Abstract
Purpose
Long-term survival for children with diffuse intrinsic pontine glioma (DIPG) is less than 10%, and new therapeutic targets are urgently required. We evaluated a large cohort of DIPGs to identify recurrent genomic abnormalities and gene expression signatures underlying DIPG.
Patients and Methods
Single-nucleotide polymorphism arrays were used to compare the frequencies of genomic copy number abnormalities in 43 DIPGs and eight low-grade brainstem gliomas with data from adult and pediatric (non-DIPG) glioblastomas, and expression profiles were evaluated using gene expression arrays for 27 DIPGs, six low-grade brainstem gliomas, and 66 nonbrainstem low-grade gliomas.
Results
Frequencies of specific large-scale and focal imbalances varied significantly between DIPGs and nonbrainstem pediatric glioblastomas. Focal amplifications of genes within the receptor tyrosine kinase–Ras–phosphoinositide 3-kinase signaling pathway were found in 47% of DIPGs, the most common of which involved PDGFRA and MET. Thirty percent of DIPGs contained focal amplifications of cell-cycle regulatory genes controlling retinoblastoma protein (RB) phosphorylation, and 21% had concurrent amplification of genes from both pathways. Some tumors showed heterogeneity in amplification patterns. DIPGs showed distinct gene expression signatures related to developmental processes compared with nonbrainstem pediatric high-grade gliomas, whereas expression signatures of low-grade brainstem and nonbrainstem gliomas were similar.
Conclusion
DIPGs comprise a molecularly related but distinct subgroup of pediatric gliomas. Genomic studies suggest that targeted inhibition of receptor tyrosine kinases and RB regulatory proteins may be useful therapies for DIPG.
INTRODUCTION
Diffuse intrinsic pontine gliomas (DIPGs) comprise 10% to 15% of pediatric CNS tumors and carry poor prognosis despite treatment with irradiation and chemotherapy.1,2 Diagnosis is generally based on neuroimaging. When biopsy or autopsy is obtained, the predominant histologic diagnosis is glioblastoma (WHO grade 4).3,4 Molecular research has been greatly limited, because biopsies usually offer no therapeutic benefit and are rarely performed.
Several groups have reported significantly different frequencies of specific copy number abnormalities (CNAs) between pediatric and adult high-grade gliomas (HGGs).5–8 Because of limited sample sizes, it is less clear whether DIPGs show the same molecular signatures as nonbrainstem pediatric HGGs, or if they also show a distinct spectrum of alterations. In studies with focused analyses of specific genes in small cohorts, approximately half of DIPGs contained TP53 mutations, and three (19%) of 16 high-grade DIPGs contained EGFR amplification.9–11 More recently, our group and others conducted genome-wide analyses of copy number imbalances from small cohorts of DIPGs and identified recurrent focal amplifications of PDGFRA. However, no other common focal alterations were consistently detected, likely because of the limited numbers of tumors evaluated in each study.6,7,12
We prospectively collected DIPG tumor samples at biopsy, when clinically indicated, or more commonly at autopsy.4 Here, we report the genome-wide analyses of copy number and gene expression signatures from this cohort of DIPGs. Our results, including recurrent focal gains in the receptor tyrosine kinase (RTK) and retinoblastoma protein (RB) pathways, suggest promising new therapeutic targets for DIPG.
PATIENTS AND METHODS
DIPG samples were obtained at autopsy or surgery with informed consent from the patients' parents or legal guardians as part of an institutional review board–approved protocol at St Jude Children's Research Hospital, as described.4 Thirty-seven DIPG samples were obtained postmortem. Seven samples were obtained before adjuvant therapy: six surgical samples and one postmortem sample from a child who died before start of therapy. There were two patient cases with matched surgical and autopsy samples. For one patient, separate autopsy samples were collected for DIPG in brainstem and tumor from a region of contiguous involvement of the cerebellum. Median age at diagnosis (6.1 years) and median survival (10.8 months) of the cohort were representative of DIPG. Matched normal tissue was collected as peripheral blood or, for autopsy cases, normal brain. Sections from matched formalin-fixed paraffin-embedded tissue reviewed by a neuropathologist (D.W.E.) showed 38 glioblastomas, one anaplastic astrocytoma, and four samples of insufficient quality for diagnostic subclassification. Magnetic resonance images of brain for all patient cases were reviewed by a pediatric neuro-oncologist (A.B.) to confirm diagnosis.4 Clinical and histologic characteristics of the low-grade gliomas (LGGs) are detailed in the Data Supplement. Nucleic acid extraction was performed as described.13 DNA was labeled and hybridized to Affymetrix SNP 6.0 arrays (Affymetrix, Santa Clara, CA), and RNA was profiled using Affymetrix Human Genome U133 Plus 2.0 arrays (Affymetrix). Details of single-nucleotide polymorphism (SNP) data analyses, validation by quantitative polymerase chain reaction and fluorescent in situ hybridization (FISH), and expression and statistical analyses are provided in the Data Supplement. Array data are deposited at www.ncbi.nlm.nih.gov/geo/, accession GSE26576.
