Abstract
Myosin Binding Protein-C slow (MyBP-C slow), a family of thick filament associated proteins, consists of four alternatively spliced forms, namely variants 1–4. Variants 1–4 share common structures and sequences, however, they differ in three regions; variants 1 and 2 contain a novel 25-residue long insertion at the extreme NH2-terminus, variant 3 carries an 18-amino acid long segment within immunoglobulin (Ig) domain C7 and variant 1 contains a unique COOH-terminus consisting of 26-amino acids, while variant 4 does not possess any of these insertions. Variants 1–4 are expressed in variable amounts among skeletal muscles, exhibiting different topographies and potentially distinct functions. To date, the regulatory mechanisms that modulate the activities of MyBP-C slow are unknown. Using an array of proteomic approaches, we show that MyBP-C slow comprises a family of phosphoproteins. Ser-59 and Ser-62 are substrates for PKA, while Ser-83 and Thr-84 are substrates for PKC. Moreover, Ser-204 is a substrate for both PKA and PKC. Importantly, the levels of phosphorylated skeletal MyBP-C proteins (i.e. slow and fast) are notably increased in mouse dystrophic muscles, even though their overall amounts are significantly decreased. In brief, our studies are the first to show that the MyBP-C slow subfamily undergoes phosphorylation, which may regulate its activities in normalcy and disease.
Keywords: MyBP-C, skeletal muscle, phosphorylation, PKA, PKC, MDX mouse
Introduction
Myosin Binding Protein C (MyBP-C) is a thick filament associated protein that has been implicated in contributing to the structural stability of sarcomeres and regulating their contractile function1. Striated muscles contain three major MyBP-C isoforms, namely cardiac, fast skeletal and slow skeletal, that are mapped to different chromosomes2, 3. An additional MyBP-C isoform was recently reported in cat jaw-closing muscles that express masticatory myosin, although its molecular characterization is still pending4, 5. The core structure of MyBP-C is composed of seven immunoglobulin (Ig) domains and three fibronectin type III (FNIII) repeats, numbered from the NH2-terminus as C1–C106. The C1 domain is flanked by a Pro/Ala rich7 motif and a conserved linker region, termed MyBP-C or M motif. Although the three isoforms share significant structural and sequence homology, cardiac MyBP-C possesses three additional features, including an Ig domain at the very NH2-terminus of the molecule, termed C0, a unique 9-residue long insertion within the MyBP-C motif containing consensus phosphorylation sites and a 28-amino acid loop in the middle of Ig domain C58.
A number of studies have demonstrated that phosphorylation of cardiac MyBP-C at four key serine residues located within the MyBP-C-motif (i.e. Ser-273, Ser-281, Ser-302 and Ser-307), contributes significantly to contractile regulation9 (also reviewed in BareField and Sadayappan, 201010). Specifically, phosphorylation of these serine residues by PKA disrupts the interaction of cardiac MyBP-C with the S2 segment of myosin11, increasing the probability of cross-bridges formation, and therefore serving as a key modulator of actomyosin interaction and force generation12, 13, 14. In addition to PKA, other kinases, including PKC-epsilon and CaMKII, are also capable of phosphorylating the same serine residues, likely in a hierarchical manner15, 16, 17, 18. Recent studies have demonstrated that phosphorylation of cardiac MyBP-C protects it from degradation under stress conditions19, whereas lack of phosphorylation is associated with the development of hypertrophy and ultimately heart failure20, 21, 22.
To date, four different MyBP-C slow transcripts have been identified in human skeletal muscle referred to as variants (v) 1–4 (accession numbers NM_002465, NM_206819, NM_206820 and NM_206821, respectively). These differ at three regions due to alternative splicing events that result in the inclusion of novel segments within the Pro/Ala rich motif, the C7 domain and the extreme COOH-terminus, encoding sequences of 25, 18 and 26 amino acids, respectively23. V1 and v2 contain the NH2-terminal insertion within the Pro/Ala rich motif, v3 carries the insertion within the C7 domain, while v1 also contains the unique COOH-terminal region. V4 does not possess any of the novel insertions.
Although extensive research has focused on the role of cardiac MyBP-C in the formation of actomyosin cross-bridges and its modulation through complex phosphorylation events, the activities and regulation of the skeletal MyBP-C isoforms have remained elusive. Herein, we employed proteomics tools, including 2D SDS-PAGE combined with an array of phosphochemical treatments and tandem mass spectrometry to examine the presence of phosphorylation on skeletal MyBP-C slow. Our studies show that similar to its cardiac counterpart, MyBP-C slow undergoes phosphorylation. Importantly, PKA phosphorylates Ser-59 and Ser-62, while PKC phosphorylates Ser-83 and Thr-84. Both PKA and PKC phosphorylate Ser-204. Moreover, the phosphorylation levels of skeletal MyBP-C are considerably increased in mouse dystrophic (MDX) skeletal muscles. Our data provide, for the first time, concrete biochemical evidence indicating that skeletal MyBP-C slow comprises a family of phosphoproteins, and suggest that similar to the cardiac isoform, phosphorylation may be essential in the regulation of its activities.
Materials and Methods
Animals
Tissue was collected from adult FVB mice (~2–4 months old; Jackson Laboratories, Bar Harbor, ME), unless otherwise noted. For experiments involving dystrophic skeletal muscles, tissue was collected from adult control C57BL/6Scsn/J and dystrophic MDX C57BL/10Scsn-Dmd mdx/J mice (~2–4 months old; Jackson Laboratories, Bar Harbor, ME). Skeletal muscles were freshly isolated for each experiment; the number of animals used was as follows: for 1-D SDS-PAGE, n(FVB)=3, for 2-D SDS-PAGE, n(FVB)=5, n(C57BL/6Scsn/J)=3 and n(C57BL/10Scsn-Dmd mdx/J)=3. All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Maryland.
1-D Gel Electrophoresis, Western Blotting and Protein Staining
Homogenates of murine extensor digitorum longus (EDL), flexor digitorum brevis (FDB), tibialis anterior (TA), gastrocnemius (gastroc), quadriceps (quad) and soleus muscles from wild type FVB or age-matched (~10 weeks old) wild type C57BL/6Scsn/J and dystrophic C57BL/10Scsn-Dmd mdx/J animals were prepared and fractionated as previously described23, 24. Samples were either subjected to western blot analysis and probed with antibodies to MyBP-C slow (300 ng/ml, Abnova, Walnut, CA), MyBP-C fast (1:1000, Abnova) and GAPDH (1 µg/ml, Ambion, Austin, TX) or stained with Pro-Q Diamond phosphoprotein gel stain (Invitrogen, Carlsbad, CA) followed by SYPRO Ruby total protein gel stain (Invitrogen), following the manufacturer’s instructions. The relative amounts of MyBP-C slow and fast were calculated by quantifying and averaging the pixel intensities of the respective immunoreactive bands in at least three independent experiments using ImageJ (version 1.41, NIH, Bethesda, MD) and Excel (Microsoft, Bellevue, WA) software. Calculated intensities were normalized to those of GAPDH and reported as percent change between control and MDX tissues. Values from at least three experiments were averaged and evaluated by Student’s t-test (p<0.01).
Generation of Antibodies
Antibodies were generated to specifically recognize the phosphorylated forms of Ser-83 and Thr-84. Peptides 5’ CANSQLSTLFVEK 3’ and 5’ CANSQLSTLFVEK 3’ containing phosphorylated Ser-83 and Thr-84 (underlined), respectively, were used to immunize rabbits for production of polyclonal antibodies (Open Biosystems, Huntsville, AL). Specific antibodies were obtained through two rounds of affinity purification, following the manufacturer’s instructions. In the first step, immune sera were purified against the appropriate phosphorylated form of the antigenic peptide. In the second step, the eluted antibodies were further purified against the non-phosphorylated form of the peptide (5’ CANSQLSTLFVEK 3’). The antibodies contained in the flow-through fraction from the second column were used as affinity purified phospho-specific antibodies at 400 ng/ml in subsequent immunoblotting experiments.
