Abstract
Even with the availability of tissue-restricted knockouts, studying the role of essential, multifunctional proteins in vivo is often complicated by the complete loss of cell viability. Here we make use of a knock-down approach to investigate the function of the essential protein PSF (Polypyrimidine tract binding protein-associated Splicing Factor) in thymocytes. PSF is an RNA- and DNA-binding protein that has roles in all aspects of RNA biogenesis from transcription to stability. We show that a 50% reduction in expression of PSF in the thymus leads to an increase in apoptosis and a corresponding decrease in thymic cellularity. Using microarrays we find that thymocytes depleted of PSF show reduced expression of several histone genes relative to wildtype littermates likely due to reduced mRNA stability. Remarkably, reduced expression of a specific one of these histone genes (H2AE from histone cluster 1) is sufficient to induce apoptosis of a cultured T cell line. Therefore, we conclude that PSF contributes to the stability of a subset of histone genes and that loss of H2AE expression in the PSF-deficient thymocytes uniquely contributes to an increase in thymic apoptosis.
Keywords: PSF, Thymus, mouse knock-down, histones, mRNA stability
Introduction
The nucleosome is comprised of 4 core histones (H2A, H2B, H3 and H4) and one linker histone H1 that together package and control the functional accessibility of DNA [1]. Each of the core histone proteins is encoded by multiple non-allelic genes that are present in clusters within the genome [2]. For example, the H2A protein is encoded by more than 10 distinct genes (i.e. H2A-M), some of which encode identical proteins while others deviate from one another by a few amino acids [2]. The multiple gene clusters are conserved across mammalian species suggesting functional importance; however, the subtle differences between these genes and their corresponding proteins has complicated efforts to define unique function and/or expression differences [3]. Thus, to date, little clear understanding has emerged regarding the functional relevance of individual histone genes.
By contrast, much is known regarding the expression of the histone genes as a family. Expression of all of the core histone genes is replication-dependent, such that high levels of expression is confined to S phase of the cell cycle by a combination of mechanisms including regulated transcription and mRNA stability [4]. Notably, the replication-dependent histone genes have the additional unique feature of being intronless and having their own pathway for 3′ end processing which involves a conserved stem loop structure instead of a canonical polyA tail [4]. This stem loop is processed and bound by specific transacting factors and contributes to the tight control of the export, translation and stability of histone mRNAs [4].
PSF (PTB-associated splicing factor) is a RNA- and DNA-binding protein that is involved in a variety of nuclear processes [5]. Among the activities attributed to PSF are regulation of transcription, alternative and constitutive splicing, polyadenylation, mRNA export and stability and the organization of nuclear architecture [5]. However, most of these functions have been discovered using in vitro systems or cell culture models, while in vivo data remains scarce. One of the few in vivo studies was performed in zebrafish and suggested a role of PSF in differentiation as well as the regulation of apoptosis [6]. This study also showed lethality of PSF knock out in fish, strongly suggesting that knock out of PSF in mice would also be lethal, thus complicating in vivo analyses in mammals.
To circumvent the potential problem of early lethality in PSF knock out mice, we have used PolII directed expression of an shRNA in order to knock-down PSF in the T cell compartment of mice. We obtained a transgenic line with reduced PSF expression and show that these mice display reduced thymic cellularity and increased apoptosis. We also noted decreased expression of several replication-dependent histones likely due to reduced mRNAs stability. Knock-down of a single histone, H2AE, was sufficient to reduce cell viability in a cell line model, suggesting that loss of this histone is at least partly responsible for the increased thymic apoptosis observed in our mice. Therefore, we conclude that PSF regulates the abundance of several replication dependent histone mRNAs and that H2AE plays a non-redundant role in cell viability and/or T cell development.
