Summary
The DNA elements responsible for centromere activity in a metazoan have been localized using the Drosophila minichromosome Dp1187. Deleted minichromosomes were generated by irradiation mutagenesis, and their molecular structures were determined by pulsed-field Southern blot analysis. Analyses of the transmission behavior of Dp1187 derivatives localized sequences necessary for chromosome inheritance within the centric heterochromatin. The essential core of the centromere is contained within a 220 kb region that includes significant amounts of complex DNA. Completely normal inheritance also requires ~200 kb on either side of the essential core. This flanking DNA predominantly contains highly repeated sequences, and the amount required for normal transmission differs among division types and between the sexes. We propose that the essential core is the site of kinetochore formation and that flanking DNA provides two functions: sister chromatid cohesion and indirect assistance in kinetochore formation or function.
Introduction
Chromosome replication, segregation, and transmission are mechanisms used by organisms and cells to ensure faithful transfer of essential genetic traits. Errors in chromosome inheritance can result in genomic abnormalities (aneuploidy) that cause a variety of human disorders, including birth defects (e.g., Down's syndrome) and cancer. Meiotic and mitotic chromosome transmission require interactions between a specific chromosomal region (the centromeric DNA) and the cellular machinery responsible for chromosome movement (kinetochores, spindle-associated microtubules, and centrioles). Chromatid cohesion and separation also are required for proper disjunction of chromosomes and sister chromatids (Miyazaki and Orr-Weaver, 1994). The term centromere has been used historically to describe a cytologically visible component of chromosomes that, among other properties, appears as a constriction and serves as the site of spindle attachment (White, 1973). Here, we use centromere to refer to the minimal DNA element sufficient to promote normal transmission, which includes kinetochore and chromatid cohesion functions.
Great progress has been made recently (Ault and Rieder, 1994) in identifying protein components of the kinetochore (Earnshaw and Tomkiel, 1992; Doheny et al., 1993), characterizing the behavior of spindle microtubules during mitosis (Mitchison and Salmon, 1992; Rieder and Salmon, 1994), and identifying some of the molecular motors responsible for chromosome movement (Yen et al., 1992; Goldstein, 1993; Middleton and Carbon, 1994). However, a complete understanding of how accurate chromosome inheritance is accomplished and regulated requires identification of the DNA sequences that act in cis to ensure chromosome pairing and movement, and determination of how these elements interact with the cellular “machines.” Molecular-genetic approaches have successfully identified and characterized the centromeric DNA and some centromere-binding proteins in the unicellular eukaryotes Saccharomyces cerevisiae and Schizosaccharomyces pombe (Clarke et al., 1993; Hegemann and Fleig, 1993). Studies utilizing these elegant and powerful systems will continue to make important contributions to our understanding of eukaryotic chromosome behavior. However, significant structural differences between the chromosomes of unicellular and multicellular eukaryotes, and the instability of yeast minichromosomes in animal cells (Allshire et al., 1987; Fitzgerald, 1987), suggest that these systems may not be appropriate models for chromosome inheritance in multicellular eukaryotes. Furthermore, multicellular organisms display diverse types of chromosome cycles and cell divisions, such as polyploidy and polyteny, DNA elimination, germline and somatic mitoses, syncytial nuclear divisions, meiosis I, and meiosis II. Understanding the developmental regulation of these processes requires study of inheritance elements in higher eukaryotes.
One important characteristic that distinguishes higher eukaryotic centromeres from those of unicellular eukaryotes is that the former are embedded in large blocks of heterochromatin (White, 1973), often megabases in length. Heterochromatic sequences also are involved in chromosome pairing during meiotic divisions (McKee and Karpen, 1990; Hawley et al., 1993) and maintenance of sister chromatid contact and kinetochore apposition during mitosis (Lica et al., 1986; Miyazaki and Orr-Weaver, 1994). Heterochromatin is sparsely populated with genes, constitutively condensed throughout the cell cycle, replicated late in S phase, and rich in tandemly repeated satellite sequences (John, 1988). The presence of repeated DNA has made molecular-genetic analyses of heterochromatin and centromeres extremely difficult (Cook and Karpen, 1994; Le et al., 1995).
Numerous studies have associated tandemly repeated satellite DNAs with centromeres, but identification of a specific role for satellite DNA in kinetochore formation or function has been lacking (Tomkiel and Earnshaw, 1993). The presence of highly repeated sequences in the immediate vicinity of the centromere (Dvorkin and Hamkalo, 1991) does correlate with cytological studies indicating that higher eukaryotic kinetochores are composed of repeated components (Brinkley et al., 1992). Analyses of rearranged mammalian chromosomes indicate that retention of alphoid satellite DNA can be correlated with chromosome stability (Tyler-Smith et al., 1993; Brown et al., 1994). Alphoid DNA inserted into ectopic chromosomal locations exhibits some properties of centromeres, but does not promote complete centromere function (Earnshaw et al., 1989; Haaf et al., 1992; Larin et al., 1994). The exact function of alphoid or other satellite DNAs in inheritance is unclear in large part because the transmission behavior of molecularly defined components has not been assayed directly. Many questions remain unanswered. How large is a functional higher eukaryotic centromere? Is it composed of repeated DNA, single-copy sequences, or both? What are the nucleotide sequences responsible for centromere function, and what are their biochemical roles?
A more complete understanding of chromosome inheritance in higher eukaryotes requires an experimental system in which chromosomal domains can be manipulated genetically and molecularly and in which structural and functional properties can be assessed. Drosophila is a genetically tractable metazoan that fulfills these requirements. More than 80 years of study provide convenient genetic and cytological assays for chromosome functions in vivo, during both mitosis and meiosis (Ashburner, 1990). Furthermore, there is a fully functional Drosophila minichromosome (Dp1187), whose relatively small size (1.3 Mb) makes it amenable to molecular analysis and manipulation (Karpen and Spradling, 1990, 1992; Tower et al., 1993). Previously we reported that restriction mapping of Dp1187 revealed a surprising amount of substructure in the centric heterochromatin; it contains alternating blocks of complex DNA and highly repeated simple satellite sequences (Le et al., 1995). Here, we define the DNA elements necessary for centromere function in Drosophila by generating Dp1187 deletions and studying their inheritance in vivo.