2-D Gel Electrophoresis and Western Blotting
The phosphorylation state of endogenous MyBP-C slow was analyzed by treatment with phospho-chemicals followed by 2-D gel electrophoresis and western blot analysis. Freshly isolated FDB muscle from wild type FVB mice was lysed using a mini-homogenizer (VWR, Radnor, PA) in 2% Chaps, 20 µM DTT (or 20 µM BME) supplemented with complete protease inhibitors (Roche, Mannheim, Germany). Homogenates were treated with one of the following: 1. calf alkaline phosphatase (1 unit/20 µg protein; Invitrogen) at 30° for 2 hours, 2. bacterial alkaline phosphatase (1 unit/20 µg protein; Invitrogen) at 30° for 2 hours, 3. a cocktail of phosphatase inhibitors, adapted from Goodall et al 201025; 1 mM β-glycerol phosphate, 1 mM sodium pyrophosphate, 20 µM sodium orthovanadate, 5 mM sodium fluoride, 100 µM EDTA and 100 µM EGTA for 1 hour at room temperature, 4. H89, a kinase inhibitor (1 µM and 10 µM; EMD Chemicals Inc., Gibbstown, NJ) for 1 hour at room temperature, 5. PKA (1 unit/20 µg protein, NEB, Ipswich, MA) in the presence of 2 mM ATP for 6 hours at 4°C, 6. peptide 6–22 of PKA (Sigma, St. Louis, MO) acting as a direct PKA inhibitor at 1 µM for 1 hour at room temperature, 7. PKC epsilon (2 units/25 µg protein, EMD Chemicals Inc.) in the presence of 2 mM ATP for 6 hours at 4°C, and 8. PKC inhibitor peptide 19–31 (1 µM, EMD Chemicals Inc.) for 1 hour at room temperature. 25 µg of total protein from treated or sham samples were diluted in DeStreak rehydration reagent (Amersham Biosciences, Piscataway, NJ) with 1.5% Bio-Lyte 5/7 Ampholyte solution (Bio-Rad, Hercules, CA) and incubated with narrow range pH 5.3–6.3 strips (Invitrogen) overnight at 4°C, according to the manufacturer’s instructions. Iso-electric focusing (IEF) in the first dimension was performed for at least 7,500 V hours. For the second dimension, each strip was equilibrated in 1xSDS buffer, followed by electrophoretic separation using a 4–12% Bis-Tris gel in MOPS running buffer (Invitrogen), according to the manufacturer’s instructions. Membranes were then subjected to western blotting using antibodies to MyBP-C slow (Abnova), as previously described24.
Homogenates of flexor digitorum brevis (FDB) and tibialis anterior (TA) muscles from age-matched (~10 weeks old) wild type C57BL/6Scsn/J and dystrophic C57BL/10Scsn-Dmd mdx/J mice were prepared as above, fractionated by 2-D gel electrophoresis and subjected to western blot analysis. To analyze MyBP-C fast, narrow range pH strips, 6.1–7.1, were used. Antibodies to MyBP-C slow (Abnova) and MyBP-C fast (Abnova) were used according to the manufacturer’s instructions.
Reverse Transcriptase-Polymerase Chain Reaction
Recombinant proteins of three distinct regions of MyBP-C slow were designed to include the three novel insertions; the NH2-terminal construct included residues 1–285 (accession number HQ848554) and contained the very NH2-terminus of MyBP-C slow, the Pro/Ala rich motif, the novel insertion, the C1 Ig domain and the MyBP-C motif, the C7-domain construct included amino acids 1–104 (accession number HQ848555) and consisted of the C7 FNIII domain with the novel insertion, and the COOH-terminal construct included amino acid residues 1016–1127 (accession number EDL21488) and contained the last Ig domain, C10 and the novel COOH-terminal insertion.
Total RNA was isolated with Trizol reagent (Invitrogen) from adult FVB murine soleus muscle and with the RNeasy kit (Qiagen, Valencia, CA) from control C57BL/6Scsn/J and dystrophic C57BL/10Scsn-Dmd mdx/J FDB and TA muscles. Approximately 5 µg of freshly prepared RNA were then used to synthesize cDNA with the Superscript First Strand Synthesis System for RT-PCR (Invitrogen). Newly synthesized cDNA was used as template for PCR amplification of the above MyBP-C slow constructs (i.e. NH2-terminal construct, 855bp; C7-domain construct, 312bp; and COOH-terminal construct, 333bp), the very COOH-terminus of MyBP-C fast (330bp) and a portion of the housekeeping gene GAPDH (240bp) with the primers listed in supplemental table 1. Following amplification, PCR products were analyzed by electrophoresis in 1% agarose gels and their authenticity was verified by sequence analysis. Semi quantitative analysis was performed using ImageJ (NIH) and Excel (Microsoft) software. The expression levels of MyBP-C slow and fast in wild type and dystrophic muscles were normalized to the expression of GAPDH and reported as percent change between samples. Values from at least three experiments were averaged and evaluated by Student’s t-test (p<0.01).
Generation of Wild Type and Mutant Recombinant Proteins
The NH2-terminal, C7 and COOH-terminal constructs of MyBP-C slow fragments containing the three novel insertions described above were introduced in the pGEX4T-1 vector at EcoR1/Xho1 sites (Amersham Pharmacia, Piscataway, NJ) to generate GST-fusion proteins. Recombinant polypeptides were expressed by induction with 1 mM isopropyl β-D-thioglycopyranoside (IPTG) for 4 hours at 30°C and purified by affinity chromatography on glutathione-agarose columns.
Tandem site-directed mutagenesis was performed as previously described26 using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to replace the PKA (Ser-59 and Ser-62) and PKC (Ser-83 and Thr-84) phosphorylation sites within the NH2-terminal construct by alanine residues. Sense and antisense oligonucleotide sets are listed in supplementary table 1. The authenticity of all constructs was verified by sequence analysis, and mutant fragments were expressed as GST-fusion proteins, as described above.
In vitro Kinase Assay
Purified recombinant proteins were concentrated using Centrifugal Filter Units (Millipore, Carrigtwohill, Co. Cork, Ireland) to a final concentration of at least 1 µg/µl and dialyzed against Buffer A containing: 20 mM Hepes, 50 mM KCl, 10 mM MgCl2, 1 mM ATP and 1 mM DTT. PKA or PKC was added to recombinant proteins at 80 U kinase/mg protein, along with 1 mM ATP27. The reaction was incubated at 4°C for 16 hours and then separated by SDS-PAGE. ProQ Diamond staining, used to identify phosphorylation, was followed by SYPRO Ruby staining for total protein.
In-Solution Digestion of Proteins and Phosphopeptide Enrichment
In-solution digestion of affinity purified recombinant proteins treated with either PKA or PKC was performed as follows. Proteins were denatured with 0.1% RapiGest SF surfactant (Waters, Milford, MA) by boiling at 95°C for 5 minutes followed by reduction with 5 mM TCEP for 30 minutes at 56°C and alkylation with 15 mM iodoacetamide for 30 minutes at room temperature (22°C) in the dark. The alkylation reaction was quenched with 30 mM DTT for 30 minutes at room temperature. After desalting using 7 kDa MWCO Protein Desalting spin columns (Thermo Fisher Scientific, Waltham, MA), trypsin (2% w/w) or GluC (2%w/w) was added for overnight digestion at 37°C. The digestion was stopped by acidification with 5% formic acid followed by 30 minutes incubation at 37°C to hydrolyze the RapiGest. RapiGest by-products were pelleted by centrifugation at 10,000×g for 10 min. Peptides were de-salted using C18 desalting spin columns (Thermo Fisher Scientific) and dried using a SpeedVac concentrator.