RESULTS
Copy Number Imbalances Distinguish DIPG From Nonbrainstem Pediatric and Adult Glioblastoma
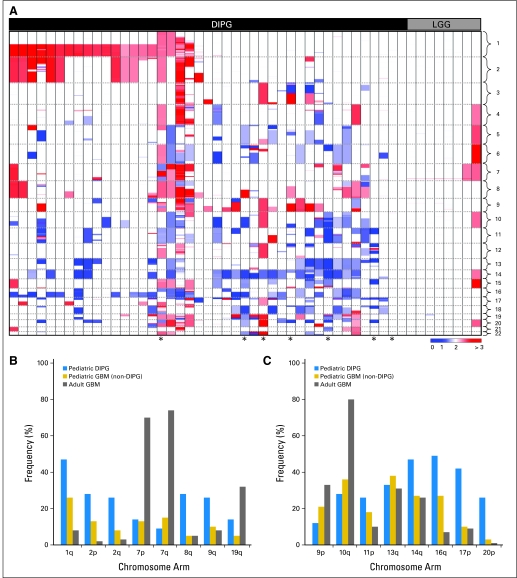
We evaluated CNAs in 43 DIPGs and eight brainstem LGGs. Forty-one (95%) of 43 DIPGs showed numerous large-scale imbalances (Fig 1A). In contrast, brainstem LGGs had relatively stable genomes, with seven (88%) of eight tumors lacking large-scale CNAs (Fig 1A). We compared the frequencies of specific large-scale imbalances in DIPG with those found in nonbrainstem pediatric and adult glioblastomas6,14 (Figs 1B, 1C; Data Supplement). Gains of chromosomes 2q, 8q, and 9q and losses of 16q, 17p, and 20p were significantly more frequent in DIPG than in nonbrainstem glioblastomas from either age group (Figs 1B, 1C; P values ranging from .05 to < .001). The frequency of chromosome 7 gain and 10q loss was similar in DIPG and nonbrainstem pediatric glioblastoma and significantly lower than in adult glioblastoma (P < .001). Gain of chromosome 1q was more common in DIPG than in adult glioblastoma (P < .001) but not significantly different between DIPG and nonbrainstem pediatric glioblastoma. Interestingly, loss of chromosomes 13q and 14q occurred at similar frequencies among all glioblastomas regardless of age group or brainstem location.
Fig 1.
Copy-number abnormalities in diffuse intrinsic pontine glioma (DIPG). (A) Heat map showing segmentation analysis of normalized data from Affymetrix SNP 6.0 arrays to identify copy-number gains (red) and losses (blue) in 43 DIPGs and eight brainstem low-grade gliomas (LGGs). Chromosome positions are indicated along y-axis and separated by dashed line. Histologic subtypes are indicated across top. Scale bar shows color gradient to indicate copy number. Comparison of frequencies of most common large-scale genomic (B) gains or (C) losses in DIPG, compared with adult14 or pediatric nonbrainstem glioblastoma (GBM).6 Large-scale gains or losses were scored when more than half of markers on chromosome arm had copy-number gains or losses, respectively. All frequencies and P values listed in Data Supplement. (*) Indicates tumors obtained before adjuvant therapy.
There were no focal deletions of CDKN2A in 43 DIPGs, in contrast to nonbrainstem pediatric HGGs5–8 (10 of 39 nonbrainstem pediatric glioblastomas6 and none of 43 DIPGs; P < .001). Loss of heterozygosity (LOH) was evaluated for 36 samples for which we had matched normal DNA (Data Supplement). Although copy-neutral LOH of chromosome 17p is common in adult glioblastoma,15 a majority of 17p losses in DIPG were detected as CNAs (18 [42%] of 43), with only five (14%) of 36 showing copy-neutral LOH of 17p (Data Supplement). Other regions with the highest frequencies of LOH were chromosomes 14q and 20p, which were also largely the result of CNAs rather than copy-neutral changes.
The average overall number of large-scale CNAs per tumor was not significantly different between DIPGs obtained before and after treatment (10.3 and 6.9, respectively). Two DIPGs with matched samples from diagnosis and autopsy showed that CNAs in the paired samples were mostly concordant. Some additional CNAs were detected at autopsy, and some CNAs present at diagnosis were absent at autopsy (Data Supplement).