Results
Gross Localization of a Dp1187 Centromere
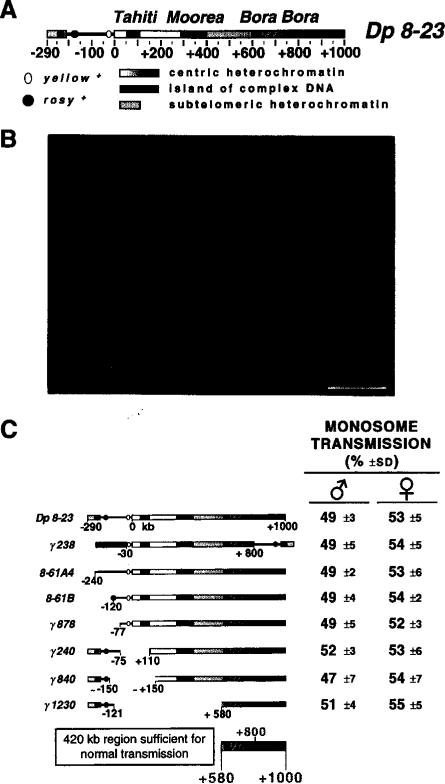
The structure of the Dp1187 derivative Dp 8-23 is shown in Figure 1A. This minichromosome is present in addition to the normal Drosophila chromosome complement and is significantly smaller than the other chromosomes, including the small fourth chromosome (Figure 1B). Furthermore, the presence or absence of Dp1187 has no effect on the viability of the organism. Pulsed-field restriction mapping of Dp 8-23 and its rearranged derivatives revealed the presence of three “islands” of complex DNA named Tahiti, Moorea, and Bora Bora (Le et al., 1995). These islands contain middle-repetitive or single-copy DNA (or both), whereas the interisland “seas”-predominantly consist of highly repeated satellite sequences.
Figure 1. The Structure of Dp1187 and Gross Localization of a Centromere.
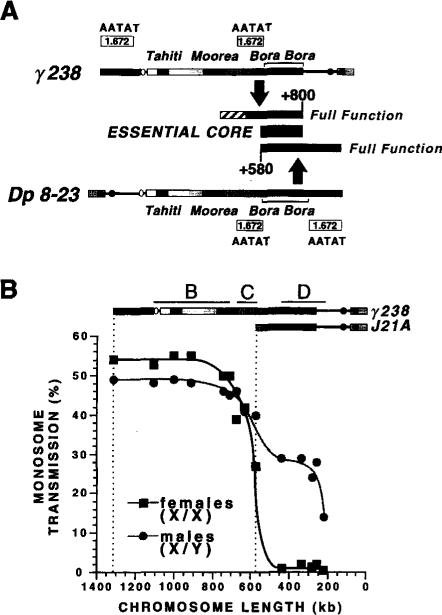
(A) Structure of Dp 8-23. Dp 8-23 contains sequences from the X-tip euchromatin (black line) and subtelomeric heterochromatin (closed box) juxtaposed to 1 Mb of X centric heterochromatin (stippled gradient, 0 to +1000 kb). Two ry+ marker genes (stippled circles) introduced by P element transposition are present in the Dp1187 derivative Dp 8-23 (Tower et al., 1993), in addition to the y+ gene (open oval) normally present in the X and Dp1187 euchromatin. The kilobase coordinates correspond to those determined for the parental Dp1187 (Karpen and Spradling, 1990, 1992); the actual size of Dp 8-23 is 1320 kb, because each PZ insertion adds 14.5 kb to the 1290 kb Dp1187 length. The centric heterochromatin contains islands of complex DNA (single-copy or middle-repetitive sequences; closed boxes, Tahiti, Moorea, and Bora Bora) separated by blocks composed predominantly of highly repeated satellite DNA (stippled blocks). At least eight enzymes failed to cut in the central region of Bora Bora (diagonal bars); this region may contain satellite DNA or an unusual chromatin structure (Le et al., 1995).
(B) Cytological visualization of Dp8-23. A metaphase squash from a third instar larval brain neuroblast is shown (bar equals 5 μm). The monosomic minichromosome (Dp) and the X, 2, 3, and 4 chromosome pairs are marked.
(C) Gross localization of a Dp1187 centromere. The monosome transmission behavior of Dp 8-23 derivatives was monitored from both male and female parents (percent ± SD; see Experimental Procedures). From these analyses we conclude that a centromere and other sequences essential to inheritance through mitosis and meiosis are located maximally between +580 and +1000, which includes Bora Bora and satellite DNA.
The generation of rearranged Dp1187 derivatives provided a method for localizing the centromeric DNA to a specific part of the minichromosome. Molecular alterations associated with chromosome instability positively identify regions necessary for normal inheritance. Deletions that do not alter transmission identity sequences dispensable for centromere function. We tested the meiotic and mitotic transmission of a series of Dp1187 derivatives with a monosome transmission assay (see Experimental Procedures). In this genetic test, recovery of a derivative in 50% of the progeny reflects normal transmission and thus normal centromere function. The normal behavior of all the Dp1187 derivatives shown in Figure 1C, including the 620 kb derivative γ1230, indicates that the entire euchromatin, subtelomeric heterochromatin and telomere (–290 to 0), and over one half of the Dp1187 centric heterochromatin (0 to +580) are dispensable for chromosome stability. Normal transmission in this in vivo assay requires chromosome stability in five types of divisions: preblastoderm nuclear mitoses; germline mitoses; meiosis I; meiosis II in the parent; as well as preblastoderm nuclear mitoses and postblastoderm somatic mitoses in the progeny. Thus, sequences necessary and sufficient for centromere activity in all five types of divisions reside in the 420 kb region from +580 to +1000, which includes the complex island Bora Bora and highly repeated satellite DNA. The stable transmission of the inversion derivative γ238 suggests that a centromere lies to one side or the other of the +800 heterochromatic breakpoint. If repeated and independently functional centromeric sequences flanked this breakpoint, the γ238 chromosome should have behaved as an unstable dicentric.
Generating γ238 Terminal Deletion Derivatives
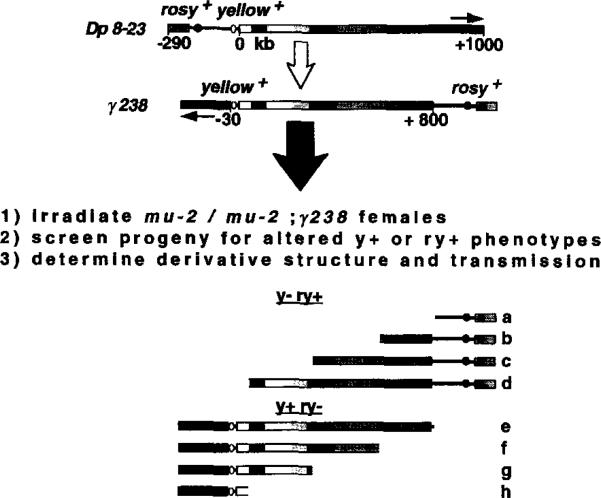
The large-scale deletion analysis reported in Figure 1C localized a centromere and other sequences sufficient for normal transmission to a 420 kb region of the centric heterochromatin. However, the use of unidirectional deletions left open the possibility that other regions of this minichromosome also could provide centromere function. Furthermore, the +580 to +1000 region of Dp1187 contains at least two distinct structural domains that could be involved in centromere function, the 230 kb island Bora Bora and a block of similar size that includes simple sequence DNA (Figure 1A).
