SUMMARY
Human liver cancer is one of the deadliest cancers worldwide, with hepatocellular carcinoma (HCC) being the most common type. Aberrant Ras signaling has been implicated in the development and progression of human HCC, but a complete understanding of the molecular mechanisms of this protein in hepatocarcinogenesis remains elusive. In this study, a stable in vivo liver cancer model using transgenic zebrafish was generated to elucidate Ras-driven tumorigenesis in HCC. Using the liver-specific fabp10 (fatty acid binding protein 10) promoter, we overexpressed oncogenic krasV12 specifically in the transgenic zebrafish liver. Only a high level of krasV12 expression initiated liver tumorigenesis, which progressed from hyperplasia to benign and malignant tumors with activation of the Ras-Raf-MEK-ERK and Wnt–β-catenin pathways. Histological diagnosis of zebrafish tumors identified HCC as the main lesion. The tumors were invasive and transplantable, indicating malignancy of these HCC cells. Oncogenic krasV12 was also found to trigger p53-dependent senescence as a tumor suppressive barrier in the pre-neoplastic stage. Microarray analysis of zebrafish liver hyperplasia and HCC uncovered the deregulation of several stage-specific and common biological processes and signaling pathways responsible for krasV12-driven liver tumorigenesis that recapitulated the molecular hallmarks of human liver cancer. Cross-species comparisons of cancer transcriptomes further defined a HCC-specific gene signature as well as a liver cancer progression gene signature that are evolutionarily conserved between human and zebrafish. Collectively, our study presents a comprehensive portrait of molecular mechanisms during progressive Ras-induced HCC. These observations indicate the validity of our transgenic zebrafish to model human liver cancer, and this model might act as a useful platform for drug screening and identifying new therapeutic targets.
INTRODUCTION
Hepatocellular carcinoma (HCC) ranks as the fifth most prevalent malignancy and the third leading cause of cancer mortalities worldwide (Villanueva et al., 2010). The neoplastic development of HCC is a complex multistage process, with hyperplastic nodules of regenerating hepatocytes in chronic inflammatory liver representing a potential first step towards HCC (Farazi and DePinho, 2006). Despite the availability of several therapies for HCC, understanding of its fundamental processes is rather limited and the ultimate clinical benefit remains negligible.
The Ras proto-oncogenes are central regulators of intracellular signal transduction pathways involved in malignant transformation. Approximately 7% of human liver cancers carry activating mutations in the KRAS oncogene, which is higher than the percentage that carry HRAS and NRAS mutations (Karnoub and Weinberg, 2008). Multiple lines of evidence have revealed the importance of extracellular signal-regulated kinase (ERK), downstream of Ras, during human hepatocarcinogenesis (Schmidt et al., 1997). Indeed, the core protein of hepatitis C virus has been shown to directly activate the Ras-Raf-MEK-ERK pathway in vitro (Hayashi et al., 2000). Although human HCC displays a low incidence of Ras mutations, activation of Ras signaling in the presence of wild-type Ras has been found in all human HCC when compared with non-neoplastic surrounding and normal livers (Calvisi et al., 2006). Many on-going clinical trials on anti-cancer drugs targeting Ras and its downstream signaling cascades in HCC are being conducted and so far the only drug approved for the treatment of advanced HCC is Sorafenib, a multi-target compound that blocks Ras-Raf-MEK-ERK and VEGF pathways (Llovet and Bruix, 2008; Villanueva et al., 2010). This demonstrates Ras signaling as an attractive target for liver cancer therapy.
Animal models have been widely used in biomedical research to understand the pathogenesis of cancer and as in vivo systems for testing new drug candidates. In recent years, several Hras-induced liver neoplasia in murine models have been reported. These models showed that Hras mutation solely caused hepatic dysplasia but was insufficient to induce HCC (Harada et al., 2004; Sandgren et al., 1989). Little evidence has determined whether liver tumorigenesis is dependent on the level of Ras activation. Moreover, the nature of Ras-induced mechanisms in liver cancer remains unclear. To date, no mouse models have intentionally utilized Kras as a driving oncogene to study liver tumorigenesis. Despite having a number of biological similarities to humans, the mouse is costly and unfeasible for large-scale studies (Sharpless and DePinho, 2006). The zebrafish (Danio rerio) is increasingly recognized as an alternative vertebrate model to study human diseases (Lieschke and Currie, 2007). It has been shown that histology and gene expression profiles of zebrafish tumors closely resemble those of humans (Lam et al., 2006; Lam and Gong, 2006; Langenau et al., 2007). Many strategies, including chemical carcinogens treatment, xenografts and transgenic approaches, have actively been applied to generate tumor models using zebrafish (Amatruda and Patton, 2008; Mizgirev and Revskoy, 2010; Spitsbergen and Kent, 2003). Over the past few years, the Ras oncogene has been used to drive tumorigenesis in several tissues, including muscle (Langenau et al., 2007), pancreas (Park et al., 2008), skin (Michailidou et al., 2009) and others (Le et al., 2007).
Motivated by these findings, we have generated a stable transgenic zebrafish model for HCC by overexpressing oncogenic krasV12 under the liver-specific fabp10 promoter using the Activator/Dissociation (Ac/Ds) transposon system. We provide evidence that a high level of krasV12 expression is crucial in driving liver tumorigenesis from hyperplasia to carcinoma by deregulating several stage-specific and common pathways that demonstrate conservation between human and zebrafish liver cancer. Further analyses revealed two important gene signatures for HCC specificity and HCC progression.
RESULTS
Generation of Tg(fabp10:EGFP-krasV12) transgenic zebrafish
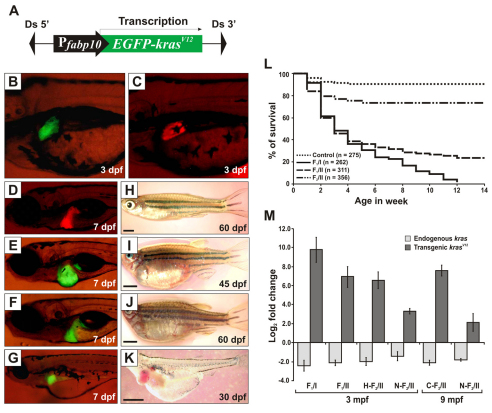
A plasmid construct was made to harbor a cDNA encoding a fusion protein of N-terminal enhanced green fluorescent protein (EGFP) and C-terminal zebrafish KrasV12 under control of the liver-specific fabp10 promoter (Fig. 1A). The construct also contained Ds transposon sequences (Emelyanov et al., 2006) and was co-injected with synthesized Ac transposase mRNA into one-cell embryos. 1 month later, 25% of F0 fish with EGFP fluorescence in the liver showed an enlarged abdomen, edema and died within 2 months. By contrast, siblings with no observable EGFP expression in the liver appeared normal and could carry the transgene insertions in germ cells. To obtain stable transgenic line, we crossed these normal F0 fish with wild-type (WT) zebrafish and screened their offspring for EGFP expression in the liver. Two founders transmitted the transgene to their progenies (F1).
Fig. 1.
Generation and characterization of Tg(fabp10:EGFP-krasV12) transgenic zebrafish. (A) Schematic diagram of the DNA construct used to generate Tg(fabp10:EGFP-krasV12) transgenic zebrafish. Ds, maize Ds transposon sequence. (B–G) Liver-specific expression of EGFP-KrasV12 in F1 transgenic fry (B,E,F,G) as compared with the Tg(fabp10:dsRFP; elaA:EGFP) transgenic line expressing RFP in liver (Korzh et al., 2008) as normal control (C,D). (H–K) Gross observation of control fish (H) and F1 krasV12 transgenic fish (I–K). Ages of the fish are indicated. Scale bars: 2 mm. (L) Kaplan-Meier survival curves of the Tg(fabp10:EGFP-krasV12) fish for three groups of heterozygous transgenic zebrafish from F1/I (n=262), F1/II (n=311), F2/II (n=356) and WT siblings as control (n=275). (M) Determination of endogenous and transgenic kras expression levels by qRT-PCR. Log2 fold changes for endogenous and transgenic kras mRNAs were calculated against an internal housekeeping gene (β-actin) using the CT method. Histological analysis was performed to confirm their neoplastic stages before qRT-PCR (H, hyperplasia; C, carcinoma; N, normal liver). Results are presented as the mean ± s.d. and each bar represents five biological replicates.