The digested peptides were further purified to enhance for phosphopeptides using either an immobilized gallium phosphopeptide enrichment kit (Thermo Fisher Scientific), according to the manufacturer’s instructions, or TiO2 STAGE Tips as detailed in Rappsilber et al28 and briefly described here. Three µL of 10 µm Titansphere beads (GL Sciences) were added to in-house made C8 (Empore, 3M, St. Paul, MN) STAGE Tips in 200 µL pipet tips. The TiO2 columns were washed with Buffer B (80% ACN, 0.1% TFA and 300 mg/mL lactic acid). Dried SCX-fractionated peptides were re-constituted in Buffer B and loaded onto the TiO2 columns. The columns were washed with Buffer B followed by Buffer C (80% ACN, 0.1% TFA) and the phosphopeptides were eluted with freshly prepared 0.5% NH4OH into tubes containing 2% TFA. To elute any peptides bound to the C8 membrane, the TiO2 columns were washed with Buffer B; this eluate was combined with the 0.5% NH4OH eluate. The gallium and TiO2 phosphopeptide enriched samples were dried in a SpeedVac concentrator followed by de-salting with C18 STAGE tips. De-salted peptides were re-suspended in 0.1% formic acid immediately prior to LC-MS/MS analysis.
Mass Spectrometry (MS)
Chromatographic separation of peptides was performed using an Xtreme Simple Nano LC system (CVC Micro-Tech Scientific) equipped with a 150 mm × 75 µm C-18 reversed-phase column (3 µm particles, 300 Å pores - Michrom Bioresources). Mobile phase compositions were as follows: A) 2% acetonitrile, 0.1% formic acid; B) 95% acetonitrile, 0.1% formic acid. Samples were injected in 0.1% formic acid using a Surveyor Autosampler (Thermo Electron). A 60 minutes LC gradient method from 5–40% solvent B at a flow rate of 0.5 µL/minute flow was used to elute the peptides into the mass spectrometer. All mass spectrometry analyses were performed using an LTQ-Orbitrap (Thermo Electron) mass spectrometer operated using Xcalibur v. 2.0.7 SR2. The mass spectrometer was equipped with a nanospray ionization source containing an uncoated 10 µm i.d. SilicaTip™ PicoTip™ nanospray emitter (New Objective, Woburn, MA). The spray voltage was 1.8 kV and the heated capillary temperature was 200°C. MS1 data for peptides 57–75 and 204–221 was acquired in the Orbitrap profile mode (lock mass not engaged) with a resolution of 60,000 at 400 m/z. MS1 data for peptide 76–89 was acquired in the linear ion trap (LTQ). All MS/MS data was acquired in the linear ion trap as follows: top five most intense ions in each MS1 scan were selected for collision-induced dissociation with dynamic exclusion enabled (repeat count 1, exclusion duration 180 sec). Other mass spectrometric data generation parameters were as follows: collision energy 35%, full scan MS mass range 400–1600 m/z, minimum signal 500 counts and isolation width 3.0 m/z.
Mass spectrometry data analysis
MS/MS spectra were searched against a mouse protein database downloaded from UniProt to include ~65,000 sequences and a custom database using Bioworks 3.3.1 SP1 with the SEQUEST algorithm. The custom database was constructed to include only those proteins known to be present in the in vitro reaction mixture; glutathione S-transferase (NCBI accession number, Caa46155), PKA (NCBI accession number, NP_032880), PKC (NCBI accession number, AAG53692) and mouse MyBP-C slow including the novel insertions (NCBI accession numbers, 8030451F13, EDL21488, HQ848554, HQ848555). Search parameters were as follows: enzyme cleavage, trypsin (for the NH2- and COOH-terminal constructs) and GluC (for the C7 construct); cleavage type, full; precursor mass tolerance, 1.5 amu or 20 ppm; fragment ion tolerance, 0.5 Da; missed cleavages, 2; modifications, Cys carbamidomethylation (+57.02 Da), Met oxidation (+15.99 Da) and Ser, Thr and Tyr phosphorylation (+79.97 Da). The peptides were filtered by the following criteria: Xcorr ≥ 1.5, 2.5, 3.0, and 3.5 corresponded to the presence of 1+, 2+, 3+, and 4+ peptides, respectively30. All MS/MS spectra of identified phosphopeptides were confirmed by manual validation.
Immuofluorescence Staining and Confocal Microscopy
Adult FDB and TA muscles of control C57BL/6Scsn/J and dystrophic C57BL/10Scsn-Dmd mdx/J mice were fixed both in situ, via whole animal perfusion-fixation and ex vivo with 2% paraformaldehyde in PBS, as previously described24, 31. Fixed tissue was embedded in PBS containing 7.5% gelatin, 15% sucrose, slowly frozen using 2-methylbutane and cryosectioned. Longitudinal sections were immunolabelled with antibodies to MyBP-C-slow (1:400, Abnova), MyBP-C fast (1:100; Abnova) and an antibody to the RhoGEF domain of obscurin (obscurin RhoGEF, 3µg/mL32).
Results and Discussion
MyBP-C slow is a family of phosphoproteins
Mammalian MyBP-C slow is a family of four isoforms differing from one another at three regions due to complex alternative splicing, that results in the inclusion of exons 3 and 4 in the Pro/Ala rich motif (NH2-terminal insertion), exon 23 in the middle of the C7 FNIII domain (C7 insertion) and exon 31 at the extreme COOH-terminus (COOH-terminal insertion), which encode novel sequences of 25, 18 and 24 amino acids, respectively (Fig. 1A and SFig. 1B). These novel insertions combine to form at least 4 distinct variants, which share structural and sequence homology (~85–95%) among humans, rats and mice23 (and our unpublished data). Variant 1 (v1, ~131 kDa) contains both the NH2- and COOH-terminal inserts, variant 2 (v2, ~129 kDa) harbors only the NH2-terminal insert, variant 3 (v3, ~128 kDa) carries the C7-domain insert, and variant 4 (v4, ~126 kDa) lacks all three inserts.
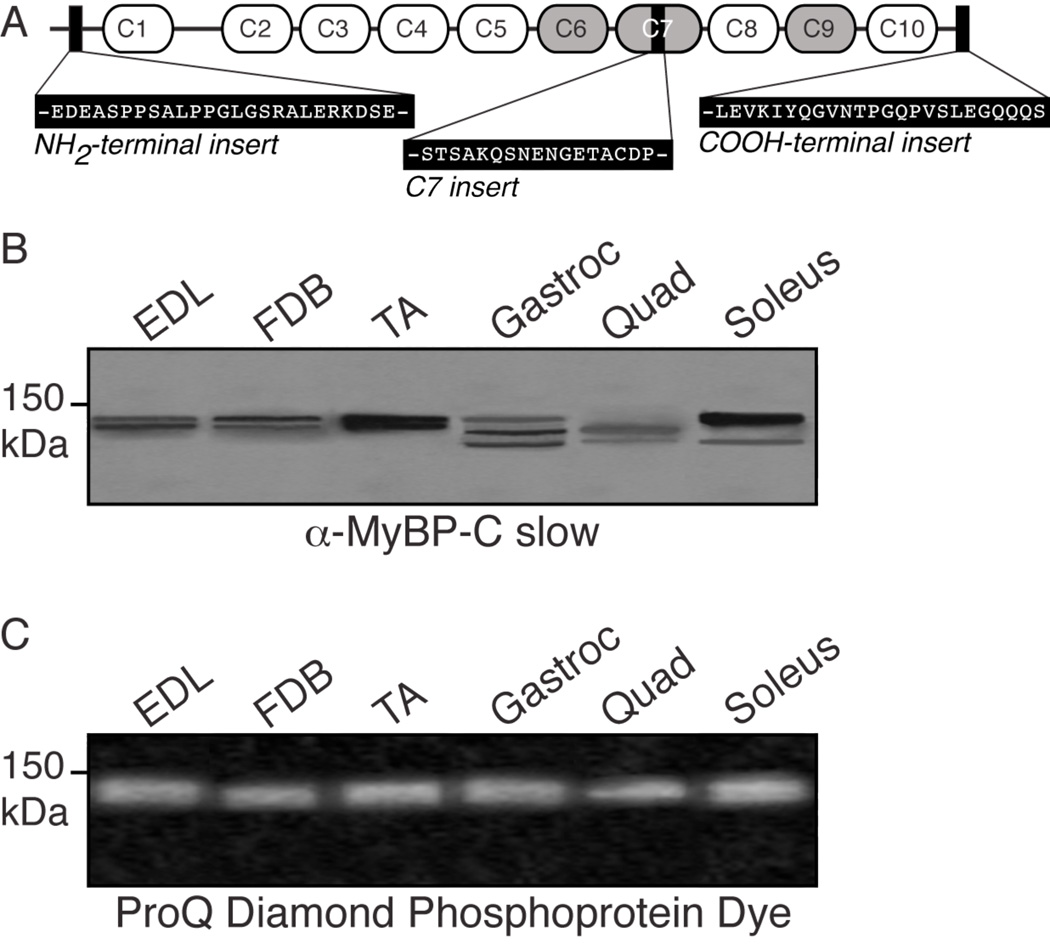
Figure 1.