A bidirectional deletion analysis was performed to determine the number, extent, and nature of Dp1187 regions able to provide centromere function. The γ238 inversion provided an excellent mutagenesis substrate, because the yellow+ (y+) and rosy+ (ry+) marker genes are on opposite sides of the centric heterochromatin and the +580 to +1000 region is split and separated (Figure 2). Terminal deficiencies (single-break events) were generated that removed either the right (ry+) or left (y+) end of γ238 (see Experimental Procedures for details). Hypothetically, recovery and stable transmission of only derivatives b, c, d, and e would support a critical role for Bora Bora in centromere function, whereas recovery and stable transmission of e, f, g, and h would substantiate an essential role for the satellite DNA. Recovery and stable transmission of b, c, d, e, f, g, and h would indicate that Dp1187 contains more than one heterochromatic region that can provide centromere function.
Figure 2. Generating Bidirectional Deletion Derivatives Using the γ238 Inversion.
The structures of Dp 8-23 and the irradiation-induced derivative γ238 are shown (Le et al., 1995; orientation of the centric heterochromatin is indicated by the stippled gradient and arrows). Terminal deficiencies were generated and analyzed as described in steps 1, 2, and 3 (see Experimental Procedures). The mutation mu2 allows terminal deficiencies to be recovered at very high frequencies after females are treated with low radiation doses (Mason et al., 1984). y- ry+ lines are candidates for harboring minichromosomes with deletions in the left end of γ238, while y+ ry- lines could contain minichromosomes that lacked the right end. Hypothetically, random induction of terminal breaks during γ238 mutagenesis could generate derivatives with the structures shown below (a–h).
The Structures of γ238 Derivatives
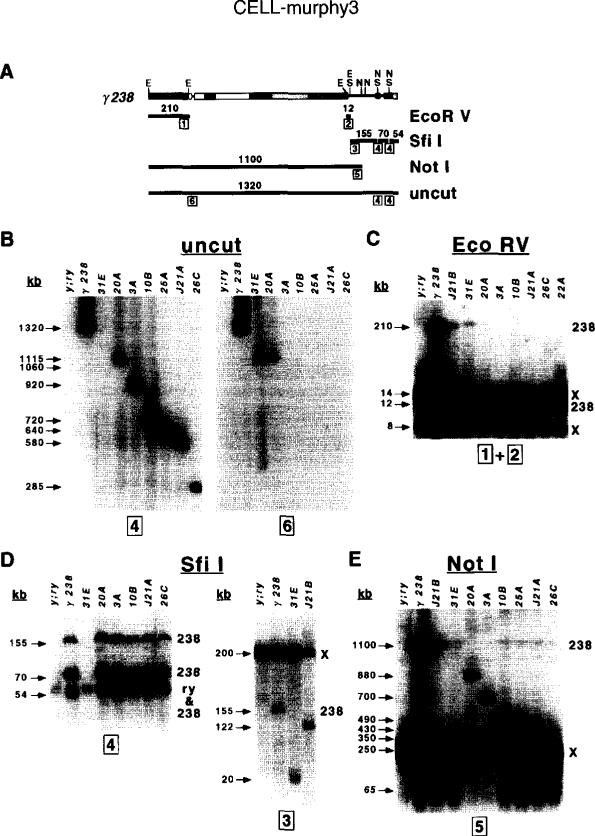
y- ry+ and y+ ry- lines were analyzed with pulsed-field Southern blot analysis (Le et al., 1995) to ascertain whether minichromosomes were present and to determine their size and structure. Regions spanning the entire parental chromosome were analyzed by hybridizing single-copy probes from different parts of γ238 (Figure 3A) to Southern blots containing uncut or restriction-digested high molecular weight embryonic DNA. The overall size of the derivatives was determined by analyzing uncut DNA (Figure 3B). The structures of the left and right ends of the derivatives were ascertained with EcoRV (Figure 3C) and Sfil digests (Figure 3D), and the amount of centric heterochromatin in each derivative was determined from the Notl digests (Figure 3E).
Figure 3. Structural Analyses of γ238 Derivatives.
(A) Restriction fragments used to analyze the structures of γ238 derivatives. The relevant restriction sites are shown above the chromosome (E, EcoRV; S, Sfil; N, Notl), while the fragments are shown as thick closed lines below. The sizes of the fragments in kilobases are indicated above each line, while the probes (1–6, in boxes) are indicated below at their chromosomal locations (see Experimental Procedures).
(B–E) Examples of pulsed-field Southern blot hybridization analyses performed on uncut, EcoRV-digested, Sfil-digested, and Notl-digested high molecular weight DNAs. See Experimental Procedures for further analyses of the results. Sizes of bands are shown on the left of each blot (in kilobases), and the genomic sources of fragments present in the parental genotype (y; ry506; γ238) are indicated on the right (238, γ238; X, endogenous X chromosome; ry, endogenous ry locus). The probes hybridized to each blot are indicated below. J21B and 31E were recovered as y+ ry- lines, while the other derivatives shown in this figure were y- ry+ lines. Minichromosomes and restriction fragments that contain large amounts of heterochromatin displayed weak signals (e.g., 210 kb fragment in EcoRV digest and uncut chromosomes >1000 kb), likely owing to incomplete transfer to membranes (Glaser and Spradling, 1994) and inefficient probe hybridization. The following run conditions were used: (B) uncut DNA, 50–110 s, 2 s ramp, 28 hr at 180V, 160 mA; (C) EcoRV digests, 1–30 s, 1 s ramp, 18 hr at 180V, 160 mA; (D) Sfil digests, 1–30 s, 1 s ramp, 24 hr at 180V, 160 mA; (E) Notl digests, 40–100 s, 2 s ramp, 22 hr at 180V, 160 mA.
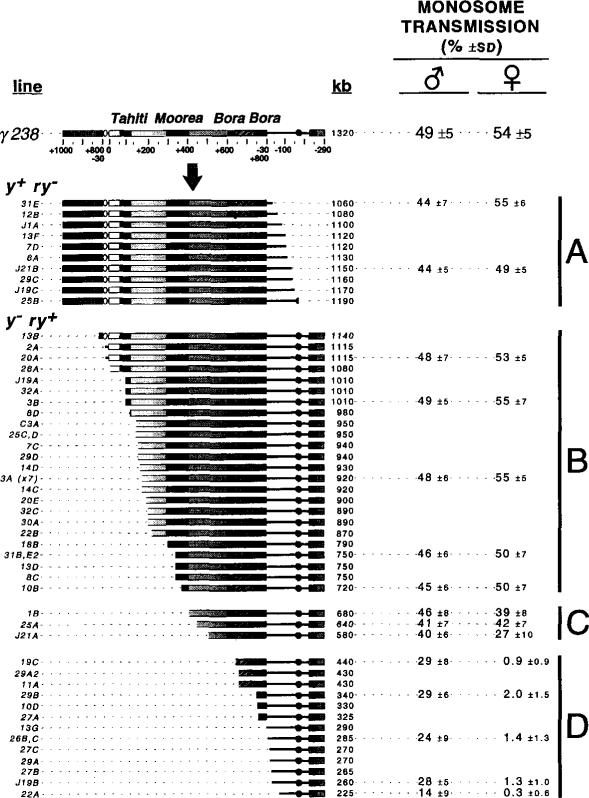
A summary of the structures of the γ238 derivatives, determined by combining the results of all the pulsed-field mapping experiments, is shown in Figure 4. A very high frequency of useful derivatives (54 independent derivatives from 25,000 irradiated chromosomes, or 0.22%) was generated during this screen (see Experimental Procedures for further discussion of frequencies). All the y+ ry- derivatives (group A in Figure 4) retained the left end of γ238 and all the centric heterochromatin; these derivatives had breakpoints restricted to the euchromatin. All the y- ry+ derivatives (groups B, C, and D in Figure 4) retained the right end of γ238. These derivatives had terminal deficiency breakpoints that were distributed randomly throughout most of the Dp1187 centric heterochromatin, with a breakpoint on average every 20 kb. No breakpoints were recovered in the +660 to +750 region of the island Bora Bora or between +510 and +650, as discussed below.