In the F1 generation of the two founders [F1/Line I (F1/I) and F1/Line II (F1/II)], EGFP expression could be detected from 3 days post-fertilization (dpf) (Fig. 1B). Higher intensity of EGFP fluorescence was observed in F1/I than in F1/II. As compared with Tg(fabp10:dsRed) fry that had normal liver morphology with liver-specific RFP expression (Korzh et al., 2008) (Fig. 1C,D), microscopic examination of the EGFP-positive F1/I (n=196) at 7 dpf revealed that the majority of transgenic larvae had different degrees of liver enlargement (Fig. 1E,F), whereas 7% exhibited a smaller liver (Fig. 1G). In F1/II (n=149), 63% of larvae showed enlarged liver and 3% had a smaller liver, whereas 34% showed no significant abnormalities that correlated with lower EGFP fluorescence. By juvenile to adult stage, fish with enlarged livers displayed abdominal swelling and progressive hemorrhages surrounding the abdominal walls (Fig. 1I,J). The health of fish with smaller livers worsened with time and most died by 1 month post-fertilization (mpf) (Fig. 1K). The effect of oncogenic krasV12 on liver development is shown in more detail in supplementary material Fig. S1.
A high level of KrasV12 expression led to early lethality and induced HCC
The Kaplan-Meier survival curves for heterozygous F1 offspring (n=262, F1/I; n=311, F1/II) showed ∼70% mortality by 30 dpf (Fig. 1L). Most of the fish that died before 30 dpf showed severe liver enlargement. By 90 days, 100% mortality was observed in F1/I. By contrast, 24% of F1/II transgenic fish survived by 90 dpf, thus enabling the maintenance of this line. Fish that survived past 90 days exhibited lower EGFP levels in the liver as compared with those that died before 90 days. F2/II (n=356), which was obtained by outcrossing, showed mendelian 1:1 EGFP segregation, indicating a single transgene insertion.
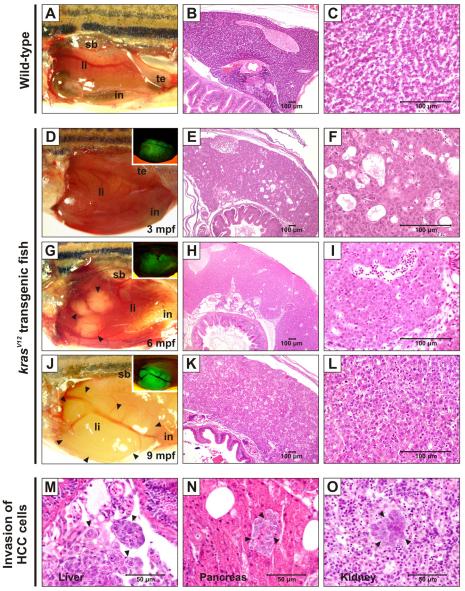
Fig. 2A–C shows normal liver morphology and histology in WT zebrafish. Approximately 58% of F1/I (n=12; all of which died at 65–90 dpf) showed macroscopic liver nodules. Histopathological analysis revealed that these fish had multiple large tumors with microscopic features of HCC (supplementary material Fig. S2). For line II, detailed histological progression of liver tumors was conducted with the F2 generation. We found no tumor protrusion in the livers of 45 transgenic fish displaying swollen belly euthanized at 3 mpf (Fig. 2D). However, 16 fish (36%) displayed moderate liver hyperplasia (Fig. 2E,F). At the later stages, liver tumors in 12/54 (22%) F2/II fish dissected at 6 mpf were observed, showing histopathological features of hepatocellular adenoma (Fig. 2G–I). By 9 mpf, malignant HCC appeared in 11/42 (26%) transgenics (Fig. 2J–L). These fish showed hemorrhage and swollen body upon death, with five of them showing invasions of tumor cells into blood vessels and internal organs (Fig. 2M–O).
Fig. 2.
Liver tumor progression in krasV12 transgenic zebrafish. Abbreviations: in, intestine; li, liver; sb, swimbladder; te, testis. (A–C) Gross morphology and histology of WT zebrafish showing normal liver and tissue architecture. (D–L) Gross morphology and histology of F2/II krasV12 zebrafish. (D,G,I) Brightfield and fluorescence (insets) images displaying the progressive stages of liver tumors at 3, 6 and 9 mpf. Various tumor protrusions are indicated by arrowheads. Corresponding histological sections are shown in the same rows. Livers were observed at 3 mpf (D) and histological appearance revealed multifocal mild-to-moderate cystic degeneration (spongiosis hepatis) and diffused moderate hepatocellular hyperplasia (E,F). Many white nodules were developed in transgenic liver at 6 mpf (G) and their histology indicated hepatocellular adenoma containing vacuolated clear cells with increased cytoplasmic glycogen (H,I). Malignant tumors were visibly observed at around 9 mpf (J) and histological analysis confirmed that the tumor was HCC grade II–III (K,L). (M–O) Invasion of HCC cells (indicated by arrowheads) into blood vessels (M) and adjacent tissues, namely pancreas (N) and kidney (O).
We employed quantitative real-time PCR (qRT-PCR) to assess the levels of endogenous and transgenic kras transcripts in the transgenic livers. Samples were collected from six groups: 3-mpf F1/I (carcinoma), 3-mpf F1/II (hyperplasia), 3-mpf N-F2/II (normal liver), 3-mpf H-F2/II (hyperplasia), 9-mpf C-F2/II (normal liver) and 9-mpf N-F2/II (carcinoma) (Fig. 1M). After normalization against β-actin, the expression levels of endogenous kras were consistent among these six groups, and were at similar levels as in matched WT controls (data not shown). By contrast, transgenic krasV12 was overexpressed in these groups. F1/I had the highest level of krasV12 expression with log2 fold change over β-actin levels (9.85±2.69), as compared with the change over β-actin in F1/II (6.95±2.24). In F2/II, the levels of krasV12 transcript compared with β-actin transcript in transgenic fish undergoing tumorigenesis from hyperplasia (6.58±1.81) to carcinoma (7.60±1.21) were higher than their age-matched (3 and 9 mpf, respectively) transgenic siblings without liver lesions (3.26±0.65 and 2.12±1.88, respectively). These data demonstrated the threshold level of krasV12 to drive liver tumorigenesis because we consistently observed that only a high level of krasV12 expression leads to liver tumors from hyperplasia to carcinoma.
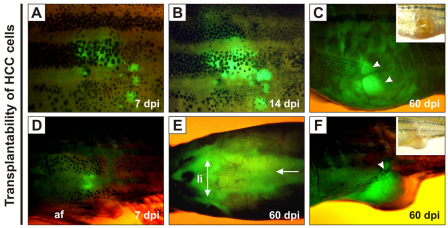
Transplantability of krasV12 liver tumors in WT recipients
Dissociated HCC cells pooled from four F2/II transgenic zebrafish were injected intraperitoneally into 25 sublethally γ-irradiated WT adult zebrafish (∼1×106 cells per recipient). At 7 days post-injection (dpi), we confirmed that all of the recipients exhibited EGFP fluorescence at the site of injection (Fig. 3A,D). Fluorescence intensified by 14 dpi and cells seemed to spread to adjacent tissues (Fig. 3B). At 60 dpi, the tumor cells distributed along the abdominal cavity and showed strong EGFP fluorescence at the abdominal region in seven of the recipients (Fig. 3E). By this time, the outgrowth of the tumor mass penetrated through the peritoneal cavity and/or abdominal wall (Fig. 3C,F), indicating capability of the krasV12 tumors for propagation and invasion in a new host.
Fig. 3.
Growth of transplanted krasV12 liver tumors in WT recipients. (A–F) EGFP-positive HCC cells were transplanted intraperitoneally into irradiated recipients. (A,D) EGFP fluorescence was observed near the sites of injection at 7 dpi. af, anal fin. (B) Transplanted cells proliferated by 14 dpi. (E) Extensive infiltration of tumor cells along the peritoneal cavity (arrow), especially in the region surrounding the liver (li; double-headed arrow), was observed at 60 dpi. (C,F) The outgrowth of EGFP-positive tumor mass (arrowheads) penetrated into the abdominal wall and/or peritoneal cavity at 60 dpi.
Differential activation of ERK, JNK and p38 MAPK pathways during krasV12 liver tumorigenesis
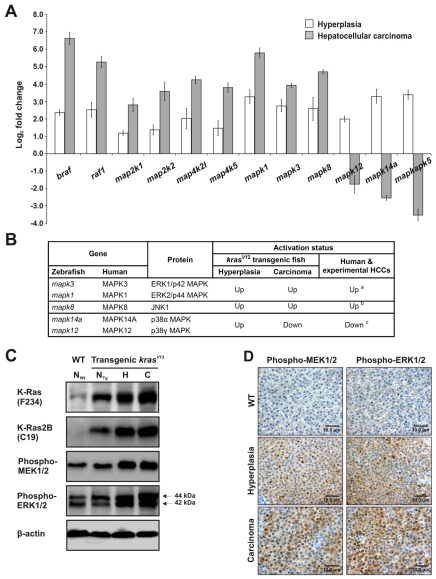
The ERK pathway, one of the most-established mammalian mitogen-activated protein kinase (MAPK) pathways, is commonly activated downstream of Ras. To determine whether the ERK pathway is activated by overexpression of krasV12, we conducted qRT-PCR to analyze the expression of various genes associated with this pathway.
