Abstract
G-quadruplexes are four-stranded nucleic acid structures whose biological functions remain poorly understood. In the yeast S. cerevisiae, we report that G-quadruplexes form and, if not properly processed, pose a specific challenge to replication. We show that the G-quadruplex-prone CEB1 tandem array is tolerated when inserted near ARS305 replication origin in wild-type cells but is very frequently destabilized upon treatment with the potent Phen-DC3 G-quadruplex ligand, or in the absence of the G-quadruplex-unwinding Pif1 helicase, only when the G-rich strand is the template of leading-strand replication. The orientation-dependent instability is associated with the formation of Rad51–Rad52-dependent X-shaped intermediates during replication detected by two-dimensional (2D) gels, and relies on the presence of intact G-quadruplex motifs in CEB1 and on the activity of ARS305. The asymmetrical behaviour of G-quadruplex prone sequences during replication has implications for their evolutionary dynamics within genomes, including the maintenance of G-rich telomeres.
Keywords: G-quadruplex, genomic instability, Pif1, recombination, replication
Introduction
The maintenance of genome stability is crucial for normal cell growth, as revealed by the numerous chromosomal rearrangements that are a hallmark of cancer cells (Stephens et al, 2011). Mechanistically, genome stability is supported by the evolution of complex repair and surveillance cellular mechanisms monitoring faithful replication and lowering mutagenic DNA damages, induced upon exposure to genotoxic agents or spontaneously generated inside cells (Aguilera and Gomez-Gonzalez, 2008). However, from an evolutionary perspective, genetic variation is a source of diversification within species, generating subtle or abrupt phenotypic variations suited for immediate adaptation and Darwinian selection to act on.
A long-standing and puzzling problem underlying the molecular mechanisms controlling the fluidity of genomes is the abundant presence in all organisms of nucleotide sequences ‘at risks’ of rearrangements (Durkin and Glover, 2007). Whether or not these sequences are detrimental or evolutionary advantageous and how they are propagated and tolerated within genomes are recurrent questions. At one end of the scale length are heterochromatin domains, such as centromeres and telomeres regions, which contain long highly repetitive sequences at risk of gross chromosomal rearrangements. On the other end of the scale length are microsatellite arrays composed of simple repeating DNA sequences of 1–10 nucleotides, which are prone to small length changes associated with human diseases. Another class of puzzling tandem repeats are minisatellite sequences which are arrays of 10–100 nucleotide motifs present in all genomes and also prone to contraction or expansion (Richard et al, 2008; present study). In the human genome, highly polymorphic G-C rich minisatellites (like hCEB1 studied here) are overrepresented in subtelomeric regions (Vergnaud and Denoeud, 2000) but minisatellites are also found in interstitial regions, near or within coding regions (Richard and Dujon, 2006). Importantly, due to their extended motif length, minisatellite arrays have great potential to carry functional information: they can be the substrate of binding proteins (Law et al, 2011) or encode polypeptides (Richard and Dujon, 2006). Thus, the addition or deletion of minisatellite repeat units in individuals of a population is a potent mechanism to create genetic polymorphisms.
The molecular mechanisms, which drive the maintenance and variation of minisatellites, remain poorly understood. It is well established that the repetitiveness of microsatellite sequences facilitates polymerase slippage and allows out-of-register homologous recombination. Repetitiveness is thought to provide a high local concentration of homologous templates favouring maintenance. Sequence composition also matters; within repeat motifs, the embedded sequence-directed determinants allowing the formation of transient non-Watson–Crick DNA structure are additional challenges. The formation of secondary structures, when DNA is single stranded, can trigger fork pausing and instability. Stable DNA secondary structures may act as structural impediments to polymerase progression and their processing yield recombinogenic intermediates (Mirkin and Mirkin, 2007). This is a well-described challenge for the subset of microsatellites that can form hairpin secondary structures and hence facilitate intramolecular slippage of the replicative DNA polymerases (Samadashwily et al, 1997).
Other nucleic acid secondary structures may also challenge replication (Mirkin and Mirkin, 2007). Among these are guanine-rich repeats, which have the capacity to adopt stable G-quadruplex structures. G-quadruplexes are composed of quartets of coplanar guanines (G-quartets) stabilized by non-canonical Hoogsteen hydrogen bonding. Then, the stacking of three G-quartets stabilized by monovalent cations (Na+ or K+) is sufficient to form a G-quadruplex (illustrated in Figure 1A). They can adopt a large variety of conformations depending on the size and sequence of intervening loops and, accordingly, G-quadruplexes will differ in stability and molecular properties (Burge et al, 2006), and their targeting thus require specific cognate ligands (De Cian et al, 2008). Experimental evidence arguing for the in vivo role of G-quadruplexes, and hence their formation have proven difficult and remains limited and heterogeneous (Maizels, 2006). G-quadruplexes regulate telomerase accessibility at the telomeres of the Stylonichia lemnae protist, as directly observed throughout the cell cycle by immunostaining (Paeschke et al, 2008). Also, recent genetic and molecular studies argue that G-quadruplexes cap compromised telomeres (Sfeir et al, 2009; Smith et al, 2011). Beside their involvement in telomere homeostasis, G-quadruplexes also appear to promote genetic instability, as suggested by the presence of G-quadruplexes motifs in the vicinity of chromosomal breakpoints in humans (Nambiar et al, 2010; De and Michor, 2011) and in C. elegans animal defective for dog-1, the homologue of the G-quadruplex-unwinding helicase FANCJ (Kruisselbrink et al, 2008). This proposal has recently been subjected to experimental challenges. Site-directed mutational analysis and G-quadruplexes stabilization by specific ligands uncovered a direct role of G-quadruplexes in regulation of gene conversion of the pilin locus in Neisseria gonorrhoeae (Cahoon and Seifert, 2009) and in the instability of the hCEB1 minisatellite in S. cerevisiae (Ribeyre et al, 2009; Piazza et al, 2010; present study).
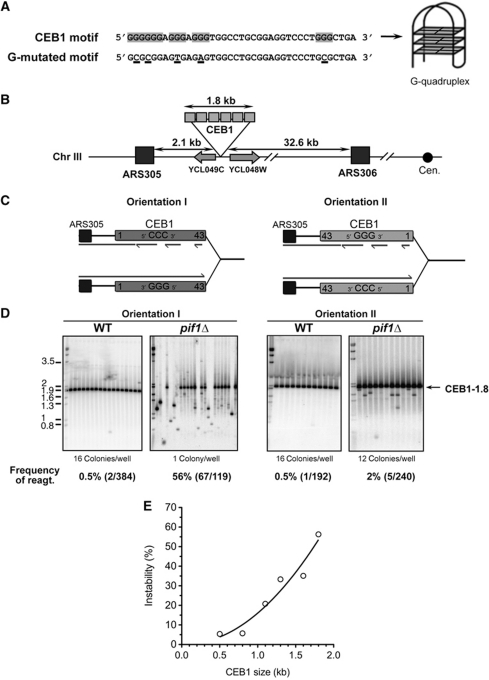
Figure 1.
Orientation-dependent destabilization of CEB1 inserted near ARS305 in pif1Δ cells. (A) Nucleotide sequence of CEB1 motifs shown in the 5′–3′ orientation. CEB1 motif: more common motif in the natural CEB1-1.8 minisatellite (Lopes et al, 2006a). The G-tracts involved in the G-quadruplex formation (Ribeyre et al, 2009) are highlighted. G-mutated CEB1 motif: sequence of the synthetic CEB1 G-mutated motif assembled in the artificial CEB1-Gmut-1.7 tandem array (Ribeyre et al, 2009). The mutated nucleotides are underlined. (B) Map of the chromosome III region in which the CEB1 arrays were inserted (see Supplementary Methods). (C) Mode of replication of CEB1 by forks emanating from ARS305 upon insertion in orientation I (left) or II (right). In orientation I, the G-quadruplex-forming strand is the template of leading-strand replication. In orientation II, the G-quadruplex-forming strand is the template of lagging-strand replication. (D) Behaviour of CEB1-1.8 in orientations I (left) and II (right) in WT (ORT6119-4 and ORT6135-36, respectively) and in pif1Δ (ORT6146-1 and ORT6136-8, respectively) cells visualized by Southern blot analysis. The number of colonies analysed per lane is indicated. The molecular ladder (in kb) allows for the size determination of the DNA fragments at a resolution of ±1 motif (40 bp). Genomic DNAs were digested with ApaI/XhoI (orientation I for WT and pif1Δ cells), ApaI/NcoI (orientation II for WT cells), and ApaI/SacII (orientation II for pif1Δ cells). Membranes were hybridized with radiolabelled CEB1 probe. The total rearrangement frequencies (%), and in parenthesis the absolute number of rearrangements over the total number of colonies analysed, are reported below the blots. Statistical analyses (P-values) of rearrangement frequencies between strains are reported in Table I. (E) Size effect of CEB1 in orientation I on its rearrangement frequency in pif1Δ cells. A non-linear, second order fit has been applied (R2=0.9133). Strain names and numerical data are reported in Supplementary Tables S1 and S2, respectively.