Adult murine skeletal muscles express multiple MyBP-C slow variants. A: Schematic representation of a “hypothetical” murine MyBP-C slow transcript consisting of tandem Ig (white ovals) and FnIII (grey ovals) domains with the three novel insertions shown (black rectangles). B–C: Protein homogenates prepared from murine skeletal muscles were labeled with anti-MyBP-C slow antibody (B) and stained with ProQ Diamond Dye that specifically detects phosphorylated proteins (C). At least three immunoreactive bands were detected, ranging in size from ~125–135 kDa that correspond to the different variants of MyBP-C slow (v1–v4; please see text). Notably, a broad band of ~125–135 kDa was detected by the ProQ Diamond phospho-dye in all skeletal muscles tested.
To study the expression pattern of MyBP-C slow in adult murine skeletal muscles, we performed western blot analysis. Homogenates from extensor digitorum longus (EDL), flexor digitorum brevis (FDB), tibialis anterior (TA), gastrocnemius (gastroc), quadriceps (quad) and soleus were separated by 1-D SDS-PAGE and probed with an antibody recognizing domain C5, which is specific yet common to all MyBP-C slow variants (Fig. 1B). Consistent with our previously published results in rat skeletal muscle23, we were able to resolve at least three immunoreactive bands across the panel of fast and slow skeletal muscles examined, ranging in size between ~125–135 kDa, and to detect at least one form of MyBP-C slow in each murine muscle. EDL, FDB and TA primarily express immunoreactive bands of ~131 and ~128–129 kDa, corresponding to v1 and v2 and/or v3, respectively (Fig. 1B, lanes 1–3). Interestingly, gastrocnemious possesses three immunoreactive bands of ~131, ~128–129, and ~126 kDa, corresponding to v1, v2 and/or v3 and v4, respectively (Fig. 1B, lane 4), with the ~128–129 kDa band being the most abundant one. Contrary to the other muscles analyzed, quadriceps and soleus contain only two bands of ~128–129 and ~126 kDa, and ~131 and ~126 kDa, representing v2 and/or v3 and v4, and v1 and v4, respectively (Fig. 1B, lanes 5 and 6). Taken together, these findings indicate that MyBP-C slow variants 1–4 are co-expressed, albeit in variable amounts and combinations, in slow and fast twitch skeletal muscles of murine (this study) and rat23 origin, where they can co-exist with MyBP-C fast within the same muscle, fiber or even sarcomere33.
Previous work has suggested that MyBP-C slow is capable of phosphorylation34, 35, 36. To test this, we analyzed protein lysates prepared from the above skeletal muscles by 1-D SDS-PAGE and stained them with ProQ Diamond, a phosphospecific dye. A broad band of ~125–135 kDa representing a mixture of phospho-proteins with the same mobility as the skeletal MyBP-C proteins, encompassing both slow and fast isoforms, was detected in all samples examined (Fig. 1C). Thus, MyBP-C slow is a multi-variant family of proteins, which are differentially expressed among skeletal muscles and may undergo phosphorylation.
MyBP-C slow is a substrate of PKA and PKC
To examine the presence of phosphorylation in MyBP-C slow, we treated protein lysates prepared from FDB muscle with a variety of phospho-specific chemicals and performed 2-D SDS-PAGE coupled with western blotting. FDB is primarily a fast twitch skeletal muscle expressing high amounts of v1 but also containing v2 and/or v3. Protein lysates were treated with a general phosphatase, a cocktail of phosphatase inhibitors or a broad-spectrum kinase inhibitor, separated by isoelectric point (pI) and molecular mass with 2-D gel electrophoresis and immuno-probed for MyBP-C slow. As the pI values of the four MyBP-C slow variants range from ~5.5–6.0, we used narrow pH strips to obtain maximal resolution of the ‘train’ of MyBP-C slow spots.
Ten immunoreactive spots (1–10) were present in sham or untreated homogenates (Fig. 2A, first panel), representing a gamut of differentially charged MyBP-C slow isoforms, resulting from variable post-translational modifications. Spots located at a higher pH (e.g. pH: 6.3) represent more positively charged species, whereas spots located at a lower pH (e.g. 5.3) represent less positively charged species. Treatment with a general phosphatase resulted in a dramatic shift of immunoreactive spots 2–10 towards a higher pH (Fig. 2A, second panel), suggesting the generation of more positively charged or less phosphorylated species. To the contrary, treatment with a cocktail of phosphatase inhibitors resulted in a major shift of spot 1 towards a lower pH, concomitant with an increase in the intensity of spots 2–9 (Fig. 2A, third panel), indicating the generation of less positively charged or more phosphorylated species. Moreover, treatment with different concentrations of a general kinase inhibitor, H89, resulted in a dose-dependent increase of the least phosphorylated species (i.e. spot 1), and a decrease in the intensity of the remaining spots (i.e. spots 2–10) (Fig. 2A, fourth and fifth panels). Collectively, these experiments demonstrate that MyBP-C slow comprises a subfamily of phosphoproteins.