Figure 4. Structures and Transmission Behavior of γ238 Derivatives.
The structures of the γ238 derivatives (line designations on the left) were determined by pulsed-field Southern blot analyses (Figure 3; see Experimental Procedures). The total size is displayed to the right of each structure in kilobases and ranged from 225 kb to 1190 kb. The monosome transmission behavior of selected derivatives was determined as before (see Figure 1C and Experimental Procedures). Groups A and B displayed normal transmission in males and females, group C was partially unstable in females, and group D was highly unstable in females and partially unstable in males.
Bora Bora Is Essential for Minichromosome Transmission
The γ238 derivatives were tested for monosome transmission efficiency to identify regions important for centromere function. Derivatives were completely stable (groups A and Bin Figure 4), moderately unstable (group C), or highly unstable (group D). The stability of groups A and B indicates that Drosophila transmits terminal deficiency derivatives well in the absence of selection, unlike mammalian or yeast cells. The structures of stable and unstable derivatives demonstrate that the complex island Bora Bora plays an essential role in Dp1187 inheritance. First, all deletions of the right end of γ238 (y+ ry- derivatives) that were recovered retained Bora Bora (group A), despite the large number of breaks generated throughout the centric heterochromatin (groups B, C, and D). Second, derivatives with deletions of the left end (y- ry+) were transmitted well only if they contained all of Bora Bora (groups B and C). Third, y- ry+ derivatives that lack part or all of Bora Bora were recovered but were highly unstable, displaying average transmission frequencies of only 0.3%–2% from young female parents (group D).
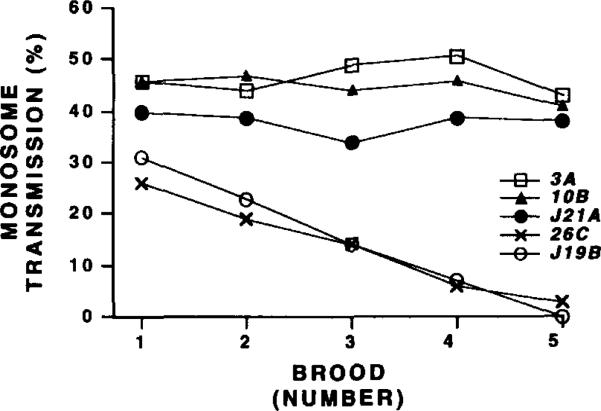
Surprisingly, derivatives that lack part or all of Bora Bora were moderately stable when transmission was measured in young males (0–2 days posteclosion, group D in Figure 4; 14%–29%). We monitored loss during additional divisions by sequentially mating males to a new brood of females every 3 days. These brooding experiments demonstrated that γ238 derivatives lacking Bora Bora are highly unstable, even in males. Group D derivatives (e.g., 26C and J19B) displayed a steady decline from 26%–32% transmission in brood 1 to 0%–2% in brood 5 (Figure 5). The brood 5 frequencies were similar to the high instability of 26C and J19B observed in young females (Figure 4). Decreased transmission during brooding is likely caused by instability in germline mitoses (see below). In contrast, larger derivatives that retained Bora Bora displayed constant transmission rates through successive broods (J21A, 10B, and 3A in Figure 5). We conclude that derivatives lacking all or some of Bora Bora are different from those that retain this complex island: they display high instability in males as well as females, a behavior expected of acentric chromosome fragments (Mather and Stone, 1933). The significance of the difference between male and female transmission of unstable derivatives will be discussed below.
Figure 5. Transmission of Acentric γ238 Derivatives Decreases During Brooding.
The monosome transmission (Y-axis, percent) of five derivatives (3A, 10B, J21A, 26C, and J19B) was determined for males mated to fresh virgin females at 2, 5, 8, 11, and 14 days after eclosion (X-axis; broods 1-5, respectively). Only chromosomes that lack Bora Bora (26C and J19B) displayed significantly reduced transmission during brooding.
The results obtained from inheritance studies of the Dp 8-23 (see Figure 1) and γ238 (see Figure 4) derivatives are compared in Figure 6A. The 420 kb required for full transmission in both sets of derivatives (stippled boxes in Figure 6A) overlap at the region between +580 and +800 (closed box). We conclude that the essential core of the centromere is contained within this 220 kb region, principally composed of the complex island Bora Bora. Stable transmission requires the presence of both ends of the +600 to +800 portion of Bora Bora and perhaps most of this complex island. Right-end deletions with breaks to the left of +800 were not recovered, and left-end deletions with breaks to the right of +600 were highly unstable, even when most of Bora Bora was present (e.g., 19C). We also conclude that the +800 to +1000 portion of Dp1187 is not essential to centromere function and that Dp1187 contains only one region that can provide normal centromere function.
Figure 6. Summary of the Regions Required for Dp1187 Transmission.
(A) Localizing the Dp1187 centromere. Summary of the results from the transmission tests of the Dp 8-23 and γ238 derivatives. For the γ238 derivatives (Figures 4 and 5), the +510 to +800 region (stippled box) promoted moderate stability, whereas another 140 kb of centric heterochromatin was necessary for full function (stippled diagonals). The sufficiency of the +580 to +1000 region (lower stippled box) was demonstrated with the Dp 8-23 derivatives (Figure 1C). Thus, the +580 to +800 region of overlap (closed box, Bora Bora) is essential to achieve stable transmission, but 200 kb of flanking heterochromatin on either side of Bora Bora provides a redundant function necessary for completely normal behavior. The left and right flanking regions both contain blocks of the 1.672 (AATAT) satellite DNA (Le et al., 1995).
(B) Comparison of the transmission behavior of γ238 derivatives in males and females. The monosome transmission of y- ry+ derivatives (Y-axis, percent) is plotted relative to length (X-axis, kilobases). Data taken from Figure 4. Note that the shape of the two curves is similar, but transmission in males is affected less by deletion of flanking heterochromatin or the removal of Bora Bora. The breakpoint distributions for groups B, C, and D (Figure 4) are indicated.
