosin head, actin binding is stabilized through numerous weak interactions. Due to the enhanced flexibility of the switch I region, ADP gets released very fast after hydrolysis. Release of ADP permits the next ATP to bind and subsequently dissociate the acto-myosin complex through suitable conformational changes at the actin-myosin interface, which would further be enhanced due to the absence of few stabilizing charged interactions. Meanwhile, since the neck region in myosin XI interacts only poorly with the converter domain, the neck flexibility is also high which might assist in faster catalysis and binding events at the head domain.
Authors' contributions
DPS carried out the study and performed the computational analysis. RS participated in the analysis and comparative sequence analysis. DPS drafted the manuscript and RS has improved to provide the final manuscript. All the authors have read and approved the final manuscript.
Supplementary Material
The class-specific residues of myosin XI are mapped over the neck region of myosin V crystal structure.
Class-specific residues (I762K and K766V) at the neck region are shown on the multiple sequence alignment.
Contributor Information
Divya P Syamaladevi, Email: dpsdevi@gmail.com.
R Sowdhamini, Email: mini@ncbs.res.in.
Acknowledgements
DPS's stay at NCBS is financially supported by NCBS and Sugarcane Breeding Institute, Coimbatore. This work is supported by Human Frontier Science Program. We also thank NCBS (TIFR) for infrastructural support.
References
- Boevink P, Oparka KJ, Cruz SS, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313X.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I. Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell. 2005;17:888–902. doi: 10.1105/tpc.104.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions J, Brach T, Kuhner S, Hawes C. Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot. 2006;57:43–50. doi: 10.1093/jxb/eri289. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin L. Stop-and-go movements of plant Golgi stacks are mediated by the acto myosin system. Plant Physiol. 1999;121:1127–1141. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW. Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 2004;37:379–390. doi: 10.1046/j.1365-313x.2003.01967.x. [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Jarve K, Paves H, Truve E. Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma. 2007;230:193–202. doi: 10.1007/s00709-006-0233-8. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Cai M, Ramachandran S. Oriza sativa myosin XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol. 2007;304:579–592. doi: 10.1016/j.ydbio.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K. Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. The EMBO Journal. 2003;22(6):1263–1272. doi: 10.1093/emboj/cdg130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos C, Huet S, Darchen F. 'Should I stay or should I go?': myosin V function in organelle trafficking. Biol Cell. 2007;99:411–423. doi: 10.1042/BC20070021. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Current Biology. 1997;7(2):112–118. doi: 10.1016/S0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- Nagai R, Hayama T. Ultra structure of the edoplasmic factor responsible for cytoplasmic streaming in Chara internodal cells. J Cell Sci. 1989;36:121–136. doi: 10.1242/jcs.36.1.121. [DOI] [PubMed] [Google Scholar]
- Milligan RA. protein _protein interactions in the rigour actomyosin complex. Proc Natl Acad Sci USA. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Kikuyama M, Sutoh-Yamamoto N, Kamitsubo E. Purification of actin based motor protein from Chara corallina. Proc Japan Acad Series B. 1994;70:175–180. doi: 10.2183/pjab.70.175. [DOI] [Google Scholar]
- Anson M. Temperature dependence and Arrhenius activation energy of F-actin velocity generated invitro by skeletal myosin. J Mol Biol. 1992;224:1029–1038. doi: 10.1016/0022-2836(92)90467-X. [DOI] [PubMed] [Google Scholar]
- Pierobon P, Achouri S, Courty S, Dunn AR, Spudich JA, Dahan M, Cappello G. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys J. 2009;96(10):4268–4275. doi: 10.1016/j.bpj.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford WH, Dupuis DE, Kennedy G, Wu J, Patlak JB, Warshaw DM. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: A molecular model for processivity. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nature Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- Higayashi-fujime S, Ishikawa R, Iwasawa H, Kagami O, Kuri-moto E, Kohama K, Hozumi T. The fastest-actin-based motor protein from the green algae, Chara and its distinct mode of interaction with actin. FEBS lett. 1995;375:151–154. doi: 10.1016/0014-5793(95)01208-V. [DOI] [PubMed] [Google Scholar]
- Ito K, Ikebe M, Kashiyama T, Mogai T, Kon T, Yamamoto K. Kinetic Mechanism of the Fastest Motor Protein, Chara Myosin. Journal of Biological Chemistry. 2007;282(27):19534–19545. doi: 10.1074/jbc.M611802200. [DOI] [PubMed] [Google Scholar]
- Ito K, Yamaguchi Y, Yanase K, Ichikawa Y, Yamamoto K. Unique charge distribution in surface loops confers high velocity on the fast motor protein Chara myosin. Proc Natl Acad Sci USA. 2009;106(51):21585–21590. doi: 10.1073/pnas.0910787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucl Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Dyke MV, Stock J. Predicting Coled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio Gene Finding in Drosophila Genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Innis CA, Shi J, Blundell TL. Evolutionary trace analysis of TGF and related growth factors: implications for site-directed mutagenesis. Protein Eng 2000. pp. 839–847. [DOI] [PubMed]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. Embo J. 2004;23:4527. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHATIF server. http://swift.cmbi.ru.nl/servers/html/index.html
- Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods in enzymology. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.2r3pre. Schrödinger, LLC; [Google Scholar]
- Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. The EMBO Journal. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin induced nucleotide release in myosin. Nature Structural Biology. 2003;10(10):826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The class-specific residues of myosin XI are mapped over the neck region of myosin V crystal structure.
Class-specific residues (I762K and K766V) at the neck region are shown on the multiple sequence alignment.