Recurrent Focal Gains of RTKs and Cell-Cycle Regulatory Genes
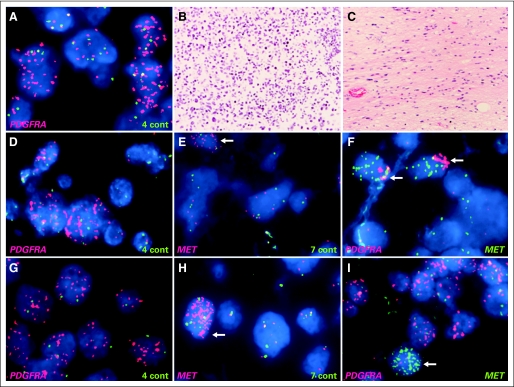
To identify candidate DIPG oncogenes and suppressor genes, we analyzed focal gains and deletions (Data Supplement). Recurrent focal gains of genes encoding RTKs or cell-cycle regulatory genes were found in 24 (56%) of 43 DIPGs (Table 1). Because there were some differences in focal gains between the matched samples collected at diagnosis and autopsy (BSG022 and BSG024) as well as the samples from the contiguous cerebellar involvement and primary DIPGs (BSG003) showing tumor heterogeneity, these paired samples were considered separately. The most common recurrent focal gains encompassed PDGFRA, occurring in 13 (30%) of 43 DIPGs. Recurrent focal gains of the RTKs MET, IGF1R, ERBB4, and EGFR were also found. Tumors with focal gains showed overexpression of the respective RTKs (Data Supplement). For PDGFRA and IGF1R, there was also overexpression in a subset of tumors without amplification, consistent with previous reports showing overexpression of these RTKs in cancer.16,17 In contrast, MET amplification was more specifically associated with overexpression (P = .001). Possible autocrine signaling was identified, with concurrent focal gains of IGF1R and its ligand IGF2 in one tumor and concurrent focal gains of MET and its ligand HGF in another tumor. Other focal CNAs affecting the RTK–Ras–phosphoinositide 3-kinase pathway included gains of KRAS, AKT1, AKT3, and PIK3CA and a focal deletion of NF1. Interphase FISH performed to evaluate focal copy number gains for PDGFRA, MET, and EGFR in 21 patient cases and IGF1R in 15 patient cases revealed substantial tumor heterogeneity. For focal gains of PDGFRA, MET, and IGF1R identified by array analysis, approximately 30% were reflected by amplification in double-minute or homogeneously staining region patterns throughout the tumor, 40% were reflected by less than 20% of tumor cells showing amplification, and 30% were not detected by FISH, indicating heterogeneity between the sample used for DNA extraction and the section used for FISH. We also identified additional tumors with limited foci of tumor cells containing amplifications of PDGFRA, MET, or IGF1R that were not detected by SNP array analysis. Histologic assessment of sections adjacent to FISH preparation showed that foci with or without PDGFRA amplification looked similar, consisting predominantly of tumor cells. Therefore, the increased sensitivity to detect amplifications by FISH compared with SNP arrays was the result of genetic heterogeneity within the tumor rather than normal tissue infiltration. Focal PDGFRA amplification was found in both solid groups of tumor cells and infiltrating tumor cells (Fig 2A). Some tumors contained amplification of more than one RTK. FISH showed examples of coamplification of PDGFRA and MET within the same tumor cells as well as an example of a heterogeneous tumor containing independent amplification of PDGFRA in one focal region and MET in a different subclone of tumor cells (Figs 2B, 2C). We did not detect any EGFR amplifications by FISH, including one patient case in which focal gain was identified by array analysis.
Table 1.