Gene expression of the Raf kinases braf and raf1, and other upstream activators of the ERK pathway, namely map2k1, map2k2, map4k2l and map4k5, as well as the downstream mapk1 and mapk3, were all upregulated in both hyperplastic liver (HL) and HCC, with higher expression levels in HCC than in HL (Fig. 4A). We also measured the transcript levels of other subfamilies of MAPKs. The level of mapk8 (JNK1) was found to increase in both stages. Although an increase in mapk12 (p38γ) and mapk14a (p38α) transcripts were observed in HL, their expressions became downregulated in HCC. A similar trend was observed in the transcript level of p38-regulated/activated protein kinase (PRAK; mapkapk5). The activation pattern of these three major MAPKs during liver tumorigenesis in our transgenic zebrafish model is consistent with those in human liver cancer and other HCC models (Fig. 4B).
Fig. 4.
Hyperactivation of the MAPK signaling pathway in krasV12 transgenic zebrafish. (A) Determination of expression levels of various kinase genes by qRT-PCR in liver hyperplasia (3 mpf) and carcinoma (9 mpf). The expression levels of these genes in each WT and transgenic liver sample were first measured and normalized with the expression level of β-actin (n=5 each). The log2 fold changes in expression in the transgenic samples as compared with matched WT sample are presented. (B) Comparison of MAPK family expression during liver tumorigenesis in krasV12 transgenic zebrafish, human liver cancer and other experimental models of HCC. a(Schmidt et al., 1997); b(Wurmbach et al., 2007; Chang et al., 2009); c(Wurmbach et al., 2007). (C) Western blots of total proteins from WT normal liver (NWt) and kras V12 transgenic liver showing normal liver morphology (NTg), hyperplastic liver (H) or HCC (C) to detect total Kras (F234), Kras2B (C19) isoform, phospho-MEK1/2 and phospho-ERK1/2 (also known as phospho-p44/42 MAPK). Arrows: double bands of phospho-ERK1/2 (44 kDa and 42 kDa). β-actin, internal control for equal loading. (D) Immunohistochemical analysis of paraffin-embedded liver sections from WT (control; top row), 3-mpf transgenic (hyperplasia; middle row) and 9-mpf transgenic (carcinoma; bottom row) fish. Sections were stained with antibodies against phospho-MEK1/2 or phospho-ERK1/2. Scale bars: 10 μm.
We then determined the protein expression level of Kras in WT normal liver (NWt) and in F2/II krasV12 transgenic zebrafish that had various liver morphologies, including normal liver (NTg), HL and HCC (Fig. 4C). Western blotting using anti-K-Ras (F234) revealed that total Kras was minimal in NWt but increasingly higher in NTg, HL and HCC. Immunoblotting with anti-K-Ras-2B (C19) confirmed that no KrasV12 protein could be detected in NWt. In NTg, KrasV12 protein was present at a much lower level as compared with HL and HCC. Because Ras signals through MEK and ERK proteins via phosphorylation, we showed that the levels of phospho-MEK1/2 and phospho-ERK1/2 proteins in NTg were similar to that in NWt but were visibly higher in HL and HCC. Indeed, immunohistochemistry of liver tumor sections showed apparently enhanced cytoplasmic and nuclear staining of phospho-MEK1/2 and phospho-ERK1/2 in HL, and this staining was intensified in HCC (Fig. 4D).
Activation of the Wnt–β-catenin pathway during krasV12 liver tumorigenesis
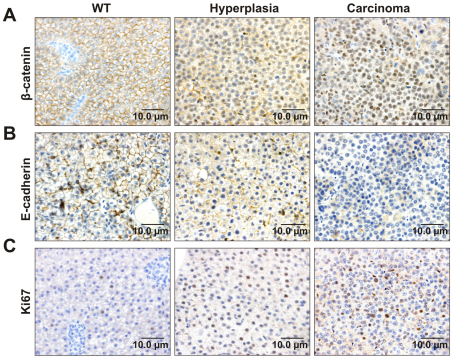
Of several signaling pathways frequently deregulated in HCC, the canonical Wnt pathway, with β-catenin as a crucial downstream component, is an important contributor to tumorigenesis (Farazi and DePinho, 2006). E-cadherin, which is a binding partner of β-catenin, also plays a critical role in liver tumorigenesis as a tumor and invasion suppressor (Wei et al., 2002). To verify activation of the Wnt–β-catenin pathway, we employed immunohistochemistry to examine the expression of β-catenin, E-cadherin and cellular proliferation marker Ki67 in krasV12 transgenic liver. β-catenin localized in the cell membrane in the WT liver (Fig. 5A). By contrast, mixed nuclear and membranous staining patterns of β-catenin were found in HL with a distinct nuclear pattern in HCC. Moreover, HCC showed no staining of E-cadherin, whereas HL retained similar E-cadherin staining in the cell membrane as in WT liver (Fig. 5B). There was little or no signal of Ki67 expression in WT liver, whereas nuclear staining of Ki67 was observed in HL and more so in HCC (Fig. 5C). These data indicated activation of the Wnt–β-catenin pathway as tumorigenesis progressed from HL to HCC in krasV12 transgenic fish.
Fig. 5.
Activation of the Wnt–β-catenin pathway during krasV12 liver tumorigenesis. Representative immunohistochemical liver sections from WT zebrafish as the control and krasV12 transgenic fish with liver hyperplasia and carcinoma are shown. (A) Immunohistochemistry for β-catenin showing an increasing nuclear localization of β-catenin during HCC progression. (B) Immunohistochemistry for E-cadherin showing loss of membranous E-cadherin expression during tumor growth. (C) Immunohistochemistry for Ki67 showing a high expression level of Ki67 in cell nuclei, which increased from hyperplasia to carcinoma.
Acceleration of liver tumor onset by loss of p53-mediated senescence in krasv12 transgenic zebrafish
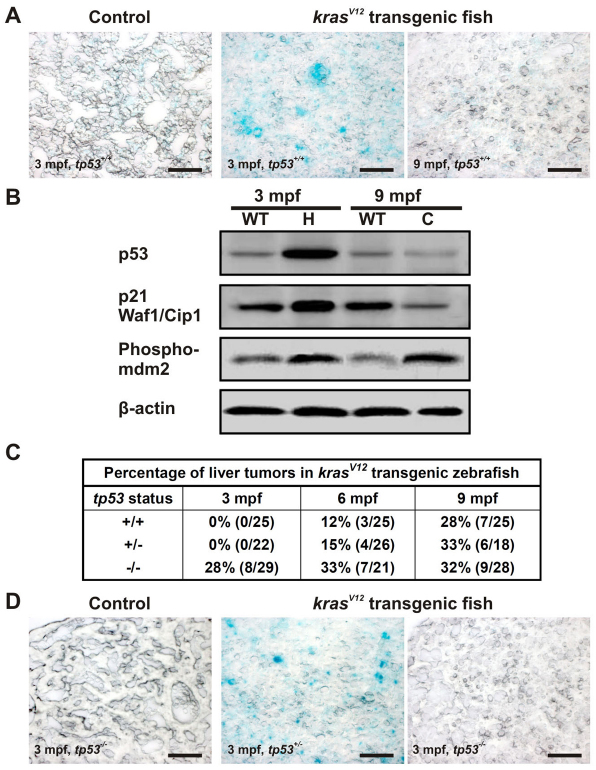
Excessive activation of Ras signaling can induce DNA damage response (DDR) by DNA replication stress due to aberrant cell proliferation that could prompt the activation of p53-induced senescence as a barrier to tumor progression induced by Ras (DiMicco et al., 2006; Vousden and Prives, 2009). We performed the senescence-associated β-galactosidase (SA-βgal) assay on liver cryosections. Intense SA-βgal signals were observed only in HL at 3 mpf, whereas little signal was detected in HCC at 9 mpf (Fig. 6A). These observations suggested that the number of cells undergoing senescence increased in the pre-neoplastic lesions but subsided during progression to the carcinoma stage. Western blotting showed that endogenous p53 increased in HL but decreased in HCC (Fig. 6B). Importantly, p21 Waf1/Cip1, a direct transcriptional target of p53 whose activation leads to senescence, was elevated in HL but decreased in HCC. The level of phosphorylated MDM2 protein, the negative regulator of p53, was found to increase in both stages (Fig. 6B). Collectively, these observations verified the response of p53 in mediating senescence in HL, resulting in latent tumor development.
Fig. 6.