Completely Normal Chromosome Stability Requires Bora Bora plus Flanking Heterochromatin
Bora Bora is clearly essential for Dp1187 transmission, but is not sufficient for completely normal inheritance. y- ry+ derivatives that contain Bora Bora but only 90–190 kb of flanking heterochromatin were moderately unstable, especially in females (group C in Figure 4). Transmission was significantly less efficient as more flanking heterochromatin was removed (female transmission for 10B is 50%, for 1B is 39%, and for J21A is 27%; these derivatives contain 230 kb, 190 kb, and 90 kb of flanking heterochromatin, respectively). This trend is most clearly visible in the graphed data presented in Figure 6B; derivatives that were stable in both males and females contained >200 kb of flanking DNA. The normal transmission behavior of γ1230 (see Figure 2) and 10B (see Figure 4) and of larger derivatives indicates that the inclusion of >200 kb of flanking heterochromatin on either side of Bora Bora is sufficient for normal inheritance (Figures 6A and 6B). Low resolution mapping suggests that 1.672 satellite DNA (AATAT repeats in Figure 6A) constitutes most of the region between Moorea and Bora Bora (+400 to +600), as well as the stretch from Bora Bora to the end of the chromosome (+830 to +1000; Le et al., 1995). We conclude that the DNA flanking Bora Bora, which contains AATAT satellite sequences, provides a redundant function that is necessary to achieve completely normal levels of transmission.
Normal Chromosome Transmission Requires Different Amounts of DNA in Different Sexes and Types of Divisions
The transmission behavior of moderately and highly unstable derivatives differed between males and females (Figure 6B). Although the shape of the two curves was similar, the magnitude of the instability for “genetically challenged” chromosomes differed between the sexes. Moderately stable y- ry+ derivatives that contain Bora Bora but limited amounts of flanking DNA were transmitted better in males than in females (e.g., J21A, 40% in males versus 27% in females; see Figure 4). As discussed earlier, sex-specific differences were most visible for the highly unstable acentric derivatives (group D in Figures 4 and 6B).
The brooding experiments (Figure 5) address one source for the instability displayed by some γ238 derivatives. If a chromosome is transmitted abnormally through germline stem cell divisions, then reduced transmission should be observed after successive matings, which cause sperm pools to be replenished with additional germline mitoses (Hannah-Alava, 1965). The highly unstable acentric derivatives, such as 26C and J19B, were remarkably sensitive to successive matings (Figure 5). Thus, at least part of the low transmission displayed by acentric fragments is caused by instability in germline mitoses. Conversely, moderately to completely stable derivatives, such as J21A, 10B, and 3A, showed similar levels of transmission in broods 1 through 5 (Figure 5). Therefore, the lower transmission (40%) displayed by J21A males is not caused by instability in germline mitoses, and loss must be restricted to preblastoderm mitoses or meiosis (or both). We conclude that the DNA sequences required for chromosome inheritance differ between the sexes and among division types.
Discussion
Structure and Function of the Bora Bora Essential Core
We have identified DNA elements necessary for chromosome transmission in a metazoan using molecular–genetic dissection of the Drosophila minichromosome Dp1187. 420 kb of centric heterochromatin must include, and may define, the fully functional centromere, since it is sufficient for completely normal transmission. A 220 kb essential core, composed predominantly of the complex island Bora Bora, is necessary to achieve good levels of stability.
Chromosome inheritance requires replication in addition to centromere function, and it is possible that parts of the essential core are also required to promote replication (e.g., origins). Identification of minimal centromeres in the yeasts S. cerevesiae (Hegemann and Fleig, 1993) and S. pombe(Baum et al., 1994) assumed that complete replication was provided by inclusion of autonomously replicating sequences (ARSs) in the minichromosomes. In our study, the 260 kb of euchromatin plus subtelomeric heterochromatin in the stable y- ry+ derivatives is likely to provide the same function. This region is large enough to contain multiple origins, based on the measured replicon lengths in embryos, cultured cells, and polytene nuclei (Blumenthal et al., 1973). The replication competence of this region is supported by the reasonable transmission of acentrics (e.g., 26C) in young males. Regardless, DNA elements required for normal segregation and transmission must contain the centromere by definition, even if additional properties and functions are present. Therefore, we conclude that the Bora Bora essential core includes the minimal region required for centromere function.
What parts of Bora Bora are required for centromere activity? Stable transmission requires the presence of both ends of the essential core. We conclude that the complex DNA (single-copy sequences, middle-repetitive sequences, or both) present in these regions (closed boxes in Figure 6A; Le et al., 1995) are likely to be essential for Dp1187 centromere function. At this time, we are unable to assess the importance of sequences in the center of Bora Bora (diagonal bars in Figure 6A). This region contains either highly repeated DNA or complex DNA that is in an unusual chromatin structure or is covalently modified (see Figure 1A). It is worth noting that no derivatives were recovered with breakpoints in the +510 to +650 and +660 to +750 regions. Perhaps the structure of this region provided protection from breakage, or recovery of such derivatives was inhibited owing to highly aberrant transmission behavior. More refined analyses are needed to assess directly the structure and importance of specific parts of Bora Bora.
What is the molecular function of the Bora Bora essential core? Normal chromosome transmission requires attachment to and movement along spindle fibers, which are mediated by the trilaminar kinetochore in Drosophila and other eukaryotes (Goldstein, 1981; Ault and Rieder, 1994). We propose that Bora Bora includes the site of kinetochore formation (Figure 7) based on the absolute requirement for this region. As described below, other functions are likely to be encoded by the nonessential portions of the centromere. Further cytological and molecular-genetic analyses will be necessary to determine whether the island is involved directly in microtubule binding and motor activity.
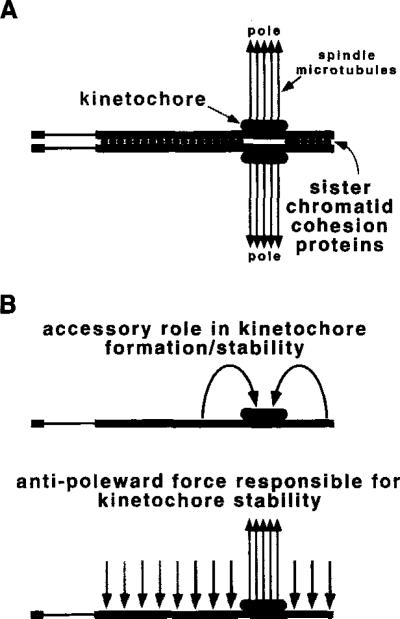
Figure 7. Models for Transmission Functions Encoded by Dp1187 Centric Heterochromatin.
The Bora Bora essential core is proposed to encode kinetochore formation (large stippled ovals), which interacts with spindle microtubules (closed arrows) to promote chromosome movement.
(A) Here the flanking heterochromatin is proposed to play a role in sister chromatid cohesion. Cytological analyses suggest that many different heterochromatic sequences can participate in this redundant function (light stippled circles), but our results define the minimal DNA sequences to be 200 kb on either side of Bora Bora (dark stippled circles).