Recurrent Focal Gains of RTKs and Cell-Cycle Regulatory Genes
| Tumor No. | RB Signaling |
RTK-Ras-PI3K-Akt Signaling Network |
PI3K Class 2PIKC2G | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDK4 | CDK6 | CCND1 | CCND2 | CCND3 | EGFR | ERBB4 | PDGFRA | MET | HGF | IGF1R | IGF2 | KRAS | NF1 | AKT1 | AKT3 | PI3KCA | ||
| BSG001T | Gain | Gain | Gain | Gain | ||||||||||||||
| BSG003T(BS) | Gain | Gain | Gain | Gain | Gain | Gain | ||||||||||||
| BSG003T(CB) | Gain | Gain | ||||||||||||||||
| BSG009T | Gain | Gain* | Gain* | |||||||||||||||
| BSG010T | Gain | |||||||||||||||||
| BSG019T | Gain | Gain | Gain | |||||||||||||||
| BSG020T | Gain | Gain | ||||||||||||||||
| BSG022TD | Gain | |||||||||||||||||
| BSG023T | Gain* | Gain | Gain* | |||||||||||||||
| BSG024T | Gain | Gain | ||||||||||||||||
| BSG024TD | Gain | Gain | ||||||||||||||||
| BSG025T | Gain* | |||||||||||||||||
| BSG027TD | Gain | |||||||||||||||||
| BSG035T | Gain | Gain | Gain | Gain | ||||||||||||||
| BSG037T | Gain | Gain | Gain | Gain* | Gain | |||||||||||||
| BSG039T | Gain | Gain | Gain | Gain | ||||||||||||||
| BSG040T | Gain | |||||||||||||||||
| BSG044T | Gain | Gain | Gain | Gain | ||||||||||||||
| BSG045T | Gain* | |||||||||||||||||
| BSG046T | Deletion | |||||||||||||||||
| BSG047T | Gain* | Gain* | ||||||||||||||||
| BSG509T | Gain | Gain* | Gain* | |||||||||||||||
| BSG529T | Gain* | |||||||||||||||||
| BSG902TD | Gain | Gain | Gain | Gain | Gain | Gain | ||||||||||||
| Frequency, % | 7.0 | 11.6 | 4.7 | 7.0 | 4.7 | 4.7 | 7.0 | 30.2 | 25.6 | 4.7 | 18.6 | 2.3 | 2.3 | 2.3 | 2.3 | 4.7 | 2.3 | 4.7 |
Abbreviations: FISH, fluorescent in situ hybridization; PI3K, phosphoinositide 3-kinase; RB, retinoblastoma protein; RTK, receptor tyrosine kinase.
Gain identified by FISH.
Fig 2.
Heterogeneity of focal amplification in diffuse intrinsic pontine glioma. (A) Fluorescent in situ hybridization showed high-level amplification of PDGFRA in focal area of tumor (PDGFRA, red; control chromosome 4, green). Focus of tumor with PDGFRA amplification and remainder of tumor lacking high-level PDGFRA amplification (data not shown) demonstrated similar histopathologic features; both consisted of densely packed tumor cells with minimal normal tissue. PDGFRA amplification was found in both solid groups of tumor cells (B: hematoxylin and eosin [HE] staining) and scattered infiltrating tumor cells (C: HE). Coamplification of PDGFRA and MET in BSG009T (D to F) and BSG037T (G to I). Cells with amplified PDGFRA (D, G: PDGFRA, red; control, green) and MET (E, H: MET, red; control, green) showed coamplification (F, I: PDGFRA, red; MET, green) of (F) both genes in same tumor cells (arrows) or (I) independent amplification in different tumor subclones (arrows).
Focal gains were also found in RB pathway regulatory components including genes encoding cyclin D1, D2, and D3 and CDK4 and CDK6 (Table 1; Data Supplement). Concurrent focal CNAs involving both the RTK–phosphoinositide 3-kinase and the RB pathways were found in nine (21%) of 43 DIPGs. Other recurrent focal alterations included focal gains in MYCN, the Notch signaling component FZD7, the stem-cell marker SOX2, TERT, MAPK-related genes including MAPK7 and MAP2K3, and WNT regulator PLAGL2 and focal deletions of LRP1B and the SHH pathway regulator PTCH1 (Data Supplement). Seven of eight brainstem LGGs showed a focal gain of 7q34 from KIAA1549 to BRAF, similar to recurrent gains reported in nonbrainstem pediatric LGG.18–21 None of the 43 DIPGs showed this characteristic focal CNA associated with LGG (Data Supplement).
Similarities and Differences in Gene Expression Signatures Between DIPG and Nonbrainstem Pediatric Glioblastoma
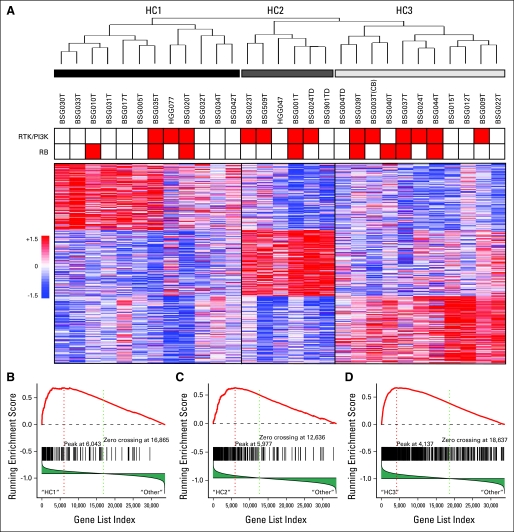
RNA quality from 27 DIPG samples was adequate to evaluate gene expression profiles. We also included expression profiles from two DIPG samples from untreated patients, one of which was obtained at autopsy, from our previous study (HGG047 and HGG077).6 Similar to our previous analysis of nonbrainstem pediatric HGG, unsupervised hierarchic clustering of DIPG showed three major expression subclasses (Fig 3A). Using gene set enrichment analysis, we found that these three subgroups were significantly similar to mesenchymal, proliferative, and proneural expression subgroups previously identified in adult and pediatric HGGs6,22 (Fig 3B; Data Supplement).