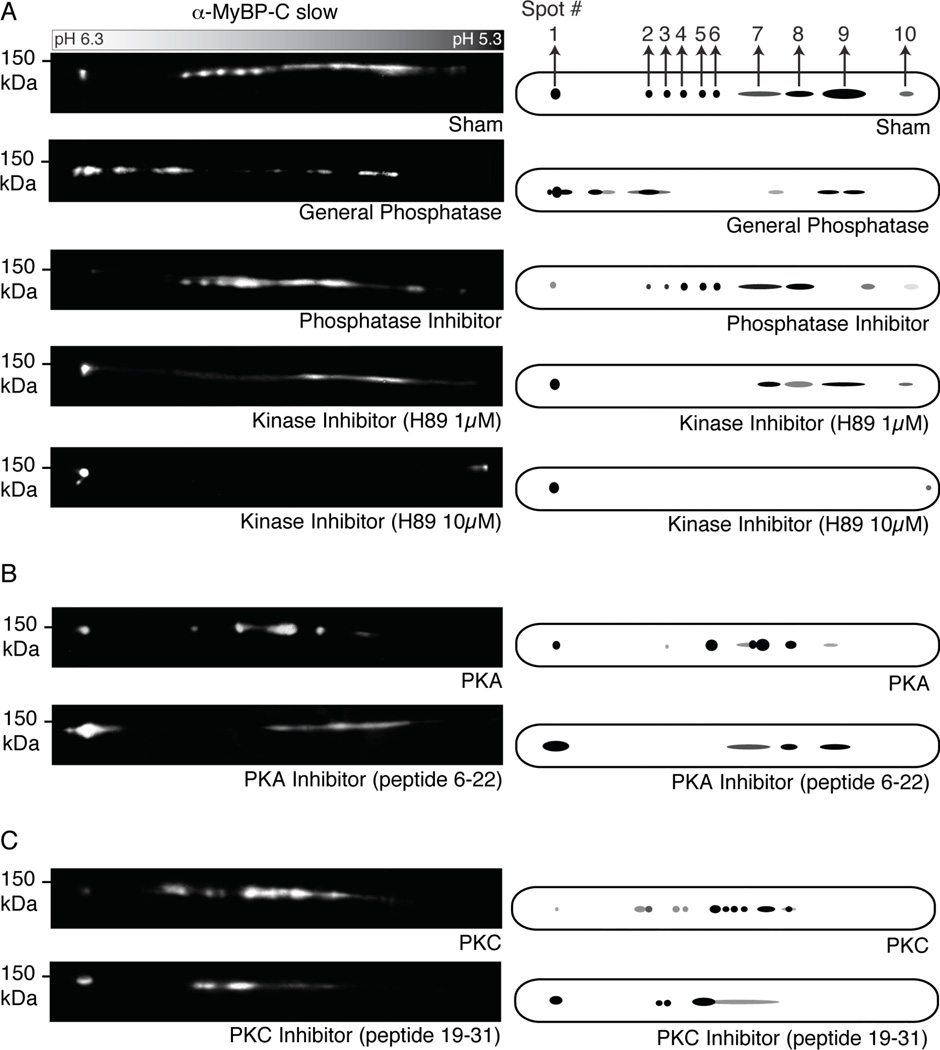
Figure 2.
Proteomic analysis of MyBP-C slow in fast twitch FDB muscle. A–C: Protein homogenates prepared from murine FDB skeletal muscle were subjected to different phospho-chemical treatments, analyzed by 2-D gel electrophoresis and immunostained with a MyBP-C slow antibody (A). Representative blots are shown on the left with corresponding cartoon representations on the right. MyBP-C slow appears to be a substrate of PKA (B) and PKC (C) in vivo.
To determine potential kinases involved in MyBP-C slow phosphorylation, we treated FDB homogenates with PKA, PKC and their specific inhibitors, and performed 2-D analysis as before. Following treatment with PKA (Fig. 2B, top panel) or PKC (Fig. 2C, top panel), immunoreactive spots 2–10 concentrated in the middle of the 6.3-5.3 pH strip, while the intensity of spot 1 was diminished. Contrary to this, after treatment with PKA (Fig. 2B, bottom panel) or PKC (Fig. 2C, bottom panel) inhibitors, immunoreactive spots 2–10 collapsed into spot 1, which increased in intensity dramatically. Taken together, these results suggest that MyBP-C slow is a substrate for PKA and PKC.
We next sought to identify specific phosphorylated residues on MyBP-C slow following activation by PKA and PKC. To this end, we produced recombinant GST fusion proteins of three distinct regions of MyBP-C slow to include the three novel insertions described in Fig. 1A and underlined in SFig. 1C. The NH2-terminal construct contains residues 1–285 (accession number HQ848554) and includes the Pro/Ala rich motif, the novel NH2-terminal insertion, the C1 Ig domain and the MyBP-C motif, the C7 construct (residues 1–104; accession number HQ848555) contains the entire C7 FNIII domain harboring the novel insertion, and the COOH-terminal construct (residues 1016–1127; accession number EDL21488) includes the C10 Ig domain along with the novel COOH-terminal sequence (Fig. 3A). Each recombinant protein (Fig. 3B) was treated with PKA or PKC and analyzed via mass spectrometry.
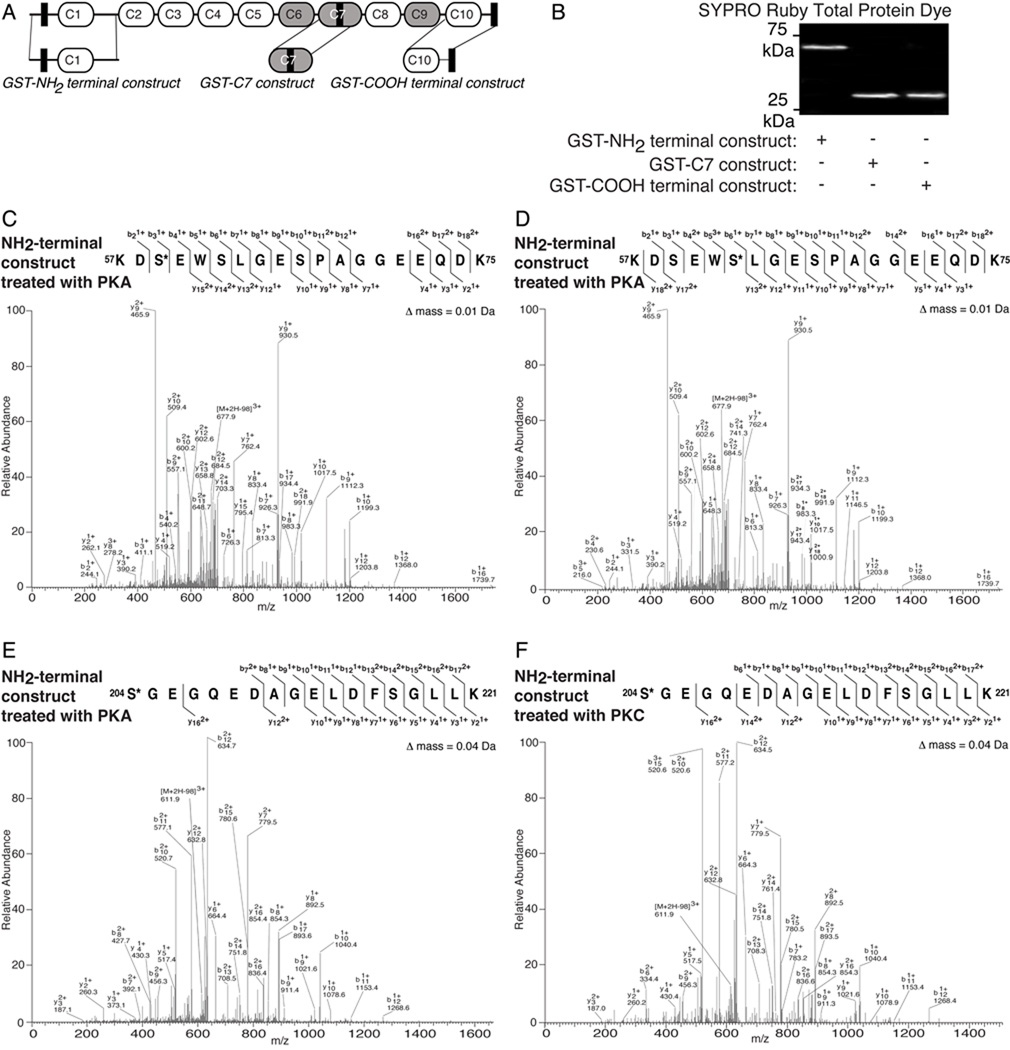
Figure 3.