Materials and Methods
Cell culture and transfections
Cell culture, transfections with morpholinos oligos (MO) and generation of stable cell lines was done as described [7]. For PSF knock down the miR30 precursor sequence was used [8] in which the sequence of the mature miRNA was replaced with a sequence targeting PSF (Fig. S1A, synthesized as PAGE Ultramer, IDT). For stable overexpression of H2AE variants the human H2AE was PCR amplified and cloned in frame with a c-terminal Flag-tag. For the MO resistant variant silent point mutations were introduced in the sequence targeted by the MO immediately downstream of the ATG. wt: atgtctggacgtggaaagcaaggcg; mut: atgtcaggtcgaggcaaacagggaggcaaagctcg. All constructs were verified by sequencing.
Cell cycle and viability assays
Growth curves were obtained by manually counting cells using exclusion of trypan blue staining to score for viability. Apoptosis and cell cycle analysis were performed with the respective kits according to the manufacturer's instruction (BD Bioscience).
Mice and analysis of thymocytes
Transgenic mice were generated in C57BL/6 background using standard procedures in accordance with IACUC protocol #2008-0216 from UT Southwestern Medical Center. The transgenic founder was bred to C57BL/6 mice and transgenic offspring was either used for analyses or for further breeding continued under IACUC protocol #802852 from the University of Pennsylvania. Transmission of the transgene was monitored by PCR. 6-8 week old littermates were used for analyses. Thymic single cell suspensions were prepared, counted, stained with CD4-PE and CD8-Fitc antibodies (BD Bioscience) and analyzed using a FACS Calibur. Hypotonic lysis and staining with Propidium Iodid was used to determine the DNA content of thymocytes; doublets were excluded during the analysis which was done using Cell Quest (BD Bioscience). For continued analysis, thymocytes were cultured in RPMI supplemented with 10% FBS, Pen/Strp/Glut, Pyruvate, non-essential amino acids and bME. To assess RNA stability Actinomycin D (Sigma) was added to a final concentration of 5ug/ml for 16h.
RNA, RT-PCR and microarray
RNA from thymocytes was prepared using RNABee (Tel-Test) using the manufacturer's protocol. An established, radioactive, low cycle RT-PCR protocol and Phosphor Imager analysis was used to determine target gene expression [9]. For microarray analysis RNA from 3 WT and 3 Tg thymi was used on Illumina MouseRef-8v2 arrays. For genes that showed a consistent change in all 3 samples (defined as p<0.1) a mean fold change was calculated as (WT1+WT2+WT3) / (Tg1+Tg2+Tg3).
Results and Discussion
Transgenic expression of an shRNA leads to knock-down of PSF in mice
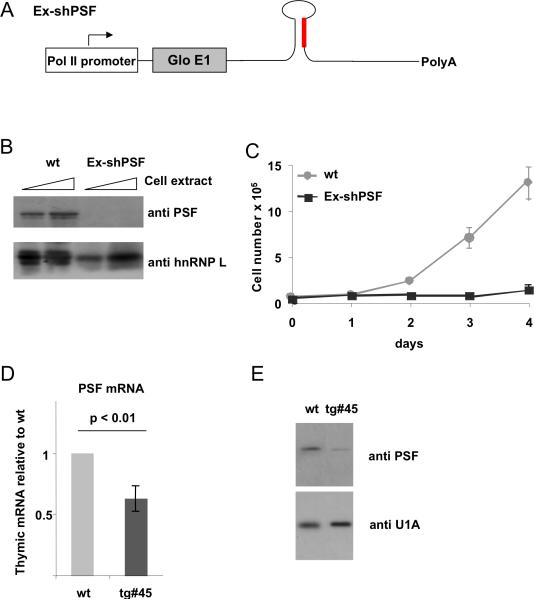
In previous work we have established a role for PSF in T cell activation using the human T cell-derived JSL1 cell line [7; 10]. In order to gain insight into further potential function(s) of PSF in T cell development in an in vivo situation we aimed at targeting PSF expression in mice. Based on previous results [6; 7; 10] we expected a complete knock out of PSF to be lethal. Therefore, we turned to an shRNA-mediated knock-down strategy to obtain partial depletion of PSF. We inserted a PSF-targeting siRNA in a sequence context mimicking endogenous miR30 that has been used previously to target gene expression in mice (Fig S1A, [8]). The sequence we used for the shRNA is targeted to a region of the coding sequence of PSF that is 100% conserved between mouse and human (Materials and Methods) and was confirmed to knock-down PSF expression using siRNAs in a pilot experiment (data not shown). The miR30-based hairpin was inserted into a variety of Pol II-based expression constructs containing an exon from the human B-globin gene and/or cDNA for GFP, and the resulting constructs were tested for their ability to knock-down a Flag-tagged version of PSF in cotransfection experiments in 293 cells (Fig S1B, C). Interestingly, knock-down efficiency differed considerably between different constructs, probably due to differential processing of the shRNA. We chose the construct with the highest knock-down efficiency (Ex-shPSF, Fig S1C and 1A) for all subsequent experiments.