Fig 3.
Unsupervised hierarchical clustering (UHC) of diffuse intrinsic pontine glioma (DIPG) showed subgroups similar to those previously identified in adult and pediatric high-grade gliomas. (A) Dendrogram of UHC using top 1,000 most variable probe sets selected using median absolute deviation scores, and heat map featuring top 150 signature probe sets of each subgroup. Three main subgroups were identified. Tumors with focal gains of components in receptor tyrosine kinase (RTK)/phosphoinositide 3-kinase (PI3K) or retinoblastoma protein (RB) pathway are indicated in red at top of heat map. (B, C, D) Gene set enrichment analysis to evaluate coordinate expression in DIPG of gene sets defining subclasses of adult and pediatric high-grade gliomas6,22 showed that DIPG subgroups identified by UHC are highly similar to subgroups previously identified. Plots of running enrichment scores showed highly significant enrichment of mesenchymal markers22 in HC1, proliferative markers22 in HC2, and pediatric proneural markers6 in HC3; 33,928 genes were analyzed. (B) 132 genes in gene set; P = 0, false discovery rate (FDR) = .00162. (C) 176 genes in gene set; P = .0457, FDR = .216. (D) 616 genes in gene set; P = 0, FDR = .0445.
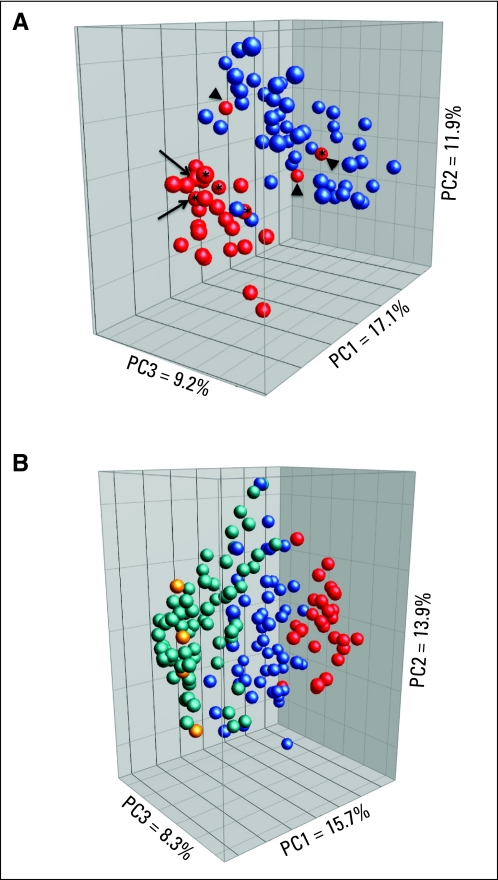
Using principal component analysis (PCA) to evaluate the relationship between gene expression signatures, a majority of DIPGs clustered separately from nonbrainstem pediatric HGGs (Appendix Fig A1, online only). This was likely not an artifact of RNA degradation associated with autopsy collection, or secondary to changes acquired after anticancer treatment, because the three DIPGs that clustered with the nonbrainstem HGGs were obtained at autopsy, and four pretreatment samples, three of which were surgical samples, clustered with the other DIPGs (Appendix Fig A1, online only). Furthermore, RNA was available from a matched pair of samples obtained at diagnosis and autopsy. Both samples clustered close together within the DIPG group, indicating that expression signatures for this sample were similar at diagnosis and autopsy. Moreover, LOH was easily detectable throughout the sample cohort and in the majority of cases was associated with copy-number loss, indicating that the expression signatures should not have been substantially diluted by normal tissue.
The most significant differences in expression between DIPG and nonbrainstem pediatric HGG were in genes that encode transcription factors and genes associated with developmental processes (P < .001). For example, there was significantly higher expression of multiple HOX family members in DIPG, including HOXA3, HOXA2, HOXD3, HOXB2, and HOXD4 (P < .001; Data Supplement). To determine if all tumors arising from the brainstem showed a distinct location-based signature, we also compared the gene expression profiles from six brainstem LGGs and 66 nonbrainstem LGGs (Appendix Fig A1, online only). Brainstem LGGs clustered with nonbrainstem LGGs and were distinct from DIPGs as well as nonbrainstem HGGs. There were no statistically significant associations between molecular features and age at diagnosis, pattern of progression (local v distant), or overall survival (Data Supplement).