The next critical issue is how G-quadruplexes are processed? In vitro several DNA helicases, such as the mammalian BLM and WRN RecQ orthologues, PIF1, and FANCJ unwind G-quadruplex structures (Sanders, 2010; Wu and Brosh, 2010). In S. cerevisiae, the Sgs1 (RecQ orthologue) and Pif1 helicases which belong to different helicase subfamilies and work in opposite directionality (3′–5′ and 5′–3′, respectively) unwind G-quadruplexes in vitro (Sun et al, 1999; Ribeyre et al, 2009). In vivo, our previous studies (and present data) clearly demonstrate that Pif1 is involved in the processing of the G-quadruplexes formed by CEB1 (Ribeyre et al, 2009; Piazza et al, 2010). Consistently, it has been recently reported that potential G-quadruplex-forming sequences are significantly enriched among the numerous Pif1 and Pif1-helicase dead (pif1-K264A) chromatin binding regions (1584 and 1153 regions, respectively), with an estimate that ∼25% (138/558) of the potential G-quadruplex-forming sequences of the S. cerevisiae genome might be processed by Pif1 (Paeschke et al, 2011). Why, at last, Pif1 binds to so many chromosomal regions and certainly not only at G-quadruplex sites remains to be elucidated.
Another issue is to determine in which circumstance G-quadruplex forms in vivo. The formation of single-stranded DNA being most favourable circumstances, the folding of DNA G-quadruplex is most likely occurring during transcription and replication (Maizels, 2006). Strong evidences that G-quadruplex forms during replication remained to be brought. In this direction, Paeschke et al (2011) recently reported that in pif1-m2 cells (proficient for the Pif1 mitochondrial function) but not in wild-type cells, a small fraction (11%) of the DNA Pol2 occupancy sites overlap regions containing potential G-quadruplex-forming sequences and that replication fork progression is slowed in hydroxyurea grown pif1-m2 cells in a regional but not in G-quadruplex site-specific manner. Here, complementary to the Paeschke et al (2011) study, we took advantage of the clearest demonstration that the minisatellite CEB1 can form G-quadruplex in S. cerevisiae cells (Ribeyre et al, 2009; Piazza et al, 2010) and specifically address whether G-quadruplexes affect replication and, if they do so, have different effects upon formation in the leading or in the lagging-strand template. The temporal program of replication, which initiates from a limited number of well-defined origins along the chromosomes being well established (Raghuraman et al, 2001), our key experimental strategy was to insert CEB1 in both orientations in an intergenic region near the early-firing replication origin ARS305 and monitor its stability. This method was previously developed to determine the leading- and lagging-strand polymerases using a reporter strand-specific mutagenic assay (Nick McElhinny et al, 2008). Importantly, the selected chromosomal context further allows to demonstrate the role of replication by modulating the activities of the replication origins. Thus, the G-quadruplex-driven tandem-repeat instability assay in S. cerevisiae enabled us to demonstrate that G-quadruplexes frequently form on the leading strand in wild-type cells treated with the specific G-quadruplex-ligand Phen-DC3 and in the absence of the Pif1 helicase.
Results
Experimental system and rationale
To determine the role of G-quadruplexes on yeast replication, we made use of the G-rich minisatellite CEB1 which is a sensitive reporter of G-quadruplexes formation and processing by monitoring its change of length during mitotic growth (Ribeyre et al, 2009; Piazza et al, 2010). Increase (expansion) or decrease (contraction) of the total number of CEB1 motifs (Figure 1A) can be visualized by Southern blot analysis of DNA extracted from individual or pooled cell colonies (Lopes et al, 2006a) and allow for the determination of a rearrangement frequency, also referred as ‘instability’.
To examine a potential instability bias upon replication by the leading or the lagging machinery, we inserted the natural CEB1-1.8 allele containing 42 slightly polymorphic 39 nt motifs arranged as direct repeats (for full sequence, see Lopes et al, 2006a) in an asymmetrical position relatively to the dynamic of replication between ARS305 and ARS306 (Supplementary Figure S1; Pavlov et al, 2002; Fachinetti et al, 2010). Specifically, CEB1 was inserted in both orientations (I and II) in an intergenic region 2.1 kb to the right of ARS305 and 32.6 kb away from ARS306, the closest downstream origin on the left arm of chromosome III (Figure 1B; Supplementary Methods). As to determine the DNA polymerases involved in leading- and lagging-strand replication, the rationale of this experimental design is that CEB1 will primarily be replicated by the fork emanating from ARS305, so that the leading- and lagging-strand templates can be unambiguously assigned (Nick McElhinny et al, 2008). In orientation I, the CEB1 G-quadruplex-forming strand will be the leading-strand template and becomes the lagging-strand template in orientation II (Figure 1C). The intergenic insertion of CEB1 was chosen to avoid transcriptional interference and all strains were grown in rich media to avoid eventual selective pressure. Furthermore, to avoid any risk of background heterogeneity, all strains were derived by transformation of the SY2209 haploid strain, previously used to examine replication dynamics (Fachinetti et al, 2010).
Orientation- and size-dependent instability of CEB1 near ARS305 in Pif1-deficient cells
To examine the role of Pif1 in the stability of CEB1 inserted near ARS305, we measured the frequency of CEB1-1.8 size variants in WT and pif1Δ cells carrying CEB1 in orientation I or II. In WT cells, CEB1-1.8 is rather stable in both orientations, showing 0.5% (2/384) and 0.5% (1/192) of rearrangements, respectively (Figure 1D; Table I). In pif1Δ cells, the orientation greatly matters. In orientation II, the frequency of CEB1-1.8 rearrangements (2.0%) is not significantly different than in WT (0.5%) (P-value pif1Δ orientation II versus WT orientation II=0.23). In sharp contrast, it is extremely unstable in orientation I. The instability reaches 56.3% (67/119) (P-value pif1Δ orientation I versus WT orientation I=1.24e−47 and P-value pif1Δ orientation I versus pif1Δ orientation II=1.69e−33) and yields a large diversity of size variants (Figure 1D; Table I), which ascertains the independent origin of these events. Importantly, when instability reaches such high levels, our measures are likely underestimates. To eliminate potential clonal events, similar size variants were counted only once (see Materials and methods), although sequencing showed that they could be different (Supplementary Figure S5). Thus, in pif1Δ cells, CEB1 in orientation I is at least 28-fold more unstable than in orientation II and than in either orientation in WT cells.
Table 1. Rearrangement frequencies of CEB1 arrays placed near ARS305 in both orientations in WT and mutant strains.
| Minisatellite | Genotype | Orientation I | Orientation II | P-value orientation I versus II |
|---|---|---|---|---|
| CEB1-1.8 | WT | 2/384 (0.5%) | 1/192 (0.5%) | NS |
| pif1Δ | 67/119 (56.3%)* | 5/240 (2%) | 1.69e−33 | |
| pif1-K264A | 27/76 (35.5%)*† | 6/192 (3.1%) | 9.77e−12 | |
| pif1Δ rad51Δ | 6/191 (3.1%)† | 0/192 | 0.03 | |
| pif1Δ rad52Δ | 1/192 (0.5%)† | 1/192 (0.5%) | NS | |
| pif1Δ rad54Δ | 17/167 (10.2%)*† | 10/191 (5.2%)* | NS | |
| sgs1Δ | 2/192 (1%)† | 2/192 (1%) | NS | |
| rrm3Δ | 0/192† | 0/192 | NS | |
| rad27Δ | 14/81 (17.3%)*† | 9/83 (10.8%)*† | NS | |
| CEB1-Gmut-1.7 | pif1Δ | 0/384 | ND | NA |
| NA, not applicable; ND, stability not determined; NS, P-value orientation I versus orientation II not significant. | ||||
| *P-value single or double mutants versus WT <0.05. | ||||
| †P-value pif1-K264A, single or double mutants versus pif1Δ <0.05. | ||||
Formally, the lack of CEB1 size variants in orientation II could be explained by the induction of massive cell lethality. To test this possibility, we examined cell viability in the WT and pif1Δ cells carrying CEB1 in either orientation. We measured cell viability in a single generation assay by separating pairs of mother and daughter cells under the microscope and monitoring colony growth (Supplementary Figure S2A). In both wild-type and pif1Δ cells, the insertion of CEB1 near ARS305 in either orientation has no effect on cell viability: over 90% of budded mother cells (a landmark of entry into S-phase) give rise to viable colonies (Supplementary Figure S2B). Therefore, induction of cell death in orientation II is not the source of the orientation effect of CEB1.