We next generated JSL1 cell lines stably expressing the Ex-shPSF construct (Fig 1A) to obtain knock-down of endogenous PSF. One clone was obtained that showed an almost complete loss of PSF expression (Fig 1B). Consistent with our previous data suggesting the knock-out of PSF to be lethal, we observed a severe defect in growth and nuclear morphology in this clone (Fig. 1C and Fig S2) thus confirming that our targeting strategy worked in the T cell line. We therefore went on to produce transgenic mice expressing the same Ex-shPSF construct under control of the Lck promoter to direct knock-down to the T cell compartment [11]. Our assumption was that due to differing levels of expression among transgenic animals we might obtain a line in which PSF knock-down is in a range to retain viability yet exhibit a phenotype. Evidence that such a window of PSF expression exists has been obtained with modest knock-down of PSF in JSL1 cells in previous studies [10].
Figure 1. Generation of mice with reduced expression of PSF in the T cell compartment.
(A) Schematic view of the Ex-shPSF construct used to generate stable cell lines and transgenic mice. (B) PSF expression in WT JSL1 cells or cells stably expressing Ex-shPSF, as determined by Western blotting whole cell extract with anti-PSF or a loading control (anti-hnRNP L). (C) Growth curve of the cell lines from B. (D) RNA from thymus was prepared from wt or Ex-shPSF transgenic line #45 (Tg#45) and PSF expression was assessed by radioactive RT-PCR, quantified by Phosphorimager analysis and expressed as % of wt (n=3). (E) Whole cell extracts of WT and Tg#45 thymocytes analyzed by Western blotting for PSF expression.
We generated several lines of transgenic founders, confirmed germline transmission of the transgene, and tested PSF expression in the thymus by RT-PCR. We obtained one line that showed a significant knock-down of PSF mRNA (Fig. 1D). This reduction in mRNA corresponded to an approximately two-fold reduction of PSF protein in thymic extracts (Fig. 1E). Together these results show that we are able to induce a modest but stable reduction of PSF expression in mouse thymocytes by PolII-mediated transgenic expression of an shRNA.
Reduced PSF expression leads to decreased thymic cellularity and increased apoptosis
Having established knock-down of PSF in thymocytes we investigated potential defects in thymic development. We first noticed a reduction in the total cellularity of transgenic thymi (Tg#45) to approximately 50% of wildtype (WT) (Fig. 2A). Three reasons could account for the reduced thymic size observed in the Tg#45 line: a block in thymic development, a defect in cell proliferation, or increased cell death. Although the total number of thymocytes was reduced in the Tg#45 line compared to WT, T cell development was largely normal in the transgenic thymus as evidenced by an only slightly changed distribution in the subpopulations of double negative (DN), double positive (DP) or single positive (CD4 or CD8 SP) cells (Fig. 2B). Similar results were obtained for the early developmental stages DN1 to DN4 (data not shown). As robust cell proliferation is required for progression from the DN to DP stage, the lack of a marked reduction of DP cells indicates that neither a significant defect in cell proliferation nor a block in development is the primary cause of reduced thymic size.