DISCUSSION
Prospective collection of DIPG samples at autopsy allowed us to perform genome-wide studies on a sizable cohort of tumors for which samples are extremely limited. This study shows the advantages and disadvantages of using postmortem specimens. CNAs in autopsy samples collected after adjuvant therapy likely present a combination of the initial mutations driving DIPG tumorigenesis as well as secondary changes induced by irradiation and/or chemotherapy. Our findings support the relevance of molecular analysis of tissue obtained at autopsy. PDGFRA amplifications have been identified both in post-treatment autopsy samples in the current study as well as in a previous report12 and in small numbers of pretreatment samples.6,7,12 The overall number of large-scale and focal gains and losses was not significantly different between samples obtained at diagnosis and autopsy and was consistent with the spectrum of CNAs observed in 71 pretreatment pediatric nonbrainstem HGGs,6 indicating that post-treatment samples did not show increased widespread genomic instability. Amplifications of a number of the genes that were identified in DIPG have been previously identified in pediatric or adult HGG.5–8,14,22,23 Thus, it seems likely that analyses of autopsy samples identify CNAs relevant to oncogenesis.
An advantage of the current study was the availability of large tissue samples obtained at autopsy that enabled the identification of small focal regions of tumor containing therapeutically relevant CNAs, which may have been missed had analysis been performed in rather small biopsies. Finally, autopsy samples allow analysis of end-stage disease, which is almost universally fatal. It is essential to identify potential therapeutic targets in this final stage of disease, even if they arise as late events in the evolution of DIPG.
There are shared molecular features between DIPG and nonbrainstem pediatric glioblastoma, including similar frequencies of some large-scale gains and losses. The proneural, proliferative, and mesenchymal gene expression subgroups originally identified in adult glioblastoma can also be readily identified in pediatric glioblastoma, regardless of tumor site.6,22
However, there were significant differences in the frequency of some large-scale genomic imbalances as well as focal deletion of CDKN2A between DIPG and nonbrainstem pediatric glioblastoma. This suggests that the selective pressure driving DIPG development and growth may be different in the brainstem compared with the tumor microenvironment outside the brainstem or that DIPG may be most similar to only a subset of nonbrainstem pediatric glioblastomas. PCA showed that gene expression signatures from DIPG seemed to comprise a distinct subgroup compared with nonbrainstem pediatric HGG. Interestingly, DIPG showed significantly higher expression of specific genes regulating developmental processes and transcription factors, with coordinate upregulation of multiple HOX family genes including HOXA1 and HOXB2, which play important roles in hindbrain development24,25 (Data Supplement). The multiple specific HOX family members that are upregulated in DIPG are distinct from those previously shown to be differentially expressed in therapy-resistant adult glioblastoma or associated with decreased survival in pediatric HGGs.26,27
Previous studies showed that LGGs arising in different regions of the brain did not cluster independently in an unsupervised analysis, but location-dependent signatures could be identified in supervised comparisons.28 Consistent with this result, brainstem and nonbrainstem LGGs did not cluster separately in a PCA comparison (Appendix Fig A1, online only); however, we could identify differential expression of signature genes previously shown to associate with LGGs arising in the posterior fossa28 within the brainstem and cerebellar LGGs (data not shown). The brainstem and nonbrainstem LGGs were also similar on the genomic level, showing minimal CNAs and recurrent gains of 7q34, including BRAF.
The identification of focal amplifications of RTKs and/or cell-cycle regulatory genes in approximately half of all DIPGs has potential therapeutic relevance. Several recent clinical trials have employed small-molecule inhibitors in the treatment of this tumor; however, the choice of targeted agents was based primarily on experience in the treatment of adult glioblastoma, not on the unique genetic characteristics of DIPG.29–31 On the basis of our genomic studies, it may be useful to integrate broad, as well as selective, inhibitors of RTKs32–35 in addition to agents that block cell-cycle progression through the G1 phase, such as selective inhibitors of CDK4 and CDK6, which inhibit intracranial tumor growth of glioblastoma xenografts.36
Although biopsy of DIPG at diagnosis has been routinely performed in some European countries,3 this is not standard practice in North America. A recent study showed that gene-specific assays can be performed in biopsy samples of DIPG tissue,37 and our results may provide a rationale for biopsy at diagnosis to determine if specific targets are amplified and to stratify treatment.