Next, we investigated the relationship between array size and stability. As illustrated in Figure 1E, the overall frequencies vary by at least one order of magnitude, from 5.4% for the shortest allele (11 repeats) to 56.3% for the longest alleles (CEB1-1.8, 43 repeats) (Supplementary Table S2). Thus, the rearrangement frequency increases steadily with the number of CEB1 motifs (P=2.78e−3, Spearman correlation test), apparently without threshold and in a more-than-linear relationship despite the underestimate of the rearrangement frequencies for the longer alleles (Figure 1E).
In sum, the above data demonstrate that Pif1 is required to stabilize CEB1 near ARS305 in an orientation-dependent manner with rearrangement frequencies tightly correlated to the number of repeats. With respect to the mode of replication, the extreme instability corresponds to the orientation in which the CEB1 G-rich strand is the template of leading-strand synthesis.
The helicase activity of Pif1 is required to stabilize CEB1
To determine if the helicase activity of Pif1 was required to stabilize CEB1-1.8, we examined strains carrying the pif1-K264A mutation that inactivates the Pif1 ATPase/helicase activity (Schulz and Zakian, 1994). We observed that CEB1 is very unstable in orientation I (27/76, 35.5%) but not in orientation II (6/192, 3.1%) (P=9.77e−12; Table I). Thus, the helicase activity of Pif1 has a role in the stabilization of CEB1 in the G-leading orientation. Compared with pif1Δ cells, the frequency of size variant in orientation I was ∼2-fold lower in pif1-K264A cells (P=5.3e−3). This may reflect the fact that the pif1-K264A mutant retains wild-type level of DNA binding in vitro (Boulé et al, 2005), which could, to a certain extent, prevent G-quadruplex formation in CEB1 (Ribeyre et al, 2009). Thus, the absence of Pif1 or loss of its helicase activity results in a CEB1 instability induction that is 28- and 11-fold, respectively, higher in orientation I than in orientation II.
Orientation-independent destabilization of CEB1 near ARS305 in rad27Δ/FEN1 cells
In a previous study, we found that the absence of Rad27/FEN1, a 5′–3′ Flap endonuclease involved in the processing of Okazaki fragments formed during lagging-strand synthesis (Kunkel and Burgers, 2008) destabilizes all types of tandem repeats (microsatellite and minisatellite), including CEB1-1.8 inserted near ARG4 (Lopes et al, 2002, 2006a; Richard et al, 2008). Compared with WT cells, the inactivation of Rad27 increases the frequency of CEB1 rearrangements in both orientations (Table I), and the difference between the orientations I (18%) and II (11%) is not significant (P=0.27). These results demonstrate that the biased behaviour of CEB1 near ARS305 is specific to the inactivation of Pif1 and, in the occurrence, not a general feature shared by another mutation perturbing replication.
Other helicases, as well as Rev1 polymerase, are not required to ensure CEB1 stability
To examine whether this orientation-dependent instability of CEB1 near ARS305 is specific to the absence of Pif1, and not mutations in other helicases, we assayed the stability of CEB1-1.8 in both orientations in strains deleted for the RRM3 or SGS1 helicase genes. Sgs1 was reported to unwind G-quadruplexes in vitro (Sun et al, 1999), and Rrm3 is, like Pif1, a member of the SF1 helicase superfamily, reported to travel with the replication fork and to promote its progression at pause sites (Ivessa et al, 2003; Azvolinsky et al, 2006). In the sgs1Δ or rrm3Δ cells, CEB1 remained stable in either orientation (Table I). Hence, the destabilization of CEB1 in orientation I is specific to the inactivation of the Pif1 helicase. Additionally, it shows that neither Pif1, nor Sgs1, nor Rrm3 seems required to ensure CEB1 stability when the G-strand is template for the lagging-strand synthesis (see Discussion). A recent study suggested that in DT40 chicken cells, REV1, a polymerase of the Y-family, promotes the replication of G-quadruplex-forming sequences during leading-strand synthesis (Sarkies et al, 2010). To address the eventual role of the orthologous REV1 gene in yeast, we constructed and assayed the instability of CEB1 near ARS305 in rev1Δ cells in the presence or absence of Phen-DC3 treatment. rev1Δ cells behaved like wild type (Table II). Thus, further studies will be required to generalize the role of Rev1 on potentially forming G-quadruplex sequences.
Table 2. Rearrangement frequencies of CEB1-1.8 in WT and mutant strains treated or not with Phen-DC3.
| CEB1-1.8 | Orientation I |
Orientation II |
||||
|---|---|---|---|---|---|---|
| Genotypetreatment | Control | Phen-DC3 | P-value (control versus Phen-DC3) | Control | Phen-DC3 | P-value (control versus Phen-DC3) |
| WT | 1/192 (0.5%) | 111/992 (11.2%) | 4.5e−8 | 1/192 (0.5%) | 57/1376 (4.1%)** | 7.1e−3 |
| rad51Δ | 2/176 (1.1%) | 14/576 (2.9%)* | NS | 2/384 (0.5%) | 16/564 (2.8%) | 0.013 |
| rad52Δ | 1/192 (0.5%) | 1/154 (0.6%)* | NS | 0/176 | 0/192* | NS |
| rad54Δ | 0/192 | 1/384 (0.3%)* | NS | 5/192 (2.6%) | 8/368 (2.2%) | NS |
| pif1Δ | ND | ND | NA | 3/191 (1.6%) | 47/480 (9.8%)* | 6.7e−5 |
| rev1Δ | 0/192 | 18/192 (9.4%) | 5.0e−6 | 0/192 | 4/192 (2.1%) | NS |
| NA, not applicable; ND, not determined; NS, not significant. *P-value versus WT <0.05. **P-value versus orientation I <0.05. | ||||||
G-quadruplex-dependent instability of CEB1 near ARS305 in WT and pif1Δ cells
Does the orientation-dependent instability of CEB1 near ARS305 depend on its capacity to form G-quadruplexes in vivo? To address this issue and to demonstrate the in vivo formation of G-quadruplexes, we conducted two independent experimental approaches.
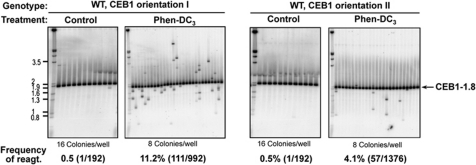
First, we asked whether or not CEB1 near ARS305 could be destabilized in WT cells upon treatment with a G-quadruplex ligand, in an orientation-dependent manner. We previously showed that the potent G-quadruplex-stabilizing compound Phen-DC3 (Monchaud et al, 2008) efficiently inhibits G-quadruplex unwinding by Pif1 in vitro, and specifically triggers instability of G-quadruplex-prone CEB1 alleles inserted near the ARG4 locus (Piazza et al, 2010). Thus to establish that G-quadruplexes impede the replication machinery, we examined the stability of CEB1-1.8 inserted in either orientation near ARS305 in WT cells grown for eight generations with or without Phen-DC3 at 10 μM. Remarkably, compared with the control samples (with 0.02% DMSO, as in the treated sample), which showed a single rearrangement out of 192 colonies (0.5%), Phen-DC3 treatment triggered instability of CEB1 in both orientations but to different extents: in orientation I, we observed 111/992 rearrangements (11.2%, P-value versus control=4.5e−8) and in orientation II, 57/1376 rearrangements (4.1%, P-value versus control=7.1e−3) (Figure 2; Table II). The difference between both orientations is highly significant (P-value=6.9e−11). These data strongly suggest that the instability of CEB1 near ARS305 depends on the in vivo formation of G-quadruplexes promoted or stabilized by the Phen-DC3 compound.
Figure 2.
Orientation-dependent induction of CEB1 instability upon treatment of WT cells with the G-quadruplex-ligand Phen-DC3. Southern blot analyses of CEB1-1.8 behaviour in ORT6119-4 (orientation I) and ORT6135-36 (orientation II) cells treated or not with 10 μM of Phen-DC3. Genomic DNAs were digested with ApaI/XhoI (orientation I) and ApaI/NcoI (orientation II). Other legends as in Figure 1D.