KrasV12-induced p53-dependent senescence in the pre-neoplastic liver. (A) Oncogenic KrasV12-induced senescence at an early stage of liver tumor development. SA-βgal staining was performed on liver cryosections from 3- and 9-mpf krasV12 transgenic and WT fish (tp53+/+). (B) Western blots of total proteins from liver hyperplasia (H), carcinoma (C) and age-matched WT liver to evaluate the levels of tumor suppressor p53 together with its target, p21 Waf1/Cip1, and its regulator, phospho-MDM2, during tumorigenesis. β-actin, internal control for equal loading. (C) Acceleration of liver tumor onset in homozygous tp53M214K mutant transgenic fish. Percentages of tumors observed from krasV12 transgenic fish with different p53 backgrounds at different time points are shown. (D) Suppression of senescence induced by oncogenic krasV12 in tp53−/− background. SA-βgal staining was performed with four biological replicates in each group. Scale bars: 50 μm.
To find out whether loss of p53 could promote tumorigenesis, the homozygous tp53M214K (tp53−/−) mutant line (Berghmans et al., 2005) was crossed with krasV12 F2/II to obtain heterozygous tp53+/− fish expressing krasV12 in the liver. This family was again crossed with the tp53−/− line to obtain mixed offspring with tp53+/− and tp53−/− background. This cohort was randomly divided and maintained in three tanks. Each tank was sacrificed at 3, 6 and 9 mpf to screen for the presence of liver tumors (Fig. 6C). Liver morphology revealed that, at 3 mpf, 28% of the tp53−/− zebrafish showed tumor protrusions, with 63% of these having confirmed HCC. By contrast, no tumors were observed in the tp53+/− and tp53+/+ siblings at 3 mpf. Notably, SA-βgal assay showed no senescence in transgenic tp53−/− fish, whereas strong signals were still detected in the transgenic fish with tp53+/− background (Fig. 6D). At 6 mpf, pre-neoplastic tumors were observed in the transgenic fish liver with tp53+/+ and tp53+/− backgrounds (12% and 15%, respectively). At 9 mpf, liver tumor incidences in the krasV12 transgenics became similar, regardless of tp53 status. No significant differences in the survival rates were noticed between these cohorts before 90 dpf. Thus, loss of p53 accelerated tumor onset but not the overall tumor incidence after 9 mpf.
Transcriptomic analyses of krasV12 liver tumorigenesis
Oligonucleotide microarray was conducted to investigate gene expression profiles in HL and HCC as outlined in supplementary material Fig. S3. By applying the selection criteria of ≥1.5 log2 fold change and P≤0.01 false discovery rate (FDR) adjustment, 1417 and 1564 differentially expressed genes having human homologs were obtained for HL and HCC, respectively (supplementary material Table S1). Because a biological process often involves a group of genes acting in concert, GSEA (Gene Set Enrichment Analysis) was used to compare predefined human gene sets from the Gene Ontology (GO) and KEGG of the Molecular Signature Database to gain biological insight into krasV12-driven liver tumorigenesis. Overall, GSEA identified 42 and 151 significant human gene sets for zebrafish HL and HCC, respectively (supplementary material Table S2). Strikingly, activation of key signaling pathways was found in HL (p53) and HCC (TLR-NFκB, JAK-STAT, insulin-IGF and TGFβ), as well as in both HL and HCC (Raf-MEK-ERK, PI3K-AKT, Wnt–β-catenin, VEGF and complement cascade). GSEA also determined the genes within each significantly enriched human set that contributed most to the enrichment score in zebrafish HL and HCC. A total of 261 genes in the 42 hyperplasia gene sets and 598 genes in the 151 HCC gene sets were obtained (supplementary material Table S2). We then compared these genes with the differentially expressed zebrafish genes identified in supplementary material Table S1 to limit the results to only the significantly up-or downregulated genes. These genes are termed zebrafish enriched genes. Hence, 173 up- and 61 downregulated zebrafish enriched genes were found in HL, whereas there were 398 up- and 99 downregulated zebrafish enriched genes in HCC. By overlapping HL and HCC genes, we identified 128, 106 and 391 HL-specific, overlapping and HCC-specific enriched genes, respectively (supplementary material Table S3).
Identification of a HCC-specific signature and a liver cancer progression signature
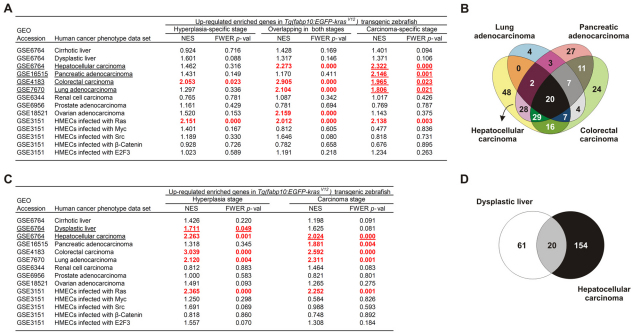
To explore whether the zebrafish stage-specific and overlapping enriched genes in HL and HCC possess Ras signature, transcriptomic profiles of human mammary epithelial cells (HMECs) infected with activated oncogenes such as β-catenin, E2F3, Myc, Ras and Src (Bild et al., 2006) were used for cross-species GSEA. The upregulated zebrafish enriched genes were associated with signatures linked to oncogenic Ras but not the other oncogenes (Fig. 7A). Next, we questioned whether these zebrafish genes were conserved throughout human HCC progression, exclusively at a particular stage (cirrhosis, dysplasia and carcinoma) (Wurmbach et al., 2007) or involved in other human tumor types. Transcriptomic profiles from different human cancers were used for the comparison. We found that the upregulated zebrafish HCC-specific enriched genes were associated with human liver, pancreatic, colorectal and lung tumors (Fig. 7A; supplementary material Table S4). Consistent with this, KRAS mutations are most frequently found in human tumors of pancreas, colon, lung and ovary (Karnoub and Weinberg, 2008). The fact that the upregulated zebrafish HCC-specific enriched genes matched to Ras signature might explain their association with these human tumor types. Next, we defined a HCC-specific gene signature (48 genes) from a subset of upregulated HCC-specific zebrafish enriched genes that were associated only with human HCC but not with the other tumor types (Fig. 7B; supplementary material Table S5).
Fig. 7.
GSEA identification of conserved gene signatures common between zebrafish and human HCC. (A) Cross-species GSEA comparisons of different human cancer transcriptomic profiles with zebrafish upregulated hyperplasia-specific, carcinoma-specific and overlapping enriched genes. Human cancer data sets were collected from the Gene Expression Omnibus (GEO) database and their access numbers are indicated. Positive normalized enrichment score (NES) indicated enrichment of the zebrafish enriched genes in the human tumor state. Results shown in red were statistically significant with family-wise error rate (FWER) P-value =0.05. Significantly matched human data sets are underlined and used for gene signature identification by overlapping the zebrafish upregulated carcinoma-specific enriched genes that were found associated with each human data set. (B) Venn diagram illustrating the identification of 48 genes specific to HCC in both human and zebrafish. The 48 genes are presented in supplementary material Table S5. Lists of zebrafish enriched genes in each significant human cancer data set used to identify HCC gene signature are shown in supplementary material Table S4. (C) The stage-specific and overlapping enriched genes in zebrafish hyperplasia and carcinoma used in cross-species GSEA comparisons with different human cancer transcriptomic profiles. (D) Venn diagram identification of 20 genes upregulated during tumor progression from hyperplasia/dysplasia to carcinoma in both zebrafish and human liver cancer. These genes are presented in supplementary material Table S8, representing the liver cancer progression gene signature. Lists of zebrafish enriched genes in each significant human cancer data set used to identify the gene signature are shown in supplementary material Tables S6 and S7.
Because each stage of krasV12 liver tumorigenesis contains both stage-specific and overlapping enriched genes, cross-species analysis was again performed to assess whether HL zebrafish enriched genes, consisting of HL-specific and overlapping genes, showed any similarity with genes expressed in early human liver tumorigenesis. Indeed, the upregulated HL enriched genes were significantly associated with human dysplastic liver (Fig. 7C; supplementary material Table S6). Similarly, cross-species analysis of HCC-enriched genes revealed that these genes were significantly associated with human HCC (Fig. 7C; supplementary material Table S7). By overlapping the upregulated HL and HCC enriched genes shown in supplementary material Tables S6 and S7, we identified a liver cancer progression gene signature, which comprises 20 genes that remained upregulated throughout human and zebrafish HCC progression (Fig. 7D; supplementary material Table S8). By contrast, the zebrafish downregulated enriched genes were not related to any tumor types and oncogene status (data not shown). Similar observations were previously reported in several KrasD12 transgenic models (Langenau et al., 2007; Sweet-Cordero et al., 2005), suggesting that the genes that are downregulated by Ras might be different between species and that Ras mostly upregulates gene expression.