However, our findings also indicate that tumor heterogeneity could create substantial challenges in using molecular diagnosis to guide personalized therapy of DIPG. We found tumor subclones containing different genomic amplifications in a subset of samples obtained at autopsy. It is not clear if this intratumoral variation occurrs at similar frequencies before therapy because of the limited numbers of pretreatment samples analyzed. Tumors constantly evolve, with subclones acquiring different mutations and competing for the greatest selective advantage. Treating a heterogeneous tumor with selective inhibitors may effectively ablate only one subpopulation within the tumor. Depending on the composition of the tumor, small-molecule inhibitors of multiple RTKs and/or cell-cycle regulatory components may have a dramatic effect on overall tumor survival or allow a rapid outgrowth of tumor cells lacking the amplification. It is noteworthy that some tumors lacking amplification of PDGFRA or IGF1R still show strong overexpression of these genes. Thus, analysis of copy-number imbalances may provide insight into critical pathways underlying DIPG growth, which may be altered by varying mechanisms in different tumors.
Supplementary Material
Acknowledgment
We thank the families who donated tissue and the pathologists and clinicians who helped collect tissue to advance diffuse intrinsic pontine glioma research. We are grateful to Jim Dalton, Department of Pathology, St Jude Children's Research Hospital, for advice on interphase fluorescent in situ hybridization methodology.
Appendix
Fig A1.
Principal component analysis (PCA) showed differences in expression signature based on tumor location for diffuse intrinsic pontine glioma (DIPG) but not for brainstem low-grade gliomas (LGGs). (A) PCA generated using 1,000 most variable probes, comparing 29 DIPGs (red), including two patient cases of DIPG obtained before treatment, as previously described (Paugh BS, Qu C, Jones C, et al: J Clin Oncol 28:3061-3068, 2010), and 51 nonbrainstem pediatric high-grade gliomas (HGGs; blue). Arrowheads indicate autopsy samples clustering with tumors outside brainstem. Arrows indicate matched diagnostic (with asterisk) and autopsy sample from same patient. (B) PCA shows DIPG (red), nonbrainstem pediatric HGGs (blue), brainstem LGGs (yellow), and nonbrainstem LGGs (green). (*) Indicates samples obtained before treatment.
Footnotes
See accompanying editorial on page 3956
Supported by Grant No. P01 CA096832 from the National Institutes of Health and by the Sydney Schlobohm Chair of Research from the National Brain Tumor Society, Cure Starts Now Foundation, Smile for Sophie Forever Foundation, Tyler's Treehouse Foundation, Musicians Against Childhood Cancer, Noyes Brain Tumor Foundation, Pediatric Low Grade Astrocytoma Foundation, and American Lebanese Syrian Associated Charities.
Presented in part at the Diffuse Intrinsic Pontine Glioma Think-Tank Conference, February 26, 2010, Toronto, Ontario, Canada; 14th International Symposium on Pediatric Neuro-Oncology, June 20-23, 2010, Vienna, Austria; Society for Neuro-Oncology Annual Meeting, November 18-21, 2010, Montreal, Quebec, Canada; and DIPG.org Pediatric Brain Cancer Symposium, March 18, 2011, Cincinnati, OH.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Barbara S. Paugh, Alberto Broniscer, Justin N. Baker, Amar Gajjar, Suzanne J. Baker
Financial support: Alberto Broniscer, Amar Gajjar, Suzanne J. Baker
Administrative support: Alberto Broniscer, Suzanne J. Baker
Provision of study materials or patients: Alberto Broniscer, James M. Olson, J. Russell Geyer, Susan N. Chi, Nasjla Saba da Silva, Justin N. Baker, Amar Gajjar
Collection and assembly of data: Barbara S. Paugh, Alberto Broniscer, Claudia P. Miller, Junyuan Zhang, Ruth G. Tatevossian, James M. Olson, J. Russell Geyer, Susan N. Chi, Nasjla Saba da Silva, Justin N. Baker, Amar Gajjar, David W. Ellison, Suzanne J. Baker
Data analysis and interpretation: Barbara S. Paugh, Alberto Broniscer, Chunxu Qu, Claudia P. Miller, Junyuan Zhang, Arzu Onar-Thomas, David W. Ellison, Suzanne J. Baker
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Broniscer A, Gajjar A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: Two challenges for the pediatric oncologist. Oncologist. 2004;9:197–206. doi: 10.1634/theoncologist.9-2-197. [DOI] [PubMed] [Google Scholar]
- 2.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: Critical review of clinical trials. Lancet Oncol. 2006;7:241–248. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 3.Roujeau T, Machado G, Garnett MR, et al. Stereotactic biopsy of diffuse pontine lesions inchildren. J Neurosurg. 2007;107:1–4. doi: 10.3171/PED-07/07/001. [DOI] [PubMed] [Google Scholar]