As a second, complementary experimental approach, we examined the behaviour of a version of CEB1 mutated for its G-quadruplex-forming sequences (sequence of the G-mutated motif is shown in Figure 1A; Ribeyre et al, 2009). For this purpose, we inserted our synthetically built G-mutated CEB1 allele of 42 motifs, named CEB1-Gmut-1.7 (for the full sequence of the minisatellite, see Ribeyre et al, 2009), near ARS305, at the same location as the natural CEB1-1.8 allele. Mutation of these motifs prevents G-quadruplex formation in vitro and stabilizes CEB1 inserted at the ARG4 locus (Ribeyre et al, 2009; Piazza et al, 2010). The CEB1-Gmut-1.7 array, inserted near ARS305 in orientation I in a pif1Δ mutant is stable (0/384), indicating that the high instability (56.3%) observed with CEB1-1.8 in this context relies on its G-quadruplex-forming potential (Table I).
Consistently and independently, the above data obtained from pif1Δ and Phen-DC3-treated WT cells strongly suggest that G-quadruplexes formed within CEB1 account for its differential instability behaviour in orientations I and II near the ARS305 replication origin.
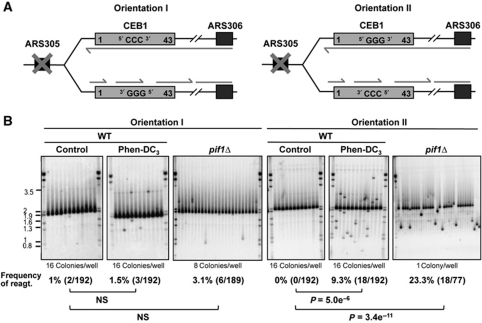
The orientation-dependent behaviour of CEB1 results from the activity of ARS305 and can be reversed
To rule out the possibility that the orientation-dependent behaviour of CEB1 could depend on other cis-acting elements than the replication directionality, we deleted the ARS305 region (Figure 3A; Supplementary Methods). In this context, it is expected that CEB1 will be mostly replicated from the closest but nevertheless distant ARS306 origin, and little from the distant and poorly active ARS300-304 origins (see Supplementary Figure S1; Poloumienko et al, 2001; Pavlov et al, 2002). It implies that in ARS305-deleted cells, the G-strand of CEB1-1.8 in orientation I will be the template of lagging-strand replication, and vice versa orientation II will be the template of leading replication (Figure 3A). If only the mode of replication across CEB1 matters, the prediction is that the orientation bias will be switched. Indeed, the deletion of ARS305 profoundly modifies the behaviour of CEB1: it is still destabilized, but in the opposite orientation. In the absence of Pif1, we observed 6/189 (3.1%) rearrangements in the CEB1 orientation I and 18/77 (23.3%) rearrangements in orientation II (Figure 3B). The difference in orientation is statistically significant (P=3.17e−6). To further establish this remarkable change of behaviour and the dependency on G-quadruplexes formation, we also examined the behaviour of CEB1-1.8 in WT cells treated or not by Phen-DC3. Consistently, the Phen-DC3 treatment stimulates the instability of the CEB1-1.8 array placed in orientation II (18/192, 9.3%, P-value versus control=5e−6) but not significantly in orientation I (3/192, 1.5%) (Figure 3B). Again, the difference in orientation is statistically significant (P=1.1e−3).
Figure 3.
The deletion of ARS305 reverses the orientation-dependent behaviour of CEB1. (A) Mode of CEB1 replication upon deletion of ARS305. CEB1 is replicated by forks emanating from ARS306. In orientation I, the G-quadruplex-forming strand is the template of lagging-strand replication whereas in orientation II, it is the template of leading-strand replication. This is the opposite of Figure 1C where CEB1 was replicated by the nearby ARS305. (B) Southern blot analysis of CEB1-1.8 behaviour placed in orientation I or II in WT cells (ORT6165-2 and ORT6166-52, respectively) treated or not with 10 μM of Phen-DC3, and in pif1Δ cells (ORT6169-2 and ORT6170-8, respectively). Genomic DNA was digested with ApaI/XhoI (orientation I) and ApaI/NcoI (orientation II). Other legends as in Figure 1D.
Together, the above data strongly support the interpretation that in the absence of Pif1, or upon stabilization of G-quadruplexes by Phen-DC3 in wild-type cells, G-quadruplexes form within CEB1 when the G-rich strand is the template for leading replication and hence destabilize CEB1.
G-quadruplex-dependent accumulation of X-shaped replication intermediates
To determine whether the presence of CEB1 near ARS305 and its orientation-dependent behaviour perturb the progression of replication, we arrested cells in G1 with α-factor, released them to progress synchronously into S-phase, and then examined DNA content doubling by FACS analysis as well as replication intermediates by two-dimensional (2D)-gel electrophoresis (see Materials and methods). Origin firing generates ‘bubble arc’ containing forks proceeding bidirectionally and ‘large Y’ molecules resulting from forks migrating asymmetrically outside of the origin fragment. Other intermediates such as X-shaped and ‘cone’ signals detected in connections with various processes such as DNA recombination (Ivessa et al, 2003; Bzymek et al, 2010) can also be visualized (Figure 4A).
Figure 4.
Recombination-intermediate formation during CEB1 replication depends on the CEB1 orientation and on the formation of G-quadruplexes. (A) Time-course analysis by 2D-gel electrophoresis of replication intermediates formed in CEB1 arrays in orientations I and II during S-phase in synchronized WT and pif1Δ cells. The WT ORT6119-4 (orientation I), ORT6135-36 (orientation II), and the pif1Δ ORT6136-8 (orientation II) cells carry the CEB1-1.8 array. Due to the very high instability of CEB1-1.8 orientation I in pif1Δ cells which yields clonal rearrangements in the cell population (see Table I), the pif1Δ strain ORT6123-1-5 used here carries the CEB1-1.6 array in orientation I. Cells were arrested in G1 by α-factor treatment, released into a synchronous S-phase, and aliquots examined by FACS to follow progression into the S-phase (see Supplementary Figure S4A). Genomic DNA was prepared from cells collected at the indicated times (min). Methods for 2D-gel analyses are described in Materials and methods. Restriction strategies and expected DNA fragment sizes for CEB1 in orientations I (HaeII/SacII) and II (HaeII/ApaI) are shown in the top panel. Membranes were hybridized with a radiolabelled CEB1-0.6 probe (Lopes et al, 2006a). A schematic representation of replication intermediates is presented in the right panel. Arrows on gel images outline abnormal X-shaped molecules (spike). The low intensity second Y arc in the pif1Δ strain with CEB1 orientation II corresponds to partially digested DNA. (B) Time-course analysis by 2D-gel electrophoresis of DNA intermediates formed in CEB1-Gmut-1.7 in orientation I during S-phase in a pif1Δ strain (ORT6157-1). Genomic DNA was digested with HaeII/ApaI. DNA fragments are revealed using a radiolabelled CEB1-Gmut probe (Ribeyre et al, 2009). Other legends as in (A). FACS analyses reported in Supplementary Figure S4B. (C) Relative time-course quantification of the spike signal normalized to the Y arc signal observed in experiments in (A) and (B). The spike signal is abundant in pif1Δ cells carrying the natural G-quadruplex-forming sequence as template of leading-strand replication (orientation I). (D) Time-course analysis by 2D-gel electrophoresis of DNA intermediates formed in CEB1 arrays in orientations I (left panel) and II (right panel) in synchronized WT cells, treated or not with 10 μM Phen-DC3. In the Phen-DC3-treated samples, the shift from the 1C to 2C peak is slightly delayed compared with the control sample incubated in the same final concentration of DMSO (0.02%) (FACS, see Supplementary Figure S4C). Thus, control and treated samples were examined at different times in order to compare cells with similar progression into S-phase. Other legends as in (A). (E) Time-course quantifications of spike signal observed in Phen-DC3-treated samples in the experiments of (D). (F) Time-course analysis by 2D-gel electrophoresis of replication intermediates in synchronized pif1Δ (ORT6123-1-5) and pif1Δ rad51Δ (ORT6139-5) strains carrying CEB1-1.6 in orientation I, and pif1Δ rad52Δ (ORT6153-2) cells carrying CEB1-1.8 in orientation I. Other legends as in (A). The spike signal observed in pif1Δ cells (A) is confirmed in this duplicate experiment and is no more observed upon inactivation of Rad51 and Rad52. FACS analyses reported in Supplementary Figure S4D. (G) Relative time-course quantification of the spike signal normalized to the Y arc signal observed in experiments of (F). The abundant spike signal in pif1Δ cells carrying the natural G-quadruplex-forming sequence as template of leading-strand replication (orientation I) depends on the presence of the Rad51 and Rad52 proteins.