Fig. 2. Reduced PSF expression leads to increased apoptosis and decreased thymic cellularity.
(A) Single cell suspensions were prepared from thymi of WT or Tg#45 animals, cells were counted and expressed as % of WT. (B) Thymocytes were stained with CD4 and CD8 antibodies and the relative proportion of double negative (DN), double positive (DP) and single positive CD4 or CD8 populations was compared for WT and Tg#45 animals. (C) The presence of dead cells as assessed by forward and sideward scatter from the cells used in B) is shown as percent of WT. (D) The DNA content of thymocytes was assessed using PI staining and FACS analysis. Dead cells (subG1), and cells in G1 and M phase were distinguished and expressed relative to WT (set at 1). (E) Thymocytes from WT and Tg mice were stained with AnnexinV to detect apoptotic cells. For all analyses the number of mice used for each genotype is shown as well as standard deviations and p-values. WT littermates were used in all cases.
By contrast, analysis of the thymocytes by forward/sideward scatter, which measures cell size and shape, suggested an increased number of dead cell in transgenic thymocytes relative to WT (Fig. 2C). To further substantiate an increase in cell death we investigated cell cycle distribution in WT and transgenic thymocytes by staining DNA with PI to distinguish dead cells (sub G1 peak) versus cells in the G1 and M phase of the cell cycle. Consistent with a loss of cell viability but not proliferation, the PI stain showed increased number of dead cells in transgenic animals, whereas the cell cycle distribution was similar to WT cells (Fig. 2D). Western blotting for 2 cell cycle markers, Cyclin A and phosphorylated Histone H3 also showed no difference between WT and Tg#45 thymocytes (data not shown). Having confirmed that there is no defect in cell cycle in the transgenic animals, we next tested whether the increased cell death in Tg#45 was due to increased apoptosis. Indeed we observed a significant increase in the staining of the transgenic thymocytes with the standard apoptosis marker AnnexinV (Fig. 2E). We therefore conclude that the reduced cellularity of the transgenic thymus is primarily due to an increase in apoptotic cell death. These data are in agreement with a report of PSF knock out in zebrafish, where loss of PSF was shown to increase apoptosis with no effect on cell proliferation (4).
Several replication dependent histone mRNAs are decreased in transgenic thymi
PSF has been primarily implicated in the regulation of transcription, splicing and mRNA stability [5]. While we previously have demonstrated a role for PSF in splicing in human T cells [7; 10], the decrease in PSF in Tg #45 was not sufficient to induce a splicing phenotype in known PSF target genes (data not shown). Therefore, we consider it likely that the increased apoptosis upon reduced PSF expression in thymocytes is due to changes in transcription and/or mRNA stability. As defects in either of these processes would result in changes in overall message level we analyzed WT and Tg#45 thymic RNA levels by expression microarrays to identify potential target genes.
Total RNA was obtained from the thymus of three WT and three Tg#45 mice and hybridized to Illumina MouseRef-8v2 arrays. We then calculated the mean fold change for genes with a consistent difference between the WT and Tg populations. Interestingly, 2 out of the 4 genes that showed at least a 1.9 fold difference in the Tg thymi encode replication dependent histones (Fig 3A). These genes, H3F and H2AE from histone cluster 1, were the two most repressed genes in the Tg#45 cells (Fig 3A). Moreover, a third replication-dependent histone cluster 1 gene (H4H) was among the next four most repressed genes in the transgenic cells (Fig 3A). The predicted difference in expression of all three of these histone genes was confirmed by our standard RT-PCR assay (Fig. 3B). Importantly, three other histone mRNAs that were predicted by the array to be unchanged between WT and Tg#45 animals were indeed shown to be expressed at similar levels by RT-PCR (Fig. 3B). Together these data confirm the validity of the array data and suggest a specific regulation of histones H4H, H2AE and H3F. We have also confirmed the predicted changes in AdamTS6 and Tensin 3, the two most upregulated genes in the Tg#45 cells (Fig. S3A). Although AdamTS6 and Tensin3 further validate the array data and may be interesting targets for future study, little is known about the function of these genes and thus we have not pursued them further herein.