To further validate the differentially expressed genes detected by microarray analyses, qRT-PCR was performed to confirm expression of 15 major genes in several individual lesions from each tumorigenesis stage and these tested genes belonged to several important signaling pathways and processes, including AKT (akt2), ERK (mapk1, mapk3, mapk8, stmn1), STAT (stat3), p53 (mdm2, tp53), TGFβ (tgfb1), WNT (nlk), angiogenesis (angpt1), cell cycle (ccnb1, nbn, nfya) and ribosome (rpl19). As shown in supplementary material Table S9, the qRT-PCR results closely paralleled the microarray data, thus confirming that liver lesions that looked similar at the histological level were also similar with respect to gene expression profiles.
DISCUSSION
A krasV12 transgenic zebrafish model for investigation of liver tumorigenesis
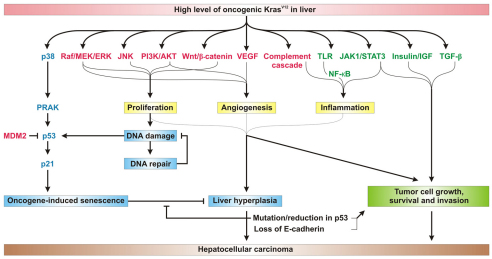
We have generated the first in vivo model of liver tumorigenesis driven by transgenic overexpression of oncogenic krasV12 in zebrafish. We provided evidence that the expression level of krasV12 is a crucial determinant of liver tumorigenesis through hyperactivation of the Ras-Raf-MEK-ERK pathway, because only the transgenic fish that exhibited higher krasV12 levels showed activation of ERK signaling and progression of liver tumor from hyperplasia to benign and invasive HCC. Although JNK and p38 MAPK were activated coordinately in HL, they were differently regulated in HCC, with the downregulation of p38 MAPK and upregulation of JNK. This is reminiscent of an earlier report that showed that hepatocyte-specific deletion of p38α promotes liver carcinogenesis with correlated activation of JNK (Hui et al., 2007). Furthermore, p38/PRAK could activate p53 in response to oncogenic Ras to mediate cellular senescence (Sun et al., 2007). Besides MAPKs, overexpression of Ras can initiate multiple signal transduction pathways implicated in tumorigenesis, as summarized in Fig. 8. We observed consistent activation of PI3K-AKT, VEGF and Wnt–β-catenin pathways as well as the complement cascade in zebrafish HL and HCC, underscoring the importance of these signaling pathways during liver tumorigenesis. It is also well known that complements act as pro-inflammatory factors as part of immune surveillance. Markiewski et al. suggested that a tumor-induced complement system could enhance tumor growth by modulation of the anti-cancer immune response (Markiewski et al., 2008). As such, our model confirms the role of complements in HCC development.
Fig. 8.
Proposed mechanism of Ras-induced liver tumorigenesis in the transgenic zebrafish model. A high level of oncogenic KrasV12 in the zebrafish liver leads to the activation of several key signaling pathways implicated in liver tumorigenesis, showing signaling pathways significantly activated in HL (blue), in both HL and HCC (red) and in HCC (green), as determined by qRT-PCR, microarray and GSEA, and western blot and/or immunohistochemistry analyses in the present study. During early tumorigenesis, the Raf-MEK-ERK, JNK, PI3K-AKT and Wnt–β-catenin pathways drive proliferation, whereas Raf-MEK-ERK, PI3K-AKT and VEGF signaling induce angiogenesis and the complement cascade induces inflammation. Collectively, crosstalk among these signaling activates the three major processes, proliferation, angiogenesis and inflammation, leading to liver hyperplasia. Another mechanism upon overexpression of KrasV12 was the deregulation of p38 MAPK signaling. In hyperplasia, p38 MAPK was upregulated, leading to the activation of PRAK and subsequently p53, which then targets the downstream p21 to mediate cellular senescence. In addition, aberrant cell proliferation could also induce DNA damage, which might further prompt the upregulation of p53. Moreover, DNA damage triggers DNA repair, enabling hyperplastic cells to overcome such damage. Thus, oncogene-induced senescence acts as a p53-dependent tumor-suppressive mechanism in the pre-neoplastic stage to guard against tumor progression. Downregulation of p38 MAPK, together with a persistently high level of MDM2 that might eventually result in a reduction in p53, could also serve as alternative mechanisms for liver tumorigenesis to bypass senescence. In neoplastic stage, activation of JAK-STAT and TLR-NFκB pathways enhance the inflammatory process. Importantly, the consistent activation of proliferation, angiogenesis and inflammation with their involved pathways, together with activation of the JAK-STAT, insulin-IGF and TGFβ pathways, are essential for contribution to tumor cell growth, survival and invasion in HCC. Loss of the key tumor suppressors p53 and E-cadherin further characterizes malignant/invasive HCC in our model.
Analysis of stage-specific liver lesions revealed the activation of DNA replication and damage elicited by excessive cell proliferation in HL only, which might result in oncogene-induced senescence (OIS). Replicative DNA damage triggered senescence through the p53 pathway. The increase of p53 then imparts its tumor suppressive effects through the induction of p21 to stall the cell cycle (Vousden and Prives, 2009). Nuclear transcription factor Y subunit α (nfya), previously proposed by Collado et al. as a marker that is specifically associated to OIS (Collado et al., 2005), was extremely up-regulated in krasV12 HL. Our list of enriched genes corresponding to HL as identified by GSEA (supplementary material Table S3) might thus yield new useful markers for the detection of senescence. Deleterious TP53 mutations enhanced hepatocarcinogenesis, whereas restoration of TP53 induced senescence that led to HCC regression (Takai et al., 2009; Xue et al., 2007). We have shown that tp53 null mutation can accelerate HCC onset marked by the abrogation of OIS in krasV12 zebrafish. However, tumor incidence did not increase in tp53−/−/krasV12 zebrafish. These observations support previous finding that TP53 mutation is a late event in human HCC (Martin and Dufour, 2008). Moreover, HCC is rarely found in tp53−/− zebrafish and Tp53 knockout mice alone (Berghmans et al., 2005; Donehower et al., 1992; Parant et al., 2010). Even in the absence of tp53, low expression of krasV12 seemed insufficient to initiate liver tumorigenesis (data not shown). This proposes that low levels of Ras require multiple mutations for neoplastic initiation in the liver.
In the sequence of events leading to HCC, we also observed the upregulation of two important transcription factors linking inflammation and cancer, namely NFκB and STAT3 (Mantovani et al., 2008). Both TLR, which is an essential upstream regulator of NFκB, and the JAK-STAT pathway were significantly activated only in krasV12 HCC. The synergistic activities of NFκB and STAT3, together with the complements, might play an important role in inflammation-mediated tumor growth. Consequently, inhibition of inflammation should be considered as a valuable strategy for HCC prevention. Furthermore, the emergence of JAK-STAT, IGF and TGFβ pathways in krasV12 HCC, and loss of E-cadherin, contributed to tumor cell growth, survival and invasion. This finding highlighted the significance of these pathways in HCC progression, which distinguishes them from the pathways that are activated in the early stage of krasV12 tumorigenesis.
Conserved gene expression signatures underlying liver tumorigenesis in humans and krasV12 transgenic zebrafish
Microarray analysis and GSEA uncovered two gene signatures, one being a HCC-specific signature and the other associated with a liver cancer progression signature. Both are upregulated in human and zebrafish liver cancer, underscoring the molecular conservation between species. Several genes in our gene signatures have been reported as prognostic markers for human HCC; for example, ANGPT1 and STMN1 in the HCC-specific signature, and APOE and CCNB1 in the HCC progression signature (Torimura et al., 2004; Wong et al., 2008; Wurmbach et al., 2007; Yokoyama et al., 2006). Interestingly, a large family of ribosomal proteins contributed to the bulk of the HCC-specific signature. Previously, Lee et al. demonstrated a consistent elevation of ribosomal proteins in human and mouse liver cancers (Lee et al., 2004). Our study highlights the possibility that ribosomal proteins might serve as evolutionarily conserved markers in HCC. By contrast, several components of ERK signaling (MAPK1, MAPK3, MAPKAPK2, MDM2, PRKCB1 and YWHAB), PI3K-AKT signaling (PI3KCA), Wnt signaling (NLK) and tumor invasion and/or metastasis (DERL1, MMP14, RUVBL2 SRC and THY1) made up the majority of genes in the liver cancer progression signature. These genes might therefore likewise play an important role in HCC progression and could serve as markers of early liver tumorigenesis. Notably, the liver cancer progression signature comprises a group of genes participating in the cell cycle and in DNA damage and repair (MCM5, NBN, RRM2 and TK1) that have not been previously reported in human HCC. The most prominent gene is nibrin (NBN or NBS1), a member of the DNA double-strand-break repair complex. NBS1 was initially known as a putative tumor suppressor protecting genome stability. However, recent findings have suggested that NBS1 is also an oncoprotein because it is overexpressed in several human cancers and leads to cell proliferation and transformation (Chen et al., 2005; Hematulin et al., 2008). Some of the most critical regulated genes in these signatures have been confirmed by qRT-PCR from several individual lesions of transgenics (supplementary material Table S9). Further investigations of these genes should provide insights into liver tumorigenesis and probably provide new therapeutic targets for HCC.