- 4.Broniscer A, Baker JN, Baker SJ, et al. Prospective collection of tissue samples at autopsy in children with diffuse intrinsic pontine glioma. Cancer. 2010;116:4632–4637. doi: 10.1002/cncr.25405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu HQ, Jacob K, Fatet S, et al. Genome-wide profiling using single-nucleotide polymorphism arrays identifies novel chromosomal imbalances in pediatric glioblastomas. Neuro Oncol. 2010;12:153–163. doi: 10.1093/neuonc/nop001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrow J, Adamowicz-Brice M, Cartmill M, et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13:212–222. doi: 10.1093/neuonc/noq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bax DA, Mackay A, Little SE, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Feng X, Koga H, et al. p53 gene mutations in pontine gliomas of juvenile onset. Biochem Biophys Res Commun. 1993;196:851–857. doi: 10.1006/bbrc.1993.2327. [DOI] [PubMed] [Google Scholar]
- 10.Louis DN, Rubio MP, Correa KM, et al. Molecular genetics of pediatric brain stem gliomas: Application of PCR techniques to small and archival brain tumor specimens. J Neuropathol Exp Neurol. 1993;52:507–515. doi: 10.1097/00005072-199309000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Gilbertson RJ, Hill DA, Hernan R, et al. ERBB1 is amplified and overexpressed in high-grade diffusely infiltrative pediatric brain stem glioma. Clin Cancer Res. 2003;9:3620–3624. [PubMed] [Google Scholar]
- 12.Zarghooni M, Bartels U, Lee E, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28:1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 13.Torchia EC, Boyd K, Rehg JE, et al. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–7934. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moschos SJ, Mantzoros CS. The role of the IGF system in cancer: From basic to clinical studies and clinical applications. Oncology. 2002;63:317–332. doi: 10.1159/000066230. [DOI] [PubMed] [Google Scholar]
- 17.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forshew T, Tatevossian RG, Lawson AR, et al. Activation of the ERK/MAPK pathway: A signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 19.Sievert AJ, Jackson EM, Gai X, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19:449–458. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar EE, Lin A, Tihan T, et al. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008;67:878–887. doi: 10.1097/NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- 22.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davenne M, Maconochie MK, Neun R, et al. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. doi: 10.1016/s0896-6273(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 25.Gavalas A, Ruhrberg C, Livet J, et al. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development. 2003;130:5663–5679. doi: 10.1242/dev.00802. [DOI] [PubMed] [Google Scholar]
- 26.Gaspar N, Marshall L, Perryman L, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 28.Sharma MK, Mansur DB, Reifenberger G, et al. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- 29.Geyer JR, Stewart CF, Kocak M, et al. A phase I and biology study of gefitinib and radiation in children with newly diagnosed brain stem gliomas or supratentorial malignant gliomas. Eur J Cancer. 2010;46:3287–3293. doi: 10.1016/j.ejca.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas-Kogan DA, Banerjee A, Kocak M, et al. Phase I trial of tipifarnib in children with newly diagnosed intrinsic diffuse brainstem glioma. Neuro Oncol. 2008;10:341–347. doi: 10.1215/15228517-2008-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollack IF, Jakacki RI, Blaney SM, et al. Phase I trial of imatinib in children with newly diagnosed brainstem and recurrent malignant gliomas: A Pediatric Brain Tumor Consortium report. Neuro Oncol. 2007;9:145–160. doi: 10.1215/15228517-2006-031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faivre S, Demetri G, Sargent W, et al. Molecular basis for sunitinib efficacy and future clinical development. Nat Rev Drug Discov. 2007;6:734–745. doi: 10.1038/nrd2380. [DOI] [PubMed] [Google Scholar]
- 33.Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer. 2010;46:1260–1270. doi: 10.1016/j.ejca.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hixon ML, Paccagnella L, Millham R, et al. Development of inhibitors of the IGF-IR/PI3K/Akt/mTOR pathway. Rev Recent Clin Trials. 2010;5:189–208. doi: 10.2174/157488710792007329. [DOI] [PubMed] [Google Scholar]
- 35.Cascone T, Gridelli C, Ciardiello F. Combined targeted therapies in non-small cell lung cancer: A winner strategy? Curr Opin Oncol. 2007;19:98–102. doi: 10.1097/CCO.0b013e328011beec. [DOI] [PubMed] [Google Scholar]
- 36.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geoerger B, Hargrave D, Thomas F, et al. Innovative Therapies for Children With Cancer pediatric phase I study of erlotinib in brainstem glioma and relapsing/refractory brain tumors. Neuro Oncol. 2011;13:109–118. doi: 10.1093/neuonc/noq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.