Fig. 3. PSF regulates the abundance of several histone mRNAs in thymocytes.
(A) Three WT and three Tg#45 thymi were analyzed by microarray. Shown are genes with the largest increase or decrease in mean expression in Tg vs. WT animals (with p<0.1 cut-off). Histone genes are highlighted. (B) Confirmation of array data by RTPCR. Three histones that were unchanged according to the microarray data (cluster 1 H1D, H2AA and cluster 4 H4) were included as controls (n=3 for each genotype). (C) Thymocytes were treated with Actinomycin D for 16h, stability of indicated mRNAs was assessed by comparing the amount of the respective mRNA pre and post Act D treatment (n=3).
Given that 3 of the 6 most repressed genes in the Tg thymi belong to the same family we wished to further investigate the mechanism and consequence of this regulation. To differentiate whether the differential expression of H4H, H2AE and H3F is more likely due to altered transcription or stability we blocked transcription in the WT or Tg#45 thymocytes with Actinomycin D and checked the presence of the respective mRNAs. Consistent with a role of PSF in regulating the stability of these histones we found a marked reduction of their mRNA expression in Tg#45 thymi versus WT 16 hrs after transcription shut-off (Fig. 3C). This was again specific, as the stability of the three unaffected histones and three unrelated genes was unchanged (Fig. 3C, S3B). Furthermore, treatment of the cells with another cytotoxic drug, Cycloheximide, had no such effect (not shown). Taken together, we conclude that PSF regulates the expression of a subset of histone genes in thymocytes by controlling the stability of these mRNAs.
Knock-down of histone H2AE in T cell induces apoptosis
To bring together the molecular and phenotypic changes in the Tg#45 thymocytes, we wished to determine if the decreased expression of H4H, H2AE and/or H3F might contribute to reduced cell viability. As mentioned above, while many genes encode the replication-dependent core histones and often differ from one another by a few amino acid substitutions, a functional difference between these genes remains largely unexplored [2; 3; 4]. We were therefore interested in establishing an experimental system that would allow us to test whether knock-down of the individual histone variants that we found regulated by PSF would cause a phenotype similar to the one observed in the thymus of our mouse model. To accomplish this we turned to the JSL1 T cell line which mimics at least certain aspects of thymic development [12], and used transfection of Morpholino-oligomers (MOs) which we have previously shown to efficiently target specific protein expression in these cells [10; 13].
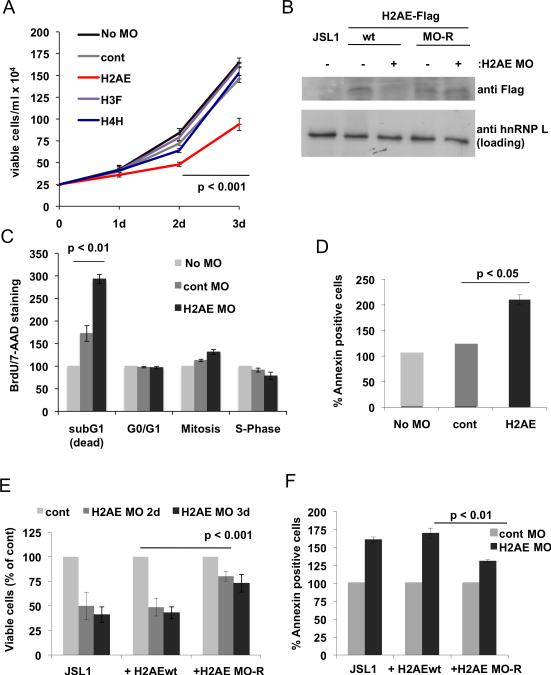
Strikingly, we found that transfection of JSL1 cells with two different MOs against H2AE is sufficient to significantly reduce the number of viable cells, whereas treatment of the cells with H3F or H4H specific MOs had no effect (Fig 4A, S4A). The lack of an effect with the latter two MOs may indicate that they fulfill redundant functions but it is also possible that the MOs were not efficient in knocking down the respective proteins, which we could not directly test as no specific antibodies exist against the individual variants. For H2AE we confirmed the efficacy of the MOs by cotransfecting a C-terminally Flag-tagged H2AE expression plasmid and the MO, which lead to a reduction of the H2AE-Flag protein (Fig 4B). As the expression plasmid contains the endogenous 5′UTR and ATG of H2AE, which are bound by the MOs, this