In WT cells synchronously released from the G1 block, DNA doubling occurs within ∼45′ as monitored by FACS (Supplementary Figure S3), and replicative Y-shaped molecules generated by forks emanating asymmetrically from ARS305 accumulate within 10–30′ without detectable difference between the two strains carrying CEB1 in either orientation (Figure 4A). CEB1 is asymmetrically placed in the HaeII–SacII or HaeII–ApaI restriction digests products (see Figure 4A), implying that the typical increase of signal within the descending part of the Y arc (corresponding to long and mostly replicated Y molecules) may mask fork arrest within CEB1. To specifically address this possibility, we used a different restriction digestion (ApaI–SpeI) to probe CEB1 replication intermediates in a different position along the Y arc (Supplementary Figure S4). Signal intensity along the ascending Y arc was regular and remained constant after having fully replicated CEB1 in WT cells, indicating that the CEB1 array per se does not impair replication fork progression (Supplementary Figure S4).
In pif1Δ mutants, Y replication intermediates appear with appropriate kinetics across CEB1 placed in either orientation (Figure 4A). Strikingly, a signal of X-shaped intermediates (see arrow in Figure 4A, right panel) accumulates in the strain carrying CEB1 in orientation I. Along the time course, the proportion of this X-spike (expressed as the X/Y signal ratio) increases between 20′ and 45′, to reach 0.8 at 45′ (Figure 4C). In sharp contrast, the X-spike is barely observed in the strain carrying CEB1 in orientation II, and never detected in WT strains, even upon longer gel exposure. Quantitatively, the X/Y signal ratio in pif1Δ cells is at least 10-fold higher in orientation I than in orientation II. These data demonstrate that pif1Δ cells accumulate aberrant intermediates migrating as X-shaped molecules during replication depending on the CEB1 orientation. Formation of X-structures correlates with very high frequency of CEB1 rearrangements. Interestingly, the formation of these X-shaped intermediates in pif1Δ cells requires intact G-triplets within CEB1, since such structures are not observed upon replication of the CEB1-Gmut-1.7 minisatellite in orientation I (Figure 4B and C; Supplementary Figure S3B).
Using a different restriction strategy (ApaI–SpeI), we probed CEB1 replication intermediates presenting forks at CEB1 in the ascending part of the Y arc. The signal intensity was stronger in the ascending than in the descending part of the Y arc in pif1Δ cells (Supplementary Figure S4). This was not the case with the CEB1-Gmut-1.7 allele. These observations suggest that replication slows down at CEB1 in the absence of Pif1 depends on the presence of intact G-triplets (Supplementary Figure S4).
Finally, we also examined the replication intermediates in WT cells treated with Phen-DC3. We observed that G1-arrested cells released into the Phen-DC3-containing media exhibited a significant delay in S-phase onset, compared with the control cells (equivalent DMSO concentration of 0.02%) that seemed unaffected, revealed both by the FACS profiles (Supplementary Figure S3C) and by the appearance of the replication intermediates on 2D gel (Figure 4D). This effect of Phen-DC3 on the S-phase entry is independent of the presence and the orientation of CEB1. Remarkably, like in the absence of Pif1, X-shaped structures accumulate in the Phen-DC3-treated WT cells during the replication of CEB1 in orientation I, but not in the opposite orientation (Figure 4E; Supplementary Figure S3D). Thus, the appearance of X-shaped intermediates dependent on the CEB1 orientation in circumstances that impair G-quadruplexes processing, strongly suggests that G-quadruplexes form during leading-strand replication and, if not resolved, trigger CEB1 rearrangements.
The formation of X-shaped molecules and CEB1 rearrangements depend on the homologous recombination pathway
On 2D gels, molecules migrating in the X-spike have been related to recombination intermediates such as double Holliday junctions (dHJ) (Bzymek et al, 2010) or convergent forks at sites of replication termination (Lopes et al, 2003). Among these two possibilities, the formation of recombination intermediates is the only one that require the activity of the Rad51 and Rad52 proteins, which play a central role in the repair of DSBs and the restart of stalled replication fork by homologous recombination (Pâques and Haber, 1999). Therefore, to distinguish between these possibilities, we deleted the RAD51 or RAD52 gene in pif1Δ cells carrying CEB1 in orientation I and then examined the replication intermediates. In contrast to pif1Δ cells, reexamined in parallel, X-shaped structures were not detected in the pif1Δ rad51Δ and pif1Δ rad52Δ double-mutant strains (Figure 4F and G). We, thus, concluded that X-shaped intermediates arising in pif1Δ cells during the replication of CEB1 in orientation I are recombination intermediates.
The formation of the CEB1 rearrangements also depends on the homologous recombination pathway
The above results raise the prediction that the occurrence of CEB1-1.8 rearrangements near ARS305 is also dependent on the activity of the homologous recombination pathway. To test this prediction, we examined the instability of CEB1 in strains deleted for the RAD51, RAD52, or RAD54 genes (Supplementary Table S1). The Rad54 protein is also involved in homologous recombination at synapsis and post-synapsis stages (Heyer et al, 2006). The behaviour of CEB1 in these various strain backgrounds is reported in Table I. In the untreated rad51Δ, rad52Δ, and rad54Δ strains, we observed no or few rearrangements, and the treatment with Phen-DC3 yielded no or few instability (Table II). Accordingly, the CEB1 instability in orientation I was abolished in the pif1Δ rad52Δ strain, and strongly decreased in the pif1Δ rad51Δ and pif1Δ rad54Δ double mutants compared with the pif1Δ strain (P-values=7.8e−34, 7.4e−28, and 1.9e−17, respectively; Table I). Thus, the remaining Rad51- and Rad54-independent rearrangements are likely produced by break-induced replication or single-strand annealing mechanisms (Pâques and Haber, 1999; Malkova et al, 2005). In orientation II, in all cases the frequencies of rearrangements were low (Table I).
Again, we examined cell viability. The absence of Rad51 has no detectable effect with CEB1 in both orientations (Supplementary Figure S2B). However, the double-mutant pif1Δ rad51Δ bearing CEB1 in orientation I exhibited a markedly reduced viability (67%) compared with pif1Δ and rad51Δ single mutants (P=7.5e−6 and 5.9e−3, respectively), whereas cells bearing CEB1 in orientation II did not (Supplementary Figure S2B). This result suggests that lesions produced in CEB1 in orientation I in the absence of Pif1 require the homologous recombination pathway to be repaired, otherwise leading to cell death or senescence.
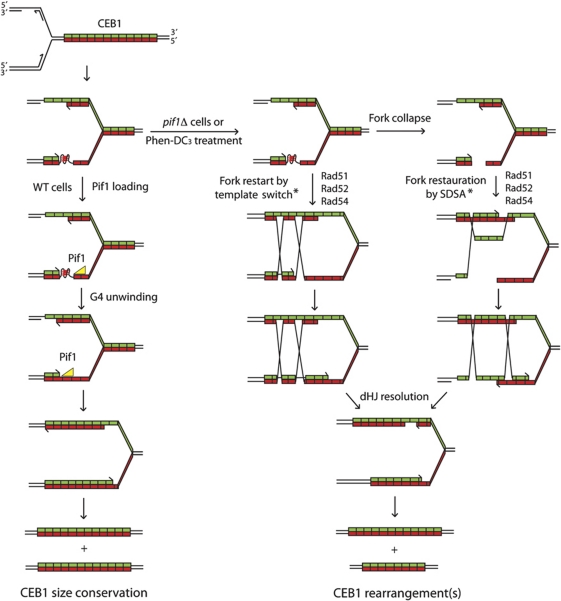
We concluded that the production of CEB1 rearrangements in orientation I generated in the absence of Pif1 or upon treatment of WT cells by Phen-DC3 and the correlated formation of the X-shaped structures during replication are fully dependent on the activity of the homologous recombination pathway. Its role is likely to allow the processing of unresolved G-quadruplex structures forming during leading-strand replication, albeit with the consequence to generate CEB1 size variations by a process of template switch or synthesis-dependent strand annealing (SDSA) (Figure 5).
Figure 5.