In summary, our study presents the overall molecular mechanisms depicting KrasV12 liver tumorigenesis, which recapitulates many of the defined features of human liver cancer. This transgenic line might thus provide a novel model for understanding the multistep nature of HCC progression as well as crosstalk between Ras and different levels of other signaling pathways, which could enable the development of successful synergistic therapies against human liver cancer. Because HCC latency is relatively long and the penetrance of HCC phenotype in krasV12 transgenics is less than 30% even in 9-month-old fish, this transgenic line might not be practical for drug screening but it could be used for enhancer screens to identify mutations that accelerate the onset of Ras-induced HCC. The data obtained from this study is also a necessary and indispensable step towards establishment of novel and more efficient models, such as inducible transgenic models of liver cancer. Furthermore, the fluorescence-tagged liver tumors would enable live imaging of tumor progression and tumor-host interaction during transplantation; these are particularly suitable to the small and relatively transparent zebrafish system. Because early Ras activation has been shown to cause premature lethality and act in a dose-dependent manner to trigger cancer development, work is underway to apply the inducible gene expression systems (Emelyanov and Parinov, 2008) to control the induction time and expression level of transgene, which will then enhance the attributes of using transgenic zebrafish as a new platform for anti-cancer drug screening to target currently undruggable pathways or several other simultaneous pathways in HCC development.
METHODS
Fish maintenance
Zebrafish were maintained according to established protocols (Westerfield, 2000). All experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the National University of Singapore.
Generation of transgenic zebrafish using the Ac/Ds transposon system
Zebrafish krasB was amplified using reverse transcription PCR with the two primers kras1 (5′-GGAGCCAAGCGGCCGC-ATGACCGAATATAAGCTTGTG-3′) and kras2 (5′-GGAAGGAAGCGGCCGCTCACATTAATGCACATTTTGTTT-TG-3′) containing the NotI restriction endonuclease excised sequence (underlined). The primers were designed based on the zgc:85725 cDNA sequence (GenBank BC078646; GI:50925043) (Strausberg et al., 2002). The amplified product was digested with NotI and cloned into the NotI site of the ET construct (Parinov et al., 2004) carrying a TAA stop codon after EGFP to terminate the translation of Kras. To produce the mutated (oncogenic) form of KrasV12 fused with EGFP, the TAA stop codon was removed, and glycine in position 12 of KrasV12 was replaced with valine using the QuikChange site-directed mutagenesis kit following manufacturer’s instructions (Stratagene) and primers Forward: 5′-CTGTACAAGTTAAGCGGCGGCATGACCGAATATAAGCTT-GTGGTCGTGGGAGCTGTAGGCG-3′ and Reverse: 5′-CGCCTACAGCTCCCACGACCACAAGCTTATATTCGGTCA-TGCCGCCGCTTAACTTGTACAG-3′. A 2.8-kb promoter of the fabp10 gene (Her et al., 2003) was digested and subcloned into a 0.6-kb miniDs construct pMDS6 (Emelyanov et al., 2006) between the NotI and SacII sites. The produced construct was then inserted with the fused EGFP-krasV12 sequence between the NotI and SacII sites. As a result, the product transcribed from the fabp10 promoter is a fusion protein of EGFP and KrasV12. Transgenic zebrafish were generated using the Ac/Ds transposon system as described previously (Emelyanov et al., 2006).
RNA isolation and qRT-PCR
Total RNA was isolated using TriZOL reagent (Invitrogen) and reverse transcribed using the SuperScript II cDNA Synthesis Kit (Invitrogen). qRT-PCR was performed with cDNA as the template using the iQ Single-Color Real-Time PCR Detection System (Bio-Rad Laboratories). Primer sequences used for amplification are as follows: β-actin forward: 5′-CCACCTTAAATGGCCTAGCA-3′, reverse: 5′-CATTGTGAGGAGGGCAAAGT-3′; kras (endogenous) forward: 5′-GTCCAGACAGGCGGTATTGT-3′, reverse: 5′-GACGCAGTTGAGGGAGAAAG-3′; krasV12 forward: 5′-CGACCACTACCAGCAGAACA-3′, reverse: 5′-GCTTTTGC-CTACGCCTACAG-3′; akt2 forward: 5′-GAACACCTTCATGAT-CCGCT-3′, reverse: 5′-CTCTGGAGCTGGATTTGGAC-3′; angpt1 forward: 5′-AACCCGAAGCCGACTTGTCC-3′, reverse: 5′-CGTCGGTCAGTTTTCGCGTC-3′; braf forward: 5′-AGCTTATGTCAGGGGCTTTG-3′, reverse: 5′-AGAGAGCGT-GCCAATAACTC-3′; ccnb1 forward: 5′-GGACTCAGACCAA-GGGCCGC-3′, reverse: 5′-GCACAGCCGGAGGTCTCCAT-3′; raf1 forward: 5′-ATGCCATACGTGTTCACAGC-3′, reverse: 5′-TCCCTTTGTGTACGGTTCCA-3′; map2k1 forward: 5′-AAAGAGCAGACCTCAAGCAG-3′, reverse: 5′-TTCAGGAGG-CAGTAGTTTGC-3′; map2k2 forward: 5′-TGCCTCATAAAGAA-CCCTGC-3′, reverse: 5′-AGGCTTACAAGCATACAGGC-3′; map4k2l forward: 5′-TGATCTGGAGGACAAGGACC-3′, reverse: 5′-AGAAAGAGCTGCGTCTCTGC-3′; map4k5 forward 5′-AGGACAGTGTTCTGGCATTC-3′, reverse: 5′-ATACAAAGGC-TCCAGCAGTG-3′; mapk1 forward: 5′-GGATGATTTGCCC-AAAGAGA-3′, reverse: 5′-GTCAGGTGAACGTTGAGGGT-3′; mapk3 forward: 5′-GAGTCGGTGAAGGGACAAAA-3′, reverse: 5′-TGATCCCGATGATGTTCTCA-3′; mapk8 forward: 5′-CTGCTGCAGATGACCATCCTTT-3′, reverse: 5′-ACAGAGCA-TATTTGAGGGGGCT-3′; mapk12 forward: 5′-GTGAAATGAC-GGGCTACGTT-3′, reverse: 5′-AGACTGTAGCTTCGCTGTGA-3′; mapk14a forward: 5′-CCCGTGCAGTATCAGAACTT-3′, reverse: 5′-CAGACTTGTGGCAGGTGTAA-3′; mapkapk5 forward: 5′-GACACAAGAACGATTTGCCC-3′, reverse: 5′-CTGGCTGATTCTGTGGAACA-3′; mdm2 forward: 5′-AACTCCCAACACAACCTTCG-3′, reverse: 5′-GGGTCTCTTC-CTGACTGCTG-3′; nfya forward: 5′-CGCGCCAAACTG-GAGGCTGA-3′, reverse: 5′-TTTACCCCAGAGGCGGGGCA-3′; nlk1 forward: 5′-GTGCCAAGTCCTGCTGAAAT-3′, reverse: 5′-AGTCGATTTGTGGAGGTTGG-3′; rpl19 forward: 5′-CTTGCGCTGTGGCAAGAAGAA-3′, reverse: 5′-TTCTCGG-GCATACGTGCGTT-3′; stat3 forward: 5′-CTGAAACCTTGA-GCGACACA-3′, reverse: 5′-AGCAGGTTGTGGAAGACCAG-3′; stmn1 forward: 5′-CTCTGAAGGGCATACTTGGACCG-3′, reverse: 5′-CTGCTTCATGAGACTTGCGTCTC-3′; tgfb1 forward: 5′-ATGATAGAATGGCTGCAGGG-3′, reverse: 5′-TGCAAGAGAGTTGCCATTTG-3′; tp53 forward: 5′-GATGGTGAAGGACGAAGGAA-3′, reverse: 5′-ACAAAGGTC-CCAGTGGAGTG-3′; zgc:194152 forward: 5′-CTTCCTCTAC-CCGCATCGTCC-3′, reverse: 5′-AACTGGCACTGCTTTC-ACGC-3′. Reactions were run in triplicate for each sample. Gene expression levels in each WT or transgenic liver sample were normalized with the expression level of β-actin as the internal control. The log2 fold changes in expression in the transgenic sample as compared with the WT sample were then calculated using the CT method (Schmittgen and Livak, 2008) following the formula: log2 fold change=–ΔΔCT=–[(CT gene of interest–CTβ-actin)transgenic sample–(CT gene of interest–CTβ-actin)WT sample]. For quantification of endogenous kras and transgenic krasV12 transcript levels, the log2 fold changes in expression were calculated using the formula: log2 fold change=– ΔCT=–[(CT gene of interest–CTβ-actin)transgenic sample] (Schmittgen and Livak, 2008).