(B) The flanking heterochromatin also could play an accessory role in kinetochore formation, stability, or both. In the model displayed on top, the flanking regions act in cis to assist kinetochore function at Bora Bora, for example by altering chromatin structure. An alternative model is displayed below, in which antipoleward microtubule-mediated forces (Rieder and Salmon, 1994) act on the flanking heterochromatin. Such forces would indirectly promote kinetochore stability at Bora Bora by creating tension (Li and Nicklas, 1995; Murphy and Karpen, 1995). Contrapoleward or interchromosomal forces are not shown, but could produce the same result. Again, 200 kb of flanking DNA would be sufficient for full function (dark stippled arrows), but this does not exclude the participation of other heterochromatic regions (light stippled arrows). In all three models, asingle chromosome in mitotic metaphase or meiosis II is shown, but the same functions could act on bivalents in meiosis I.
The Function of Flanking DNA
Full transmission also requires 200 kb of flanking heterochromatic DNA on either side of the Bora Bora essential core (Figure 6A). The two domains of flanking DNA contain AATAT satellite sequences and are functionally redundant. The failure to recover y+ ry- derivatives lacking Bora Bora indicates that the flanking domains, including the AATAT satellite, are not sufficient for centromere activity. The presence of the AATAT satellite at noncentromeric sites in other Drosophila chromosomes supports this conclusion (Lohe et al., 1993).
We propose that the flanking satellite DNA is responsible for sister chromatid cohesion (Figure 7A) and the assembly/stability of a fully functional kinetochore (Figure 7B). The involvement of flanking repeats in sister chromatid cohesion or kinetochore function (or both) is supported by preliminary cytological analyses of larval neuroblasts; derivatives with <200 kb of flanking heterochromatin (e.g., J21A) displayed high levels of mitotic nondisjunction and moderate levels of chromosome loss (K. Cook and G. H. K., unpublished data). Sister chromatid cohesion and separation are essential to mitotic and meiotic disjunction, and numerous cytological observations indicate that most of the heterochromatin can participate in cohesion (Miyazaki and Orr-Weaver, 1994).
The flanking heterochromatin could affect kinetochore function at a distance. It could play an accessory role in optimizing kinetochore formation, for example by modifying the chromatin structure or topology of Bora Bora (Figure 7B, top), as has been suggested for the S. pombe centromere (Allshire et al., 1994; Marschall and Clarke, 1995). Alternatively, flanking regions could be responsible for stabilizing or regulating interactions with the spindle, for example by involvement with forces (e.g., the so-called polar wind; Rieder and Salmon, 1994; Fuller, 1995) that maintain tension at the kinetochore (Li and Nicklas, 1995). In support of this model, we have demonstrated that multiple regions throughout Dp1187 interact with the no distributive disjunction (nod) kinesin-like gene to promote transmission in Drosophila females, including the centromere flanking DNA (Murphy and Karpen, 1995). nod protein is localized to chromosomes and is thought to provide an antipoleward force necessary to maintain the normal configuration of achiasmate chromosomes during meiotic metaphase (Afshar et al., 1995). Regardless of the role(s) played by the flanking heterochromatin, our studies of Dp1187 define the minimal amount required for full function to be ~200 kb (dark stippled circles and arrows in Figure 7).
Transmission of Acentric Fragments
γ238 derivatives that lacked some or all of Bora Bora and the rest of the centric heterochromatin were recovered and maintained. How can apparently acentric fragments be transmitted at all? The recovery of acentric fragments that contained the right end of γ238, but not the left end, suggests that acentric transmission is sequence dependent. Possible mechanisms include weak holocentric activity, alternative centromere activity of a specific region, amplification, or passive inclusion in daughter nuclei via attachment of telomeres to the nuclear membrane.
What are the properties of acentric transmission and loss? The brooding experiments indicate that loss occurs during germline stem cell mitoses in males. The relatively high initial rate of male acentric transmission suggests that they are significantly more stable in preblastoderm and meiotic divisions. The male-female difference in transmission (~30% versus ~2%) is likely due to increased loss in female meiosis, since both sexes have very similar preblastoderm and germline mitoses. Cytological analyses of acentric transmission in different tissues, sexes, and developmental stages will contribute to our understanding of normal inheritance. For example, acentric stability in preblastoderm divisions would suggest that early, rapid divisions in Drosophila and perhaps other metazoans (Gilbert, 1991) utilize more “promiscuous” mechanisms for chromosome transmission.
Different Requirements for cis-Acting Inheritance Elements
Centric chromosomes containing less than 200 kb of flanking DNA exhibited lower transmission in females than in males (27% versus 40% for J21A). Normal inheritance (50% transmission) in the genetic monosome transmission assay requires stability in five types of divisions (see Results). A stable derivative must be inherited normally in all of these divisions, but the division(s) in which an unstable derivative acts aberrantly cannot be determined from this genetic assay. Brooding experiments demonstrated that reduced transmission for J21A in males is not due to instability in germline mitoses and must be restricted to preblastoderm mitoses or meiosis (or both), unlike the acentrics. We conclude that division types differ in their requirements for flanking DNA. Specifically, the amount of flanking DNA required for normal transmission in male germline mitoses is less than in preblastoderm mitoses or meiosis (or both), and transmission in females is more sensitive to the amount of flanking DNA. Meiosis I differs significantly in Drosophila males and females (Ashburner, 1990) and is the most likely division responsible for sex-specific differences in minichromosome transmission. Similarly, studies in S. pombe also have revealed that larger amounts of centromeric sequences are required for normal segregation through meiosis than through mitosis (Clarke et al., 1993).
Why do the DNA sequences required for chromosome inheritance differ between the sexes and among division types? We propose that males and females, and mitosis and meiosis, differ in the types or quantities of frans-acting factors that promote one or more of the functions proposed for the Bora Bora flanking DNA (Figure 7). For example, the nod gene interacts with flanking DNA and is required for transmission from females but not from males, possibly accounting for sex-specific differences in transmission (Murphy and Karpen, 1995). Cytogenetic analyses of different sexes and tissues are required to understand the diverse requirements for centromeric sequences throughout development.
Comparison with Centromeres in Other Organisms
The overall organization of the Drosophila centromere described here is formally similar to that reported for S. pombe; normal segregation in this yeast requires a single-copy central core plus adjacent repeated sequences (Baum et al., 1994). The central core is proposed to be the site of spindle attachment. Nondisjunction and loss increase as more S. pombe flanking repeats are deleted (Clarke et al., 1993; Baum et al., 1994), similar to the behavior of Dp1187 derivatives reported here (e.g., J21A). However, the S. pombe essential core is significantly smaller (4–7 kb), and the flanking repeats are middle-repetitive, not satellite, DNA (Clarke et al., 1993). The very small size (125 bp) and composition of the S. cerevisiae minimal centromere (Hegemann and Fleig, 1993) appears to be quite different from both S. pombe and Drosophila. The functional components of the mammalian centromere have yet to be determined definitively. Numerous studies point to the importance of alphoid satellite repeats, but suggest they are not sufficient to promote inheritance (Haaf et al., 1992; Brown et al., 1994; Larin et al., 1994). The similarities between the Drosophila and S. pombe centromeres suggest a model for mammals. Existing data are consistent with alphoid DNA playing a role analogous to the flanking repeats (Figure 7). Other unidentified sequences, even complex DNA, may play an essential role in mammalian centromere function (e.g., spindle attachment). Direct comparisons of centromeres in multicellular eukaryotes require more knowledge of structural and functional components. The Dp1187 derivatives described here provide a manipulate system for further genetic, molecular, cytological, and biochemical studies of in vivo meiotic and mitotic chromosome inheritance in metazoans.