MyBP-C slow is phosphorylated by PKA and PKC in vitro. A: Schematic representation of the same “hypothetical” protein shown in Fig. 1A, highlighting recombinant MyBP-C slow constructs. Recombinant polypeptides are denoted as GST-NH2 terminal construct, GST-C7 construct and GST-COOH terminal construct and detected by SYPRO ruby total protein stain (B). C–F: Phosphorylation events mediated by PKA and PKC were identified in the GST-NH2-terminal construct via mass spectrometry analysis using LTQ-orbitrap. MS/MS scans of peptides carrying phosphorylated Ser-59 by PKA (C), Ser-62 by PKA (D) and Ser-204 by PKA (E) and PKC (F), identified by Bioworks-SEQUEST, with Xcorr scores of 4.471, 4.757, 4.142 and 4.475, respectively, and manually validated are shown.
Tandem LC-MS/MS analysis identified five phosphorylated residues within the NH2-terminal construct; for details regarding data acquisition please refer to Materials and Methods and Table 1. Following PKA activation, a peptide encompassing amino acids 57–75 (accession number, HQ848554) was identified multiple times with at least one phosphorylated site at Ser-59 (Fig. 3C) or Ser-62 (Fig. 3D) with cross correlation (Xcorr) values of 4.471 and 4.757, respectively (Table 1). Thus, phosphorylation may occur on either one or both residues within the peptide. An additional peptide including residues 204–221 (accession number, HQ848554) was also identified several times with one phosphorylated site at amino acid Ser-204, following treatment with either PKA or PKC with Xcorr values of 4.142 (Fig. 3E) and 4.475 (Fig. 3F), respectively (Table 1). Moreover, after activation by PKC, a peptide containing amino acids 76–89 (accession number, HQ848554) was identified with at least one phosphorylation site at Ser-83 (SFig. 1A) or Thr-84 (SFig. 1B) with Xcorr values of 2.83 and 2.94, respectively (Table 1), suggesting that phosphorylation may occur on either one or both residues within the peptide. Given the relatively low Xcorr values obtained for Ser-83 and Thr-84, we generated polyclonal antibodies specific to their phosphorylated forms, which, following affinity purification, were used to probe adult murine FDB lysates. An immunoreactive band of ~150 kDa was specifically detected by both phospho-antibodies (SFig. 1A–B, insets), indicating that Ser-83 and Thr-84 are substrates of PKC in vivo.
Table 1.
Identification of the phospho-peptides present in the NH2-terminal construct (amino acids 1–285) of MyBP-C slow following in vitro phosphorylation as determined by LC-MS/MS.
| Sequence | Residue # |
Phosphorylated Residue |
Charge | Method Used |
Theoretical Mass |
Observed Mass |
Xcorr |
|---|---|---|---|---|---|---|---|
| Treated with PKA | |||||||
| KDSEWSLGESPAGGEEQDK | 57–75 | Ser-62 | 3 | LTQ-orbitrap | 2128.87 | 2128.86 | 4.757 |
| KDSEWSLGESPAGGEEQDK | 57–75 | Ser-59 | 3 | LTQ-orbitrap | 2128.87 | 2128.86 | 4.471 |
| SGEGQEDAGELDFSGLLK | 204–221 | Ser-204 | 3 | LTQ-orbitrap | 1931.87 | 1931.83 | 4.142 |
| Treated with PKC | |||||||
| SGEGQEDAGELDFSGLLK | 204–221 | Ser-204 | 3 | LTQ-orbitrap | 1931.87 | 1931.83 | 4.475 |
| QNANSQLSTLFVEK | 76–89 | Thr-84 | 2 | LTQ | 1659.183 | 1658.779 | 2.94 |
| QNANSQLSTLFVEK | 76–89 | Ser-83 | 2 | LTQ | 1659.183 | 1658.779 | 2.827 |
Taken together, these findings are consistent with the fact that wild type recombinant NH2-terminal construct (amino acid residues 1–285) was readily stained with ProQ Diamond dye following treatment with either PKA or PKC (SFig. 1D). Consistent with this, mutant forms of it, in which either the PKA (Ser-59, Ser-62 and Ser-204) or the PKC (Ser-83, Thr-84 and Ser-204) phosphorylation sites were tandemly mutated to Ala failed to stain with ProQ Diamond dye, following treatment with PKA or PKC, respectively (SFig. 1D and data not shown).
Of the five phosphorylatable residues, Ser-59 is located within the novel NH2-terminal insertion and is therefore present in v1 and v2. On the contrary, Ser-62, Ser-83 and Thr-84 are located within the Pro/Ala rich region and are present in all four variants. Notably, Ser-59 and Ser-83 are conserved among mouse (accession number, HQ848554), rat (accession number, NM_001100758) and human (accession number, NM_002465), while Ser-62 is conserved in mouse and rat, but replaced by a Thr in human. Conversely, Thr-84 is replaced by a Gln in both rat and human. Ser-204 is located within the MyBP-C motif following the C1 domain. The MyBP-C motif of the slow isoform is ~95% homologous between mouse, rat and human, and Ser-204 is conserved among the three species. Interestingly, Ser-204 aligns with Thr-272 of cardiac MyBP-C that precedes the cardiac specific insertion within the MyBP-C motif, which contains consensus phosphorylation sites. Immediately following Thr-272 is Ser-273, a known phosphorylation site and target of both PKA and PKC16.
Collectively, these findings provide the first biochemical evidence of phosphorylation of MyBP-C slow by PKA and PKC, suggesting a novel mechanistic regulation of the activities of MyBP-C slow in the formation of actomyosin cross-bridges within skeletal muscles.
Skeletal MyBP-C slow and fast exhibit increased levels of phosphorylation in dystrophic skeletal muscles
Using 1- and 2-D SDS-PAGE combined with western blot analysis, we further examined the expression pattern and phosphorylation profile of skeletal MyBP-C slow and fast in dystrophin-deficient (MDX) murine skeletal muscles in comparison to their normal counterparts (for mouse strain information please see the Materials and Methods section). Equal loading of protein lysates was ensured by measuring protein concentration and probing for GAPDH (Fig. 4A bottom panels). 1-D SDS-PAGE demonstrated that in addition to the ~131 and 128–129 kDa bands, representing v1 and v2 and/or v3, respectively, an immunoreactive band of ~126 kDa was also present in the MDX TA and FDB muscles. This lower band likely corresponds to v4, which is absent from normal tissues (Fig. 4A, top panel, lanes 2 and 4, arrowheaed). Alternatively, it could represent a proteolytic fragment of MyBP-C slow v1 and v2 and/or v3. Moreover, there was a significant decrease in the intensity of the immunoreactive band representing v2 and/or v3 in the MDX TA muscle, but not in the MDX FDB muscle (Fig. 4A, top panel, lane 4, arrow). Consistent with this, the total amounts of MyBP-C slow were lower in TA (~35% decrease), but not FDB (a ~9% increase was observed presumably due to expression of v4) MDX muscle (Fig. 4B, black bars). Conversely, the levels of MyBP-C fast were dramatically reduced in both FDB (~90% decrease) and TA (~30% decrease) MDX muscles, however this decrease was more pronounced in the MDX FDB tissue (Fig. 4B, white bars). These changes in the amounts of the slow and fast isoforms were specific to protein levels, as their mRNA levels remained unchanged between wild type and MDX tissues (SFig. 2A and B).