Gross morphology and histological analyses
Transgenic zebrafish were dissected to expose the abdominal area and then observed under the Nikon SMZ1600 stereomicroscope for gross liver morphology. Fish were then fixed in either Bouin’s fixative (750 ml picric acid, 250 ml 37–40% formalin and 50 ml acetic acid) or 10% neutral buffered formalin (Sigma) for at least 2 days, dehydrated through a series of graded ethanol solutions, washed in clearing agent (Fisher Scientific) and embedded in paraffin blocks. Liver sections were stained with H&E for morphology analysis. Criteria for histological examination of zebrafish liver tumors were assessed as described previously (Lam et al., 2006).
Liver tumor transplantation
Adult WT zebrafish were γ-irradiated with 25 Grays and recovered for 2 days before transplantation. Liver tumors were aseptically dissected from 1-year-old krasV12 transgenic donors, washed twice with PBS and gently homogenized. Tumor cells were suspended in Hanks’ balanced salt solution to a concentration of 1×106 cells per 10 μl. 10 μl of cell suspension was injected intraperitoneally using a 25-μl Hamilton syringe into immobilized previously irradiated recipients. Mock recipients were injected with 10 μl of Hanks’ balanced salt solution.
Western blot analysis
Total proteins were isolated from samples using lysis buffer (10 mM Tris-HCl, pH 7.4, and 1% SDS) containing Complete Protease Inhibitor Cocktail (Roche). Protein concentrations were determined using the Bradford dye following the manufacturer’s instructions (Bio-Rad Laboratories). 20 μg of proteins were loaded and separated in a 10% SDS-PAGE and transferred to PVDF membrane. Immunodetections were performed using the following antibodies: K-Ras (F234), which detects ubiquitously expressed K-Ras (Santa Cruz Biotechnology); K-Ras-2B (C19), which detects the C-terminus of the K-Ras-2B splice variant (Santa Cruz Biotechnology); anti-phospho-MEK1/2 (Ser217/221; Cell Signaling Technology); anti-phospho-ERK1/2 (p44/42 MAPK; Thr202/Tyr204; Cell Signaling Technology); anti-p53 (Cell Signaling Technology); anti-phospho-MDM2 (Cell Signaling Technology); anti-p21 Waf1/Cip1 (Santa Cruz Biotechnology); and anti-β-actin (Sigma). All antibodies were used at a working dilution of 1:1000. Signals were detected using chemiluminescence (Pierce Biotechnology) and exposure to X-ray film (Kodak).
Immunohistochemistry analysis
Immunostaining was performed on 5-μm sections of formalin-fixed and paraffin-embedded tissues. Sections were first boiled for antigen retrieval in 10 mM citrate buffer pH 6.0 for 10 minutes, treated with 3% hydrogen peroxide for 10 minutes at room temperature, and blocked using the SuperBlock Blocking Buffer (Pierce Biotechnology) for 30 minutes. Sections were then incubated in a humidified chamber at 4°C overnight with antibodies against phospho-MEK1/2 (Ser217/221; Cell Signaling Technology), phospho-ERK1/2 (Thr202/Tyr204; Cell Signaling Technology), β-catenin (Abcam), E-cadherin (Abcam) or Ki67 (Sigma) at working dilutions of 1:50 to 1:100. Next, sections were washed with PBS and incubated in either anti-mouse or anti-rabbit secondary antibody for 1 hour at room temperature. Signals indicating peroxidase activity were visualized using the Metal Enhanced DAB Substrate Kit (Pierce Biotechnology) following the manufacturer’s instructions. The sections were then counterstained with hematoxylin, dehydrated and permanently mounted for microscopic examination.
Senescence-associated β-galactosidase assay
Liver cryosections of WT and transgenic zebrafish were prepared by fixing the tissues in 4% paraformaldehyde overnight at 4°C. After fixation, tissues were washed several times with PBS before incubating in 30% sucrose at 4°C overnight. Samples were then embedded and frozen at −20°C for 30 minutes, and sectioned into 10-μm thickness using Leica cryostat microtome. Sections were dried at 46°C for 2 hours and continued with the SA-βgal assay. SA-βgal expression was determined using the staining kit supplied by Cell Signaling Technology.
Zebrafish oligonucleotide microarray analyses
Microarray construction and hybridization were performed as described previously (Lam et al., 2006; Lam et al., 2009a; Lam et al., 2009b). Detailed workflow for microarray data and GSEA is presented in supplementary material Fig. S3.
Statistical analysis
Kaplan-Meier curves were computed using the survival distribution of each test group. The log-rank test was used to compare significant differences in death rates between the two transgenic lines. Statistical analysis was performed by a Student’s t-test for direct comparisons between WT and transgenic groups. Based on Bonferroni’s correction for multiple comparisons, P-values of <0.01 were considered statistically significant.
TRANSLATIONAL IMPACT.
Clinical issue
Human liver cancer, the most common of which is hepatocellular carcinoma (HCC), is a major cause of cancer death worldwide. Human HCCs have a complex molecular pathology, with heterogeneous morphology and genetics, but recent work has indicated that ubiquitous activation of the Ras-ERK pathway is present in nearly all HCCs. Thus, targeting Ras signaling has emerged as a potential strategy to treat advanced HCC. However, the mechanisms of Ras-induced liver tumorigenesis remain poorly understood, and in vivo models that enable investigations of the central role of Ras in liver cancer are lacking.
Results
In this paper, the authors establish a model of liver cancer by generating a transgenic zebrafish line, Tg(fabp10:EGFP-krasV12), that uses a strong hepatocyte-specific promoter to target oncogenic krasV12 expression to the liver. Fusing krasV12 to EGFP allows visualization of tumor development in zebrafish from early stages. The authors show that liver tumorigenesis in this system is driven only by a high level of krasV12 expression through activation of the ERK pathway, as well as by loss of E-cadherin expression and nuclear accumulation of β-catenin (indicating activation of canonical Wnt signaling). krasV12 transgenic tumors show progressive features from hyperplasia to invasive HCC, which is accompanied by a loss of the p53-dependent senescence response; the absence of p53 accelerates tumor onset. Microarrays and gene set enrichment analyses (GSEAs) of hyperplastic lesions and HCCs delineate several other pathways that are enriched during Ras-driven liver tumorigenesis. Finally, cross-species comparisons identify two conserved gene signatures accounting for HCC specificity and HCC progression in both zebrafish and human liver cancer.
Implications and future directions
This krasV12 transgenic zebrafish is the first in vivo model in which it is possible to address molecular mechanisms underlying Ras-driven liver tumorigenesis that recapitulates typical hallmarks of human HCC. The high incidence and consistent pattern of cancer in this model, coupled with low maintenance costs of zebrafish, allow systematic study of liver cancer progression from hyperplasia to carcinoma stages. Importantly, the two HCC gene signatures identified in this study might be useful as prognostic markers and/or potential therapeutic targets in human HCC. In addition, using this model to better understand crosstalk between Ras and other signaling pathways might contribute to the development of successful synergistic therapies for human liver cancer. This model provides a new platform that can be used for high-throughput screening of anti-cancer drugs to treat human liver cancer in the future.