Experimental Procedures
Drosophila Stocks and Chromosomes
YSX·YL, In(1)EN, y (attached X and Y referred to here as X^Y), and all marker genes are described in Lindsley and Zimm (1992). Dp1187 and derivatives in Figure 1 were described previously (Karpen and Spradling, 1990, 1992; Tower et al., 1993; Le et al., 1995). All stocks were crossed into a y; ry506 background.
Neuroblast Squash
Standard squash methods were used (Le et al., 1995) without colchicine but with hypotonic treatment, and chromosomes were stained with DAPI. The image was digitized with a Princeton Instruments cooled CCD mounted on a Zeiss Axiophot microscope, contrast enhanced using IP Labs Spectrum (Signal Analytics) and Adobe Photoshop for the Macintosh, and printed on a Phaser IIDX (Tektronix).
Monosome Transmission Assay
Single male or female animals carrying one copy of a particular minichromosome (see Figure 1B; genotype is y; ry506; Dp) were crossed to three y- ry- flies of the opposite sex (X^Y, y/0; ry506 males or X^Y, y/X^Y, y; ry506 females). The X^Y chromosome was used to suppress variegation of the y+ and ry+ genes observed for some Dp1187 derivatives. Monosome transmission is the percent of X^Y-containing F1 progeny that received the appropriate minichromosome-specific genetic markers (y+ or ry+ or both). Average transmission ± SD was calculated from 7–59 individual flies (median was 17), each producing 50–365 progeny (median was 116). SD2 = n Σ x2 – (Σ x)/n (n–1), where n is the number of individual flies assayed and x is their transmission. The results from individual flies were given equal weight regardless of the number of progeny produced. Higher variation among individual flies was seen with unstable derivatives (e.g., 26C), suggesting that meioses within an individual germline are not independent events. Meioses occurring In the same germline would behave similarly if mitotic instability resulted in germline clones. Dp1187 transmission was slightly higher in females than the 50% predicted for normal mono-some transmission, perhaps owing to weak meiotic drive.
γ238 Mutagenesis Screen
X, y/X, y; mu2 ry506; γ238, y+/- ry+ virgin females were exposed to low levels of γ-irradiation (500 rads) and crossed to X^Y, y/0; ry506 males. X^Y/X F1 female progeny were scored for the presence of genetically altered derivatives (y- ry+ or y+ ry- phenotypes). The extra Y chromosome suppressed the strong position effect on y+ expression exhibited by γ238 and ensured that phenotypic selection was correct and efficient. The frequency of Dp1187 heterochromatic breaks in the γ238 screen (0.22%; see text) was very similar to the frequency of euchro-matic terminal deficiencies recovered from mu2 females (Mason et al., 1984). Clusters recovered from the same bottle (e.g., 3A, a cluster of seven; Figure 4) were counted as one independent event, since they were likely to result from premeiotic events. Four translocations were also recovered (data not shown). In comparison with this y238 screen, higher levels of irradiation in a mu2+ genotype produced less than 0.01% Dp1187 deletion derivatives (Le et al., 1995).
Molecular Analysis of Derivatives
Protocols for pulsed-field electrophoresis and Southern blot hybridization have been described previously (Le et al., 1995). The probes are as follows: probe 1, sc133KK1.4, sc133SS1.0, sc133SG8.0 (mixed); probe 2, sc133RK2.6; probe 3, TG1BP11.5; probe 4, ryH7.2; probe 5, 12.1BH9.0; probe 6, probe 1 plus sc101XH3.7 and sc101RX4.0. The positions of the probes relative to the Dp1187 map are as follows: probe 1, –19.6 to –30; probe 2, –30 to –32.6; probe 3, –40 to –51.5; probe 5, -70 to –79; probe 6, 0 to –7.7 and –19.6 to –30. The ry sequences constitute 7.2 kb of the 14.5 kb PZ elements inserted at –185 and –245. See Le et al. (1995) for further description of probes.
Structural analyses showed that the derivatives fall into three basic classes. The analysis of uncut DNA showed that the overall sizes of the y+ ry- derivatives (e.g., J21B and 31E) were all greater than 1000 kb, but less than the 1320 kb observed for the parental γ238 chromosome (Figure 3B). Furthermore, they all hybridized with probe 6, but not with ry sequences (probe 4), as expected for deletions of the right end of γ238. Analysis of EcoRV-digested DNA (Figure 3C) revealed that the left-end (probe 1, 210 kb fragment in Figure 3A) and the right γ238 breakpoint (probe 2,12 kb fragment) remained intact, suggesting that no alterations had occurred in the centric heterochromatin. This was confirmed by the presence of an 1100 kb Notl fragment (Figure 3E) in J21B and most other y+ ry- derivatives, the same size band seen in γ238 (note that uncut and Sfil analysis of 31E indicated that it is deleted for the region homologous to probe 5). Sfil digests indicated that the y+ ry- derivatives were deleted for portions of the right end. Hybridization of Sfil digests with probe 3 (Figure 3D, right) allowed us to map the terminal breakpoints 20 kb (31E) and 122 kb (J21B) to the right of the –40 Sfil site, consistent with the uncut analysis.
The y- ry+ derivative J21A represents the second class, which were deleted for the left end of γ238 (including 20A, 3A, 10B, and 25A). Uncut J21A DNA (Figure 3B) hybridized with ry sequences (probe 4) but not probe 6 (right), indicating a total size of 580 kb and loss of the left end. The left-end EcoRV fragment (210 kb) also was absent in J21A (Figure 3C), although the right γ238 breakpoint (12 kb fragment) was present. Sfil digests (Figure 3D) confirmed that the right end was unaltered in J21A. Notl digests (Figure 3E) revealed a terminal breakpoint in the centric heterochromatin 360 kb to the left of the –100 Notl site. The placement of this breakpoint was consistent with the results of the uncut analysis (360 kb plus 190 kb to the right of the Notl site equals 580 kb). Note that further analysis of 20A indicates a single terminal break just to the left of probe 6.