A model for the replication of the CEB1 sequence upon formation of a G-quadruplex in the leading-strand template. CEB1 is schematically represented as a green (C-strand) and red (G-strand) array. The replication fork moves from left to right, implying that the G-rich strand of CEB1 is the template for leading-strand replication. Each motif has a certain probability to form a G-quadruplex, which will in turn stuck the replicative polymerase (Polε). To be noted, in contrast to hairpins, G-quadruplexes fold only in the G-rich strand without an equivalent counterpart in the opposite strand (i-motif forms only at unphysiological acidic pH; Gehring et al, 1993). In a WT context (left part), Pif1 loads in the downstream single-stranded DNA and, while translocating in the 5′–3′ orientation, unwind the G-quadruplex. Note that, due to the presence of Polε upstream of the G-quadruplex, the loading and thus the activity of potential 3′–5′ helicases (like Sgs1) will require further processing (removal of the polymerase and 3′–5′ resection). Thus, in this context, the G-quadruplex is a substrate only for 5′–3′ helicases. Once the G-quadruplex is unwound, the replication can proceed normally. If the G-quadruplex persists, in Pif1-deficient cells or upon G-quadruplex stabilization by Phen-DC3 (right part), fork is restarted and the G-quadruplex is bypassed or removed by a template-switch mechanism. Alternatively, an accidental single-strand break of the leading-strand template or the G-quadruplex cleavage can convert the stalled fork into a broken fork. Fork will then be restored by a synthesis-dependent strand annealing (SDSA) mechanism. Both of these SDSA and template-switch mechanisms involve a Rad52-dependent Rad51 nucleofilament formation followed by a Rad54-dependent dsDNA invasion step during which CEB1 motifs can be misaligned (denoted by an *). The subsequent extension and reannealing of a misaligned invading strand will lead to the formation of the CEB1 rearrangements. Depending on the resolution of the double Holliday junction (dHJ), one or both alleles will be rearranged.
Structure of the CEB1 rearrangements produced upon Phen-DC3 treatment in either orientation
To further explore the behaviour of CEB1 near ARS305 with respect to its orientation, we also asked whether the size variants obtained in the Phen-DC3-treated cells carrying the polymorphic CEB1-1.8 allele in either orientation were of different types. To do so, we first examined the size distribution of the variants. Compared with the parental CEB1-1.8 array containing 42 repeats, the size distribution of the variants, ranging from 3 to ≈200 repeats (median orientation I: 38 and median orientation II: 39), is not significantly different in both orientations and includes a majority of contraction events (78% in orientation I and 65% in orientation II, not significantly different). Having screened a large number of cells, we could also determine the sequence of 12 contraction events in each orientation, and compared them with the parental array, and with each other. No de novo mutation was found. As schematically represented in Supplementary Figure S5, all sequenced contractions retained their tandem array structure with full-length CEB1 motif, and are different from each other even if they contain the same number of motifs (for examples: D4 and DD1 (21 motifs) in orientation I or D4, D5, and C3 (13 motifs) in orientation II). As previously observed in rad27Δ, pif1Δ, and wild-type cells carrying CEB1 near ARG4 and treated by Phen-DC3 (Piazza et al, 2010), we recovered three categories of rearrangements (simple and double deletions, as well as complex reshuffling of the parental motifs) in similar proportions. Qualitatively, looking at the average size of the rearrangements and the motif shuffling, we did not detected notable difference between the events produced in orientations I and II. This analysis gives no evidence that the rearrangements produced by the Rad51/Rad52 pathway are differently initiated in orientations I and II.
Discussion
Using complementary genetic, chemical, and physical analyses, we provide unprecedented insights into the in vivo consequence of DNA G-quadruplex secondary structures formation on leading-strand replication and minisatellite instability. We show that (i) Yeast cells deficient for G-quadruplex processing, either by interaction with the Phen-DC3 G-quadruplex ligand or by inactivated for the Pif1 helicase, frequently rearrange the reporter G-quadruplex-prone CEB1 minisatellite inserted in close proximity to the early-firing ARS305 in an unexpected orientation-dependent manner. (ii) The instability is dependent on the G-quadruplex-forming sequence of CEB1 since site-directed mutations of the G runs abolish the instability. (iii) The instability is almost exclusively observed in orientation in which the G-quadruplex-forming sequence is the template of the leading-strand synthesis (see Figure 1). (iv) This orientation effect relies on the cis-effect of ARS305. Deletion of ARS305 not only modifies the orientation bias but also reverses it because, in the absence of ARS305, the same region becomes replicated in the opposite direction from ARS306. (v) The orientation-dependent behaviour correlates with the formation of abnormal X-shaped intermediates during replication in CEB1. Formation of these intermediates, as well as the rearrangements, is dependent on the activity of the Rad51/Rad52 homologous recombination pathway.
Model for G-quadruplex formation and its interference with leading-strand synthesis
In which biological circumstances G-quadruplexes fold in vivo remain unknown. G-quadruplexes have been implicated in the regulation of DNA- and RNA-related processes such as telomere maintenance, genome rearrangements, transcription, splicing, and translation (Maizels, 2006; Kruisselbrink et al, 2008; Paeschke et al, 2008; Cahoon and Seifert, 2009; Reinhold et al, 2010). Theoretically, the folding of a G-quadruplex DNA may initiate either from duplex or from single-stranded DNA. A large number of natural proteins have been found to specifically interact with G-quadruplex structures (Wu and Brosh, 2010). They can play a sequence-specific role in destabilizing a target duplex DNA and/or acts to shift the equilibrium in favour of the G-quadruplex-folded state. Alternatively, the intramolecular folding of G-quadruplexes on single-stranded DNA either during transcription or during replication might be easier without the competition from the complementary strand which in duplex would favour standard Watson–Crick base pairing. With this view, our mechanistic interpretation of the formation of G-quadruplexes and its specific interference with leading-strand replication, up to the ultimate formation of the rearrangement products, is illustrated in Figure 5. Globally, it addresses three major questions: how the G-quadruplex may form during S-phase, how defective G-quadruplex resolution leads to a recombinogenic lesion, and finally how the repair of these recombinogenic lesions generates the variety of rearrangements depicted in Supplementary Figure S5.
Along this path, the first issue is where single-stranded DNA might form during replication to promote the formation G-quadruplexes? During normal replication, the leading strand is synthesized continuously and the lagging strand copied in short ∼200 bp Okazaki fragments which are then joined together after removal of their 5′ ends by the Rad27/FEN1 Flap endonuclease (Kunkel and Burgers, 2008). During the unwinding of the DNA duplex by the MCM complex at the replication fork, single-stranded DNA may transiently forms between the leading Polε polymerase and the fork and/or between the lagging Okazaki fragments which are RNA primed and successively elongated by the Polα and Polδ polymerases (Kunkel and Burgers, 2008). In the case of CEB1, the sequence context may compromise the ability of the replicative polymerases to remain processive. We, thus, propose that the repeativeness of the array would provide aggravating opportunities to further slow down replication and ultimately create the more drastic G-quadruplex obstacle. It is known that G-quadruplexes can hinder DNA polymerases progression in vitro (Weitzmann et al, 1997). Replication pause site can be visualized in 2D gel as a discrete spot signal along the Y arc that can be localized using different restriction digests. Here, in most 2D gels (Figure 4), we observed that the distribution of signal along the Y arc was homogenous, suggesting the lack of a specific site for replication pausing within CEB1. However, to be noticed, using the most asymmetric ApaI/SpeI restriction digestion that places CEB1 at the distal end of the Y arc, we noted a slight accumulation of replication intermediates within the hereof compacted CEB1 region (see Supplementary Figure S4). Hence, whether or not the wild-type CEB1 sequence itself perturbs the dynamic of Polε elongation remains to be further explored. Of concern, the sensitivity of 2D gel may not be sufficient to detect low frequency pausing events distributed along the CEB1 array. Differently, the coupling between the replicative helicase and polymerase as well as the coupling between the leading- and lagging-strand synthesis may also be impaired, leaving additional opportunities to create single-stranded gap regions or more complex branched molecules. Thus, as illustrated in Figure 5, we propose that intramolecular G-quadruplex folds on the leading strand between CEB1 and the fork duplex.