Supplementary Material
Acknowledgments
This work was supported by the Biomedical Research Council of Singapore. We also thank Thomas Look for providing tp53 mutant zebrafish.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
A.T.N., A.E., S.P. and Z.G. conceived and designed the study. A.T.N., A.E. and C.H.V.K. performed the experiments. A.T.N., A.E., C.H.V.K., J.M.S., S.H.L., S.P. and Z.G. analyzed and interpreted the data. J.M.S., S.H.L. and S.M. supported techniques. A.T.N., A.E., C.H.V.K., S.P. and Z.G. wrote the manuscript. A.E., S.P. and Z.G. supervised the study.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.007831/-/DC1
REFERENCES
- Amatruda J. F., Patton E. E. (2008). Genetic models of cancer in zebrafish. Int. Rev. Cell Mol. Biol. 271, 1–34 [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R. D., Wienholds E., Neuberg D., Kutok J. L., Fletcher C. D., Morris J. P., Liu T. X., Schulte-Merker S., Kanki J. P., et al. (2005). tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 102, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild A. H., Yao G., Chang J. T., Wang Q., Potti A., Chasse D., Joshi M. B., Harpole D., Lancaster J. M., Berchuck A., et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357 [DOI] [PubMed] [Google Scholar]
- Calvisi D. F., Ladu S., Gorden A., Farina M., Conner E. A., Lee J. S., Factor V. M., Thorgeirsson S. S. (2006). Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 130, 1117–1128 [DOI] [PubMed] [Google Scholar]
- Chang Q., Chen J., Beezhold K. J., Castranova V., Shi X., Chen F. (2009). JNK1 activation predicts the prognostic outcome of the human hepatocellular carcinoma. Mol. Cancer 8, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., Su Y. N., Chou P. C., Chiang W. C., Chang M. C., Wang L. S., Teng S. C., Wu K. J. (2005). Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 280, 32505–32511 [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., et al. (2005). Tumour biology: senescence in premalignant tumours. Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- DiMicco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P. G., Bensimon A., et al. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr, Butel J. S., Bradley A. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- Emelyanov A., Parinov S. (2008). Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 320, 113–121 [DOI] [PubMed] [Google Scholar]
- Emelyanov A., Gao Y., Naqvi N. I., Parinov S. (2006). Trans-kingdom transposition of the maize dissociation element. Genetics 174, 1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi P. A., DePinho R. A. (2006). Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687 [DOI] [PubMed] [Google Scholar]
- Harada N., Oshima H., Katoh M., Tamai Y., Oshima M., Taketo M. M. (2004). Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 64, 48–54 [DOI] [PubMed] [Google Scholar]
- Hayashi J., Aoki H., Kajino K., Moriyama M., Arakawa Y., Hino O. (2000). Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology 32, 958–961 [DOI] [PubMed] [Google Scholar]
- Hematulin A., Sagan D., Eckardt-Schupp F., Moertl S. (2008). NBS1 is required for IGF-1 induced cellular proliferation through the Ras/Raf/MEK/ERK cascade. Cell Signal. 20, 2276–2285 [DOI] [PubMed] [Google Scholar]
- Her G. M., Chiang C. C., Chen W. Y., Wu J. L. (2003). In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 538, 125–133 [DOI] [PubMed] [Google Scholar]
- Hui L., Bakiri L., Mairhorfer A., Schweifer N., Haslinger C., Kenner L., Komnenovic V., Scheuch H., Beug H., Wagner E. F. (2007). p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat. Genet. 39, 741–749 [DOI] [PubMed] [Google Scholar]
- Karnoub A. E., Weinberg R. A. (2008). Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh S., Pan X., Garcia-Lecea M., Winata C. L., Pan X., Wohland T., Korzh V., Gong Z. (2008). Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev. Biol. 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. H., Gong Z. (2006). Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle 5, 573–577 [DOI] [PubMed] [Google Scholar]
- Lam S. H., Wu Y. L., Vega V. B., Miller L. D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K. R., Lee S., Mathavan S., et al. (2006). Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat. Biotechnol. 24, 73–75 [DOI] [PubMed] [Google Scholar]
- Lam S. H., Krishna Murthy Karuturi R., Gong Z. (2009a). Zebrafish spotted-microarray for genome-wide expression profiling experiments: data acquisition and analysis. Methods Mol. Biol. 546, 197–226 [DOI] [PubMed] [Google Scholar]
- Lam S. H., Mathavan S., Gong Z. (2009b). Zebrafish spotted-microarray for genome-wide expression profiling experiments: array printing and hybridization. Methods Mol. Biol. 546, 175–195 [DOI] [PubMed] [Google Scholar]
- Langenau D. M., Keefe M. D., Storer N. Y., Guyon J. R., Kutok J. L., Le X., Goessling W., Neuberg D. S., Kunkel L. M., Zon L. I. (2007). Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 21, 1382–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le X., Langenau D. M., Keefe M. D., Kutok J. L., Neuberg D. S., Zon L. I. (2007). Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl. Acad. Sci. USA 104, 9410–9415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Chu I. S., Mikaelyan A., Calvisi D. F., Heo J., Reddy J. K., Thorgeirsson S. S. (2004). Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 36, 1306–1311 [DOI] [PubMed] [Google Scholar]
- Lieschke G. J., Currie P. D. (2007). Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- Llovet J. M., Bruix J. (2008). Molecular targeted therapies in hepatocellular carcinoma. Hepatology 48, 1312–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., Balkwill F. (2008). Cancer-related inflammation. Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- Markiewski M. M., DeAngelis R. A., Benencia F., Ricklin-Lichtsteiner S. K., Koutoulaki A., Gerard C., Coukos G., Lambris J. D. (2008). Modulation of the antitumor immune response by complement. Nat. Immunol. 9, 1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Dufour J. F. (2008). Tumor suppressor and hepatocellular carcinoma. World J. Gastroenterol. 14, 1720–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou C., Jones M., Walker P., Kamarashev J., Kelly A., Hurlstone A. F. (2009). Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis. Model. Mech. 2, 399–411 [DOI] [PubMed] [Google Scholar]
- Mizgirev I., Revskoy S. (2010). Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat. Protoc. 5, 383–394 [DOI] [PubMed] [Google Scholar]
- Parant J. M., George S. A., Holden J. A., Yost H. J. (2010). Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis. Model. Mech. 3, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S., Kondrichin I., Korzh V., Emelyanov A. (2004). Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 [DOI] [PubMed] [Google Scholar]
- Park S. W., Davison J. M., Rhee J., Hruban R. H., Maitra A., Leach S.D. (2008). Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 134, 2080–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandgren E. P., Quaife C. J., Pinkert C. A., Palmiter R. D., Brinster R. L. (1989). Oncogene-induced liver neoplasia in transgenic mice. Oncogene 4, 715–724 [PubMed] [Google Scholar]
- Schmidt C. M., McKillop I. H., Cahill P. A., Sitzmann J. V. (1997). Increased MAPK expression and activity in primary human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 236, 54–58 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- Sharpless N. E., DePinho R. A. (2006). The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 5, 741–754 [DOI] [PubMed] [Google Scholar]
- Spitsbergen J. M., Kent M. L. (2003). The state of the art of the zebrafish model for toxicology and toxicologic pathology research-advantages and current limitations. Toxicol. Pathol. 31, 62–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R. L., Buetow K. H., Greenhut S. F., Grouse L. H., Schaefer C. F. (2002). The cancer genome anatomy project: online resources to reveal the molecular signatures of cancer. Cancer Invest. 20, 1038–1050 [DOI] [PubMed] [Google Scholar]
- Sun P., Yoshizuka N., New L., Moser B. A., Li Y., Liao R., Xie C., Chen J., Deng Q., Yamout M., et al. (2007). PRAK is essential for ras-induced senescence and tumor suppression. Cell 128, 295–308 [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A., Mukherjee S., Subramanian A., You H., Roix J. J., Ladd-Acosta C., Mesirov J., Golub T. R., Jacks T. (2005). An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 37, 48–55 [DOI] [PubMed] [Google Scholar]
- Takai A., Toyoshima T., Uemura M., Kitawaki Y., Marusawa H., Hiai H., Yamada S., Okazaki I. M., Honjo T., Chiba T., et al. (2009). A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene 28, 469–478 [DOI] [PubMed] [Google Scholar]
- Torimura T., Ueno T., Kin M., Harada R., Taniguchi E., Nakamura T., Sakata R., Hashimoto O., Sakamoto M., Kumashiro R., et al. (2004). Overexpression of angiopoietin-1 and angiopoietin-2 in hepatocellular carcinoma. J. Hepatol. 40, 799–807 [DOI] [PubMed] [Google Scholar]
- Villanueva A., Minguez B., Forner A., Reig M., Llovet J. M. (2010). Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu. Rev. Med. 61, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K. H., Prives C. (2009). Blinded by the light: the growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- Wei Y., Van-Nhieu J. T., Prigent S., Srivatanakul P., Tiollais P., Buendia M. A. (2002). Altered expression of E-cadherin in hepatocellular carcinoma: correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology 36, 692–701 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (4th edn). Eugene, OR: University of Oregon Press [Google Scholar]
- Wong Q. W., Lung R. W., Law P. T., Lai P. B., Chan K. Y., To K. F., Wong N. (2008). MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 135, 257–269 [DOI] [PubMed] [Google Scholar]
- Wurmbach E., Chen Y. B., Khitrov G., Zhang W., Roayaie S., Schwartz M., Fiel I., Thung S., Mazzaferro V., Bruix J., et al. (2007). Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45, 938–947 [DOI] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S. W. (2007). Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y., Kuramitsu Y., Takashima M., Iizuka N., Terai S., Oka M., Nakamura K., Okita K., Sakaida I. (2006). Protein level of apolipoprotein E increased in human hepatocellular carcinoma. Int. J. Oncol. 28, 625–631 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.