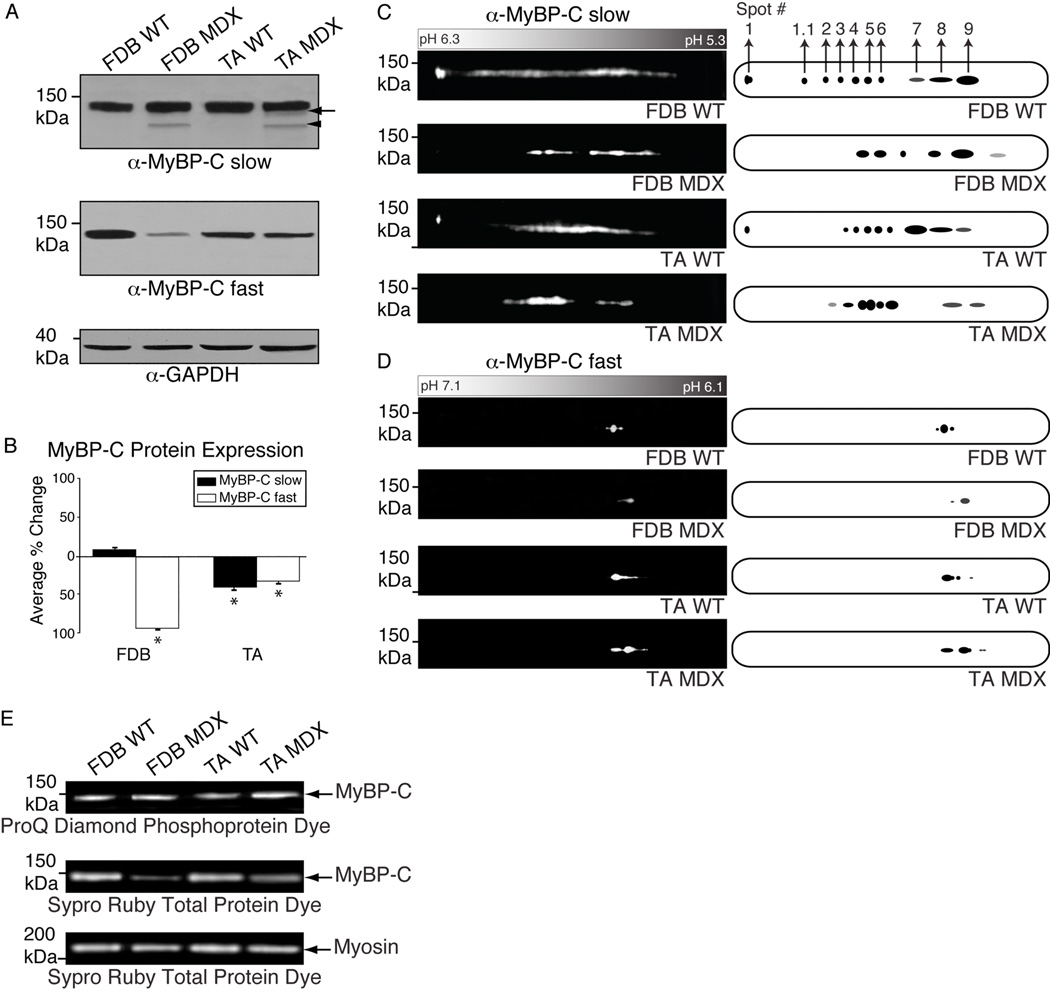
Figure 4.
Examination of the expression profile of MyBP-C slow and fast in dystrophic (MDX) skeletal muscles. A: Protein homogenates prepared from FDB and TA skeletal muscles from wild type (WT) and MDX mice were separated by 1-D SDS-PAGE and probed with antibodies to MyBP-C slow (A, top panel) and fast (A, middle panel). An immunoreactive band of ~125 kDa representing MyBP-C slow v4 (arrowhead) was detected in dystrophic FDB and TA, but not wild type, tissues. Moreover, the ~128–129 kDa band, corresponding to v2 and/or v3, is significantly diminished in MDX TA muscle (arrow). Similarly, the amounts of MyBP-C fast are considerably reduced in dystrophic FDB and TA muscles. Equal loading of protein lysates was confirmed by immunostaining for GAPDH (A, bottom panel). B: Densitometric analysis of immunoreactive bands was used to estimate the relative amounts of MyBP-C slow (black bars) and fast (white bars) in wild type and dystrophic muscles; differences in expression levels are reported as percent changes (p<0.01; Students t-test). C–D: The same homogenates were analyzed by 2-D SDS-PAGE and immunostained for MyBP-C slow (C) and fast (D). A distinct shift of both MyBP-C slow and fast immunoreactive spots toward a more phosphorylated state was observed in dystrophic muscles. E: Consistent with this, the ~125–135 kDa band, encompassing skeletal MyBP-C slow and fast, was of higher intensity in homogenates prepared from dystrophic FDB and TA muscles compared to the band detected in the respective normal muscles after staining with ProQ Diamond phosphospecific dye (E, top panel), albeit the total amounts of skeletal MyBP-C protein were significantly reduced, as indicated by Sypro Ruby staining (E, middle panel); myosin staining with Sypro Ruby was used as a loading control (E, bottom panel), indicating equivalent levels between wild type and dystrophic muscles.
Next, we employed 2-D gel electrophoresis to study the phosphorylation state of MyBP-C slow and fast in dystrophic and wild type muscles. The 2-D footprint of MyBP-C slow exhibited a shift towards a lower pH in MDX FDB and TA muscles compared to their wild type counterparts (Fig. 4C), as evidenced by the lack of spot 1 and the increased intensity of spots 1.1–9, indicating higher levels of phosphorylation of MyBP-C slow in MDX muscles. Similar analysis of MyBP-C fast by 2-D SDS-PAGE demonstrated that unlike the slow (this study) and cardiac37 isoforms, which resolve into 10 and 9 spots, respectively, the fast isoform shows a single major immunoreactive spot either flanked or followed by two less prominent spots in normal FDB and TA muscles, respectively (Fig 4D, panels 1 and 3). Interestingly, we observed a shift, albeit less dramatic, of the MyBP-C fast spots towards a lower pH in MDX FDB and TA muscles, too (Fig. 4D, panels 2 and 4). Taken together, these results suggest that the two skeletal isoforms of MyBP-C exhibit higher levels of phosphorylation in dystrophic MDX muscles, as compared to their wild type counterparts. Consistent with this, the ~125–135 kDa band, encompassing the slow and fast skeletal MyBP-C isoforms, detected with the ProQ Diamond dye in lysates prepared from dystrophic FDB and TA muscles, was of higher intensity compared to the one detected in the respective wild type muscles (Fig. 4E, top panel). Importantly, the total amounts of the skeletal MyBP-C isoforms are significantly reduced in both FDB and TA MDX muscles compared to normal tissues, as our immunoblots indicated (Fig 4B), and Sypro Ruby staining further confirmed (Fig. 4E, middle panel). It should be noted that equal amounts of lysates were analyzed from dystrophic (MDX) and wild type muscles as determined by measuring protein concentration and evaluating the expression levels of other proteins (e.g. myosin, Fig. 4E, bottom panel) by Sypro Ruby total protein dye.
Given the changes we observed in the expression and phosphorylation levels of MyBP-C slow and fast in dystrophic compared to healthy skeletal muscles, we set forth to examine their subcellular distribution, using immunofluorescence combined with confocal microscopy. The localization of MyBP-C slow appeared diffuse in ~10% of dystrophic TA fibers (SFig. 2D1–D3), whereas the localization of MyBP-C fast was unaffected (SFig. 2F1–F3). Control TA fibers exhibited regular striations for both MyBP-C slow (SFig. 2C1–C2) and fast (SFig. 2E1–E3). There was no detectable difference in the localization of MyBP-C slow or fast between MDX and wild type FDB muscle (data not shown).
Conclusions
Our data provide for the first time concrete biochemical evidence that MyBP-C slow comprises a complex family of phosphoproteins that are substrates for PKA and PKC in skeletal muscle. Furthermore, we show that in dystrophic muscle, MyBP-C slow and fast exhibit increased levels of phosphorylation compared to normal tissues. Thus, our results suggest that, similar to its cardiac counterpart, phosphorylation may be essential in regulating the activities of MyBP-C slow in skeletal muscle.