The next critical issue is how G-quadruplexes are processed? In vitro several DNA helicases, such as the mammalian BLM and WRN RecQ orthologues, PIF1, FANCJ unwind G-quadruplex structures (Sanders, 2010; Wu and Brosh, 2010). In S. cerevisiae, the Sgs1 and Pif1 helicases unwind G-quadruplex in vitro (Sun et al, 1999; Ribeyre et al, 2009). The present data demonstrate the in vivo role of Pif1 to ensure CEB1 stability during replication but we found no effect of inactivating the Sgs1 or Rrm3 helicases. It suggests that Sgs1 or Rrm3 may have no role in unwinding G-quadruplexes in vivo, act on different G-quadruplex substrates that remain to be uncovered, or show functional redundancies with each other or with the other helicases of S. cerevisiae. Our current mechanistic model is illustrated in Figure 5. Consistent with the recent report of Pif1 binding to chromosomal regions containing G-quadruplex potentially forming sequences (Paeschke et al, 2011), we propose that Pif1 unwinds the G-quadruplexes that form during leading-strand replication either by its direct recruitment to the G-quadruplex structure or indirectly, as being part of the ‘replisome or fork-movement machinery’ in order to resolve such impediments. The 5′–3′ directionality of Pif1 would play an important role in this process (Figure 5). In contrast, in the absence of Pif1 or in Phen-DC3-treated wild-type cells, the G-quadruplexes formed by CEB1 will remain unprocessed and channelled into the recombinational-repair pathway, otherwise leading to cell death.
The present observations that factors of the homologous recombination pathways involved in the nucleofilament formation (Rad51 and Rad52) and strand invasion (Rad51 and Rad54) are required for the production of the accumulation of X-shaped intermediates, in correlation with the production of complex CEB1 rearrangements, demonstrate that unresolved G-quadruplexes formed in the leading-strand template lead to a recombinogenic substrate. The 2D-gel properties of the identified repair intermediates suggest that they represent X-shaped structures typical of unresolved joint molecules likely forming between the sister chromatids, but whether it arises from an initiating nick, a single-strand gap, or a double-strand break remains to be determined. As illustrated in Figure 5, out-of-register Rad51-dependent strand invasion can occur within the array and multiple cycles of invasion and destabilization events can yield the large variety of CEB1 rearrangements (Supplementary Figure S5). In Phen-DC3-treated WT cells as well as in pif1Δ cells, the frequency of rearrangements increased with the size of the CEB1 array (Figure 1; Ribeyre et al, 2009) likely reflecting the increased probability of longer arrays to form one or several G-quadruplex structures. Alternatively, lesions in small alleles may be more frequently repaired in proper register or be resected into the non-repeated flanking regions, leading to the preferential restoration of the parental sequence.
Does G-quadruplexes form in the lagging-strand template?
The stability of CEB1 inserted near ARS305 in orientation II (G-strand being the lagging template) was surprising for two reasons. First, single-stranded DNA inherently forms on the lagging strand between elongating Okazaki fragments. Second, Pif1 involvement on the lagging-strand replication has been suggested by the genetic interactions of PIF1 with DNA2, POL32, CDC9, POL3, and RAD27 (Budd et al, 2006; Chang et al, 2009) and investigated in vitro on reconstituted Okazaki fragment processing machineries (Pike et al, 2009). Several interpretations can be invoked. It may indicate that G-quadruplexes do not fold on the template strand during lagging-strand synthesis. Alternatively, if G-quadruplexes form, it may reveal that other helicases than Pif1 are specialized to resolve them on the lagging strand. So far, we have tested deletion of the SGS1, RRM3, MPH1, and DNA2 (in combination with the PiF1 deletion) genes and found no effect of inactivating these candidate helicases (Ribeyre et al, 2009; Table I). Differently, it may also reveal the higher tolerance of the lagging-strand replication machinery to bypass the block by priming DNA synthesis downstream, although downstream priming on the leading strand is also possible (Heller and Marians, 2006).
To further address the possibility that CEB1-G-quadruplex may be differently processed on the leading and lagging strand, we also examined the behaviour of CEB1 in WT cells treated with Phen-DC3 in either orientation. Again, the instability of CEB1 was significantly more pronounced in orientation I than in II, but quantitatively the bias was less pronounced in WT-treated cells than in the absence of Pif1 (∼3–8-fold versus 28-fold, respectively; see Tables I and II and Figure 3) and importantly, it results from a slight increase of instability in orientation II compared with untreated WT cells. Mechanistically, it raises the possibility that CEB1-G-quadruplexes are processed by Pif1 in that strand, but in a dispensable manner due to the presence of other redundant G-quadruplex-processing activities.
Implication on the localization of G-quadruplex-forming sequences within genome
The ‘at risk’ destabilization of CEB1 according to the direction of replication raises the question where the potential G-quadruplex-forming sequences are located within the yeast genome relative to the position of the origins of replication. To cross these features, we stringently mined the sequence of the yeast genome with Quadparser (Huppert and Balasubramanian, 2005) for G-quadruplexes with loops size ⩽9 nt and then examined the position of the 50 best potential G-quadruplex-forming sequences relatively to the experimentally validated closest early- and middle-firing 5′ and 3′ ARS (Fachinetti et al, 2010). The methods and the source of information are described in Supplementary Methods. The resulting information is reported in Supplementary Table S3. If to be maintained, the G-quadruplex-forming sequences are preferentially located on the lagging-strand template, we expected to find them closer to the ARS in 5′ than the ARS in 3′. Clearly, no bias towards a particular location on one side of the interorigin distance exists (P=0.19 Mann–Whitney–Wilcoxon test). Thus, we conclude that in the S. cerevisiae genome the location of single G-quadruplex potentially forming sequences is not correlated with the replication directionality. Another evolutionary feature may reside in the arrangement of G-quadruplex-forming sequences in arrays like in CEB1. Indeed, whereas no negative selective pressure seems to apply for single G-quadruplex-forming sequences in the human genome (except in exonic coding sequence) (Huppert and Balasubramanian, 2005), G-quadruplex-prone tandem arrays are among the most unstable (Weitzmann et al, 1997), and the number of motifs in these arrays inversely correlates with the stability of the G-quadruplexes determined in vitro in nine vertebrate genomes (Bacolla et al, 2008). This correlation is rationalized in S. cerevisiae by the fact that except the subtelomeres and telomere repeats, interstitial tandem arrays of potential G-quadruplex-forming sequences are absent and that the majority of rearrangements produced upon the Phen-DC3 treatment in WT cells, and even more in pif1Δ-treated cells are extensive contractions (Piazza et al, 2010), all tending to eliminate potentially unstable G-quadruplex arrays, unless counteracted by functional selective pressure. Interestingly, along this line, despite the potential drawback of the G-quadruplex-forming repeats which we report here, it is remarkable that telomeric repeats in most eukaryotic phyla are TTAGGG or other potentially forming G-quadruplex sequences (Tran et al, 2010) and that both the subtelomeric repeats and the single-stranded overhang are oriented such that the G-rich strand is replicated by the lagging-strand machinery (Makovets et al, 2004; Gilson and Geli, 2007). Together with the utility of these G-quadruplex sequences to be able to form potentially useful stable structures in normal or pathological situations (Smith et al, 2011) such predominant evolution of telomeric repeats may result from the need to protect chromosome ends from damageable contractions and overall being replicated by the safer lagging-strand machinery is an adaptation to the challenge of leading-strand DNA replication.
Materials and methods
Media and strains
Media were prepared as previously described (Piazza et al, 2010). Strain was derived by transformation of the SY2209 S. cerevisiae strain of the W303 background (Fachinetti et al, 2010) and listed in Supplementary Table S1. Methods of strain construction are described in Supplementary Methods.
Measurement of CEB1 instability
CEB1 instability during vegetative growth was measured by Southern blot analysis as previously described (Ribeyre et al, 2009; Piazza et al, 2010). Briefly, cells were suspended at a density of 2 × 105 cells/ml into 5 ml of rich YPD medium, grown for eight generations at 30°C with agitation, and then plated as individual colonies on YPD media incubated at 30°C. Individual colonies or colony pools were analysed by Southern blot using the following genomic digestions: ApaI/XhoI for CEB1-1.8 (orientation I), ApaI/NcoI or ApaI/SacII for CEB1-1.8 (orientation II), and ApaI/SacII for CEB1-Gmut-1.7. The membranes were hybridized with radiolabelled CEB1-0.6 or CEB1 Gmut probes. Signals were detected with a Typhon PhosphorImager (Molecular Dynamics). To measure CEB1 instability upon Phen-DC3 treatment, cells were grown for eight generations in liquid SC media containing 10 μM of Phen-DC3 or, as a control, grown in the equivalent concentration of DMSO (0.02%) (Piazza et al, 2010). For pools of genomic DNA of 8, 12, or 16 colonies/well, a rearrangement is counted if the intensity of the rearranged minisatellite, quantified with ImageQuant software and normalized to its size, corresponds to 1/8 or 1/12 or 1/16 of the total amount of signals measured in the lane. Rearrangements migrating at the same size are considered as early clonal events, counted only once, and the remaining counts removed of the total number of colonies analysed. Sequencing of several of these same-sized alleles allowed us to confirm their clonal origin in most cases. However, in mutant strains with very high CEB1 instability (i.e., pif1Δ with CEB1 orientation I), the probability of obtaining two independent rearrangements of the same size is quite high. Consequently, the removal of all rearranged alleles of the same size may lead to an underestimation of the rearrangement frequency.