The final class of derivatives was quite unusual. Uncut analysis (Figure 3B) of the y- ry+ derivative 26C revealed that it was substantially deleted (285 kb) and had lost the left end (no hybridization with probe 6). As with other y- ry+ derivatives, the left-end EcoRV fragment (210 kb, Figure 3A) was absent. Additionally, the right γ238 breakpoint (EcoRV, 12 kb fragment) also was absent in 26C, suggesting that the entire centric heterochromatin was deleted. Analysis of Notl digests (Figure 3E) placed a terminal breakpoint 65 kb to the left of the –100 Notl site (at –35), consistent with removal of the entire centric heterochromatin. Sfil digests (Figure 3C) confirmed that the right end was unaltered and placed the 26C terminal breakpoint between the –40 Sfil site and probe 2 (located at –30), consistent with the Notl digest.
Acknowledgments
Correspondence should be addressed to G. H. K. We thank Huong Le, Deborah Duricka, Kimberly Langhammer, Janice Wahlstrom, and Josh Bonkovsky for assistance in the generation and analysis of the Dp 8-23 and γ238 derivatives and Kevin Cook for sharing results prior to publication. Special thanks go to Janice Wahlstrom for producing the neuroblast squash displayed in Figure 1B and to Dr. Jim Mason for providing the mu2 ry chromosome. We also thank Kevin Cook, Kathryn Donaldson, Susan Forsburg, Mike McKeown, and Detlef Weigel for comments on the manuscript. We gratefully acknowledge support from the Pew, Drown, Mathers, and Paul Stock Foundations to G. H. K. and from a Lucille P. Markey Fellowship and a Chapman Fellowship to T. D. M. The Dp 8-23 derivatives were generated with support from National Institutes of Health grant R01 HG00747. This work was taken from ongoing thesis work of T. D. M. in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Biology, University of California at San Diego.
References
- Afshar K, Barton NR, Hawley RS, Goldstein LSB. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995;81:129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Cranston G, Gosden JR, Maule JC, Hastie ND, Fantes PA. A fission yeast chromosome can replicate autonomously in mouse cells. Cell. 1987;50:391–403. doi: 10.1016/0092-8674(87)90493-4. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor Press; Cold Spring Harbor, New York: 1990. [Google Scholar]
- Ault JG, Rieder CL. Centrosome and kinetochore movement during mitosis. Curr. Opin. Cell Biol. 1994;6:41–49. doi: 10.1016/0955-0674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan V, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schlzosaccharomyces pombe centromere. Mol. Biol. Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness OS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp. Quant. Biol. 1973;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Brinkley B,R, Ouspenski I, Zinkowski RP. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992;2:15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- Brown KE, Barnett M,A, Burgtof C, Shaw P, Buckle VJ, Brown WR. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum. Mol. Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- Clarke L, Baum M, Marschall LG, Ngan VK, Steiner N,C. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harbor Symp. Quant. Biol. 1993;58:687–695. doi: 10.1101/sqb.1993.058.01.076. [DOI] [PubMed] [Google Scholar]
- Cook KR, Karpen GH. A rosy future for heterochromatin. Proc. Natl. Acad. Sci. USA. 1994;91:5219–5221. doi: 10.1073/pnas.91.12.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorkin N, Hamkalo BA. The distribution of α-satellite sequences in African green monkey chromosomes. J. Cell Biol. 1991;115:92a. [Google Scholar]
- Earnshaw WC, Tomkiel JE. Centromere and kinetochore structure. Curr. Opin. Cell Biol. 1992;4:86–93. doi: 10.1016/0955-0674(92)90063-i. [DOI] [PubMed] [Google Scholar]
- Earnshaw W,C, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Fitzgerald HM. Yeast centromeres. Yeast. 1987;3:187–200. doi: 10.1002/yea.320030306. [DOI] [PubMed] [Google Scholar]
- Fuller M. Riding the polar winds: chromosomes motor down east. Cell. 1995;81:5–8. doi: 10.1016/0092-8674(95)90364-x. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. Third Edition Sinauer Associates Incorporated; Sunderland, Massachusetts: 1991. [Google Scholar]
- Glaser RL, Spradling AC. Unusual properties of genomic DNA molecules spanning the euchromatic-heterochromatic junction of a Drosophila minichromosome. Nucl. Acids Res. 1994;22:5068–5075. doi: 10.1093/nar/22.23.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- Goldstein LS. With apologies to Scheherazade: tails of 1001 kinesin motors. Annu. Rev. Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Haal T, Warburton PE, Willard HF. Integration of human α-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell. 1992;70:681–696. doi: 10.1016/0092-8674(92)90436-g. [DOI] [PubMed] [Google Scholar]
- Hannah-Alava A. The premeiotic stages of spermatogenesis. Adv. Genet. 1965;13:157–226. doi: 10.1016/s0065-2660(08)60049-8. [DOI] [PubMed] [Google Scholar]
- Hawley RS, Irick H, Zitron AE, Haddox DA, Lohe A, New C, Whitley MD, Arbel T, Jang J, McKim K, Childs G. There are two mechanisms of achiasmate segregation In Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 1993;13:440–467. doi: 10.1002/dvg.1020130608. [DOI] [PubMed] [Google Scholar]
- Hegemann J,H, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- John B. The biology of heterochromatin. In: Verma RS, editor. In Heterochromatin: Molecular and Structural Aspects. Cambridge University Press; Cambridge: 1988. pp. 1–147. [Google Scholar]
- Karpen GH, Spradling AC. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990;63:97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larin Z, Fricker MD, Tyler-Smith C. De novo formation of several features of a centromere following introduction of a Y alphoid YAC into mammalian cells. Hum. Mol. Genet. 1994;3:689–695. doi: 10.1093/hmg/3.5.689. [DOI] [PubMed] [Google Scholar]
- Le MH, Duricka D, Karpen GH. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics. 1995 doi: 10.1093/genetics/141.1.283. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Lica LM, Narayanswami S, Hamkalo BA. Mouse satellite DNA, centromere structure, and sister chromatid pairing. J. Cell Biol. 1986;103:1145–1151. doi: 10.1083/jcb.103.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J. Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Strobel E, Green MM. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc. Natl. Acad. Sci. USA. 1984;81:6090–6094. doi: 10.1073/pnas.81.19.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K, Stone LHA. The effects of X-radiation upon somatic chromosomes. J. Genet. 1933;28:1–24. [Google Scholar]
- McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-z. [DOI] [PubMed] [Google Scholar]
- Middleton K, Carbon J. KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an In vitro yeast kinetochore. Proc. Natl. Acad. Sci. USA. 1994;91:7212–7216. doi: 10.1073/pnas.91.15.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W, Orr-Weaver T. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the nod.+ klnesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel JE, Earnshaw W,C. Structure of the mammalian centromere. In: Vig BK, editor. Chromosome Segregation and Aneuploidy. Springer-Verlag; Berlin: 1993. pp. 13–29. [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyter-Smith C, Oakey RJ, Larin Z, Fisher RB, Crocker M, Affara NA, Ferguson SM, Muenke M, Zuttardi O, Jobling MA. Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nature Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- White MJD. Animal Cytology and Evolution. Cambridge University Press; Cambridge: 1973. [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]