Cardiac MyBP-C regulates the formation of actomyosin cross-bridges and thus force generation through dynamic phosphorylation events that take place within its NH2-terminus13–14, 38. In agreement with this, murine models engineered to contain non-phosphorylatable Ala or phosphomimetic Asp residues in the place of the phosphorylatable Ser residues, exhibited depressed cardiac function or cardioprotection against ischemia/reperfusion injury, respectively19, 39. Taken together, these observations clearly indicate the importance of phosphorylation on cardiac MyBP-C to maintain normal sarcomeric structure and contractile function. Based on our findings here, it is reasonable to propose similar roles for phosphorylation of MyBP-C slow during normalcy and disease. Future work will focus on the effect(s) of phosphorylation of MyBP-C slow in maintaining sarcomeric structure and modulating contractile activity in skeletal myofibers.
Supplementary Material
Acknowledgements
The authors wish to thank Drs Stefani Thomas (University of Maryland, Baltimore) and George Tsaprailis (University of Arizona) for helpful discussions on mass spectrometry and Dr. Chris Ward (University of Maryland, Baltimore) for providing us MDX mice. In addition, we thank the University of Maryland Greenebaum Cancer Center Proteomics Shared Service for conducting the Lc-Ms/Ms studies and database searches as well as for providing assistance with interpretation of the mass spectrometry data. Our research has been supported by grants to A.K.K from the National Institutes of Health (R01 AR52768) and to M.A.A. from the National Institutes of Health (F32 AR058079).
Footnotes
Supporting Information Available:
This material is available free of charge via the Internet at http://pubs.acs.org.”
References
- 1.Oakley CE, Chamoun J, Brown LJ, Hambly BD. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int J Biochem Cell Biol. 2007;39(12):2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Gautel M, Castiglione Morelli MA, Pfuhl M, Motta A, Pastore A. A calmodulin-binding sequence in the C-terminus of human cardiac titin kinase. Eur J Biochem. 1995;230(2):752–759. [PubMed] [Google Scholar]
- 3.Weber FE, Vaughan KT, Reinach FC, Fischman DA. Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur J Biochem. 1993;216(2):661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
- 4.Kang LH, Rughani A, Walker ML, Bestak R, Hoh JF. Expression of masticatory-specific isoforms of myosin heavy-chain, myosin-binding protein-C and tropomyosin in muscle fibers and satellite cell cultures of cat masticatory muscle. J Histochem Cytochem. 2010;58(7):623–634. doi: 10.1369/jhc.2010.955419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang LH, Hoh JF. Regulation of jaw-specific isoforms of myosin-binding protein-C and tropomyosin in regenerating cat temporalis muscle innervated by limb fast and slow motor nerves. J Histochem Cytochem. 2010;58(11):989–1004. doi: 10.1369/jhc.2010.956847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc Natl Acad Sci USA. 1990;87(6):2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer JF, Harris SP. Species-specific differences in the Pro-Ala rich region of cardiac myosin binding protein-C. J Muscle Res Cell Motil. 2009;30(7–8):303–306. doi: 10.1007/s10974-010-9207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda M, Koshida S, Sato N, Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J Mol Cell Cardiol. 1995;27(10):2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- 9.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97(11):1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48(5):866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RH. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86(1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 12.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453(3):254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 13.Colson BA, Bekyarova T, Locher MR, Fitzsimons DP, Irving TC, Moss RL. Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ Res. 2008;103(3):244–251. doi: 10.1161/CIRCRESAHA.108.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colson BA, Locher MR, Bekyarova T, Patel JR, Fitzsimons DP, Irving TC, Moss RL. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J Physiol. 2010;588(Pt 6):981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohamed AS, Dignam JD, Schlender KK. Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch Biochem Biophys. 1998;358(2):313–319. doi: 10.1006/abbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Zhao Q, Du Y, Yuan C, Solaro RJ, Buttrick PM. PKCepsilon increases phosphorylation of the cardiac myosin binding protein C at serine 302 both in vitro and in vivo. Biochemistry. 2007;46(23):7054–7061. doi: 10.1021/bi700467k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlender KK, Bean LJ. Phosphorylation of chicken cardiac C-protein by calcium/calmodulin-dependent protein kinase II. J Biol Chem. 1991;266(5):2811–2817. [PubMed] [Google Scholar]
- 19.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc Natl Acad Sci U S A. 2006;103(45):16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119(11):1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 21.Decker RS, Decker ML, Kulikovskaya I, Nakamura S, Lee DC, Harris K, Klocke FJ, Winegrad S. Myosin-binding protein C phosphorylation, myofibril structure, and contractile function during low-flow ischemia. Circulation. 2005;111(7):906–912. doi: 10.1161/01.CIR.0000155609.95618.75. [DOI] [PubMed] [Google Scholar]
- 22.Jacques AM, Copeland O, Messer AE, Gallon CE, King K, McKenna WJ, Tsang VT, Marston SB. Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. J Mol Cell Cardiol. 2008;45(2):209–216. doi: 10.1016/j.yjmcc.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Ackermann MA, Kontrogianni-Konstantopoulos A. Myosin binding protein-C slow: an intricate subfamily of proteins. J Biomed Biotechnol. 2010;2010:652065. doi: 10.1155/2010/652065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20(12):2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodall MH, Wardlow RD, 2nd, Goldblum RR, Ziman A, Lederer WJ, Randall W, Rogers TB. Novel function of cardiac protein kinase D1 as a dynamic regulator of Ca2+ sensitivity of contraction. J Biol Chem. 2010;285(53):41686–41700. doi: 10.1074/jbc.M110.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borzok MA, Catino DH, Nicholson JD, Kontrogianni-Konstantopoulos A, Bloch RJ. Mapping the binding site on small ankyrin 1 for obscurin. J Biol Chem. 2007;282(44):32384–32396. doi: 10.1074/jbc.M704089200. [DOI] [PubMed] [Google Scholar]
- 27.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 29.Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem. 2006;281(16):10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 30.Thomas SN, Wan Y, Liao Z, Hanson PI, Yang AJ. Stable isotope labeling with amino acids in cell culture based mass spectrometry approach to detect transient protein interactions using substrate trapping. Anal Chem. 2011;83(14):5511–5518. doi: 10.1021/ac200950k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14(3):1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, Bloch RJ. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581(8):1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurasawa M, Sato N, Matsuda A, Koshida S, Totsuka T, Obinata T. Differential expression of C-protein isoforms in developing and degenerating mouse striated muscles. Muscle Nerve. 1999;22(2):196–207. doi: 10.1002/(sici)1097-4598(199902)22:2<196::aid-mus7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 34.Lim MS, Walsh MP. Phosphorylation of skeletal and cardiac muscle C-proteins by the catalytic subunit of cAMP-dependent protein kinase. Biochem Cell Biol. 1986;64(7):622–630. doi: 10.1139/o86-086. [DOI] [PubMed] [Google Scholar]
- 35.Vikhliantsev IM, Podlubnaia ZA. Phosphorylation of sarcomeric cytoskeletal proteins--an adaptive factor for inhibiting the contractile activity of muscle during hibernation. Biofizika. 2003;48(3):499–504. [PubMed] [Google Scholar]
- 36.Vikhliantsev IM, Alekseeva IuA, Shpagina MD, Udal'tsov SN, Podlubnaia ZA. Properties of C protein from skeletal and cardiac muscles of the ground squirrel Citellus undulatus at various stages of hibernation. Biofizika. 2002;47:701–705. [PubMed] [Google Scholar]
- 37.Yuan C, Sheng Q, Tang H, Li Y, Zeng R, Solaro RJ. Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am J Physiol Heart Circ Physiol. 2008;295(2):H647–H656. doi: 10.1152/ajpheart.00357.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. Journal of molecular biology. 1999;286(3):933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 39.Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Robbins J. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation. 2009;119(9):1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.