Statistical analysis
The rearrangement frequencies measured on Southern blots have been compared using the two-tailed Fischer's exact test. The correlation between the instability and the size of CEB1 has been tested using the Spearman correlation test. Size distributions of the rearrangements have been compared using the two-tailed Mann–Whitney–Wilcoxon median comparison test. All the statistical tests have been performed using R 2.12.0. An α cutoff of 0.05 has been applied for each test.
Sequencing of CEB1 rearrangements
The sequencing of a sample of CEB1 contractions was performed as previously described (Lopes et al, 2006a). Colonies that carry a CEB1 contraction visualized by Southern blot were isolated, genomic DNA extracted and the CEB1 region was PCR amplified and sequenced using flanking primers. Compared with the sequence of the progenitor polymorphic CEB1-1.8 allele, the origin of the motifs was defined by the most parsimonious interpretation to describe the reshuffling of the parental motifs (see Supplementary Figure S5).
Physical analyses of replication intermediates by 2D gels
After an O/N preculture at 25°C in YPD or SC medium, cells were diluted to 107 cells/ml in fresh medium and grown for 2 h at 25°C. Cell synchronization was performed on diluted cells (107 cells/ml) by adding α factor (Euromedex) to a final concentration of 3 μg/ml for about 130 min at 25°C. In the case of Phen-DC3 or DMSO regimen, 10 μM of Phen-DC3 or the equivalent concentration of DMSO was added 15 min before the end of α factor synchronization. The release was performed at 30°C in YPD, or in SC medium containing either Phen-DC3 at 10 μM, or 0.02% DMSO. At each time point, 1 ml of cells was taken for FACS analysis (≈107 cells) and 200 ml cultures were arrested by addition of 2 ml of 2% sodium azide for DNA extraction. Before DNA extraction, cells were washed with cold water and treated with Psoralen (Sigma). In all, 5 ml of cold water was added to the pellet and cells were transferred in culture dishes that contain six wells, in ice. In all, 300 μl of Psoralen solution (0.2 mg/ml in ethanol 100%) was added and cells were incubated for 5 min in the dark. Cells were irradiated for 10 min with UV light of 365 nm. The light source was mounted at a distance of 6–7 cm above the culture dish, always in ice. The procedure is repeated three times with a total irradiation time of 40 min. Cells were then transferred in a 50-ml Falcon tube in ice, and washed with cold water. DNA extraction was performed as described in ‘Method 3’ DNA Extraction Procedures by Lopes et al (2003). 2D-gel electrophoresis was carried out as originally described by Brewer and Fangman (1987). DNA was transferred to GeneScreen Plus® Hybridization Transfer Membrane (Perkin-Elmer) in 10 × SCC by capillarity blotting. Quantification was done using ImageQuant 5.2 (Molecular Dynamics) as previously described (Lopes et al, 2003).
Cell viability assay
To examine cell viability, cells taken from a fresh patch on YPD plate were suspended to a final concentration of 5 × 104 cells/ml in 5 ml of YPD, and grown O/N at 30°C. The exponentially growing culture was sonicated (Bioblock Scientific sonicator, Vibra Cell 75041) for 5 s, and cells spread on YPD plate at room temperature. Using a micro-dissector (Singer Instruments), budded cells were isolated on a grid and regularly examined to separate mother and daughter cells. After 3 days of plate incubation at 30°C, the cells forming colonies were examined to determine the number of mother–daughter pairs giving rise to 2, 1, or 0 outgrowth.
Supplementary Material
Acknowledgments
We thank Wolf-Dietrich Heyer and Jean-Baptiste Boulé for critical reading of the manuscript; Gael Millot for advises on statistical analysis; Valérie Borde, Thelma Capra, Rodney Rothstein, Arturo Londono-Vallejo, and members of our laboratories for helpful discussions. The AN team is supported by the Ligue Nationale contre le Cancer (LNCC; Equipe Labellisée EL2007.LNCC/AN and EL2010.LNCC/AN). AP is supported by a graduate student fellowship from the ‘Ministère de l'Education Nationale, de la Recherche et de la Technologie’. BK was supported by the 2010 Massachussets Institute of Technology—Institut Curie exchange program fellowship.
Author contributions: JL, AP, RB, MF, and AN designed the experiments. JL and AP performed the genetic experiments. JL, RB, and AC performed the 2D-gel analyses. JL, AP, RB, BK, AC, MF, and AN analysed the data. MPTF provided the Phen-DC3 molecule. JL, AP, RB, MF, and AN wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguilera A, Gomez-Gonzalez B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD (2008) Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res 18: 1545–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulé JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL (2006) Evidence suggesting that Pif1 Helicase functions in DNA replication with the Dna2 Helicase/Nuclease and DNA Polymerase ∂. Mol Cell Biol 26: 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34: 5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Seifert HS (2009) An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Luke B, Kraft C, Li Z, Peter M, Lingner J, Rothstein R (2009) Telomerase is essential to alleviate pif1-induced replication stress at telomeres. Genetics 183: 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18: 950–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL (2008) Targeting telomeres and telomerase. Biochimie 90: 131–155 [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M (2010) Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring K, Leroy JL, Gueron M (1993) A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 363: 561–565 [DOI] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2006) Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439: 557–562 [DOI] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP (2006) Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res 34: 4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33: 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M (2008) Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 18: 900–905 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I et al. (2011) ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143: 367–378 [DOI] [PubMed] [Google Scholar]
- Lopes J, Debrauwere H, Buard J, Nicolas A (2002) Instability of the human minisatellite CEB1 in rad27Δ and dna2-1 replication-deficient yeast cells. EMBO J 21: 3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Ribeyre C, Nicolas A (2006a) Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol Cell Biol 26: 6675–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Liberi G, Foiani M (2003) Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell 12: 1499–1510 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006b) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Maizels N (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 13: 1055–1059 [DOI] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH (2004) Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE (2005) RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol 25: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP (2008) Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie 90: 1207–1223 [DOI] [PubMed] [Google Scholar]
- Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC (2010) Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res 39: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA (2008) Division of labour at the eukaryotic replication fork. Mol Cell 30: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JK, Zakian VA (2011) DNA replication through G-Quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ (2008) Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol 15: 598–604 [DOI] [PubMed] [Google Scholar]
- Pâques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel TA (2002) Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell 10: 207–213 [DOI] [PubMed] [Google Scholar]
- Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A (2010) Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res 38: 4337–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JE, Burgers PM, Campbell JL, Bambara RA (2009) Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem 284: 25170–25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloumienko A, Dershowitz A, De J, Newlon CS (2001) Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol Biol Cell 12: 3317–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Reinhold WC, Mergny JL, Liu H, Ryan M, Pfister TD, Kinders R, Parchment R, Doroshow J, Weinstein JN, Pommier Y (2010) Exon array analyses across the NCI-60 reveal potential regulation of TOP1 by transcription pausing at guanosine quartets in the first intron. Cancer Res 70: 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Dujon B (2006) Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol Biol Evol 23: 189–202 [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B (2008) Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72: 686–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily GM, Raca G, Mirkin SM (1997) Trinucleotide repeats affect DNA replication in vivo. Nat Genet 17: 298–304 [DOI] [PubMed] [Google Scholar]
- Sanders CM (2010) Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J 430: 119–128 [DOI] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE (2010) Epigenetic instability due to defective replication of structured DNA. Mol Cell 40: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA (1994) The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou MP, Abramowitz L, Schultz DC, Johnson FB (2011) Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18: 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Bennett RJ, Maizels N (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res 27: 1978–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PL, Mergny JL, Alberti P (2010) Stability of telomeric G-quadruplexes. Nucleic Acids Res 39: 3282–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnaud G, Denoeud F (2000) Minisatellites: mutability and genome architecture. Genome Res 10: 899–907 [DOI] [PubMed] [Google Scholar]
- Weitzmann MN, Woodford KJ, Usdin K (1997) DNA secondary structures and the evolution of hypervariable tandem arrays. J Biol Chem 272: 9517–9523 [DOI] [PubMed] [Google Scholar]
- Wu Y, Brosh RM Jr (2010) G-quadruplex nucleic acids and human disease. FEBS J 277: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.