result strongly suggest that the endogenous mRNA is also susceptible to MO-mediated block of translation. Moreover, one of the MO we used to knock-down H2AE was directed primarily against the 5′UTR of H2AE (H2AE2 MO), which is not conserved among histone variants. Thus we have minimized potential cross reactivity with other histone variants. Finally, the specific role of H2AE was further supported by the finding that treatment of the cells with MOs against 3 other H2 variants did not show any effect on the cell number (Fig S4B).
Fig. 4. Knock down of H2AE leads to increased apoptosis in T cells.
(A) Growth curve of JSL1 T cells transfected with the indicated Morpholino-oligomers (MOs). Data represent mean values of three independent transfections. (B) Stable cell lines were generated expressing either wt H2AE-Flag or a version with several silent point mutations (MO-R) in the region targeted by the MO. These cells were transfected with the H2AE MO and expression of H2AE-Flag was analyzed by Western blotting. (C) Cell cycle distribution of JSL1 cells 48h after transfection with the indicated MOs as assessed by BrdU/7-AAD staining. Shown is the percentage relative to control (untransfected) cells (n=3). (D) AnnexinV staining of cells as in C). (E) Cells transfected with the H2AE MO were counted at the indicated time points and expressed as percent of control MO transfected cells of the same genotype (n=3). (F) Apoptosis was analyzed by Annexin V staining 48h post MO transfection.
We next investigated the cause for the reduction of viable cells by the H2AE MO by staining with BrdU and 7-AAD, which label newly synthesized and total DNA respectively to indicate cell cycle. This analysis revealed only a small alteration in cell cycle distribution but a significant increase in cell death (Fig. 4C). This was confirmed by AnnexinV staining which showed an increase in apoptosis by both H2AE specific MOs (Fig. 4D, S4C). To further verify a specific effect of H2AE, we performed a rescue experiment by generating cell lines stably expressing wt H2AE-Flag or a version that was rendered resistant to MO treatment (MO-R) by several silent point mutations. We obtained clones with similar expression levels and confirmed knock-down of the wt but not the mutant H2AE after MO treatment (Fig. 4B, S4D). Expression of the MO-resistant H2AE led to a partial rescue of viability upon MO treatment as observed by an increased number of viable cells as well as a reduction in apoptosis when compared to either the parental T cell line or the clone overexpressing wt H2AE (Fig 4E,F).
In summary, knock-down of H2AE in the JSL1 cells specifically decreases the number of viable cells and increased apoptosis. This phenotype exactly mimics the phenotype we observe in the PSF-deficient thymus in which expression of H2AE is reduced. We therefore conclude that the increased apoptosis and decreased cell number in the thymus of mice with reduced expression of PSF is at least partly mediated by the loss of H2AE expression. Importantly, these results, along with other specificity controls above, strongly argue for a non-redundant role of H2AE in determining cell viability. While future studies will be required to understand the mechanistic basis of the essential role of H2AE, our results provide an important precedent for gene-specific functions within the histone gene repertoire.
Finally, our data demonstrate that PSF regulates the stability of a subset of histone genes. The concerted regulation of several histone mRNAs suggests a common mechanism that enables PSF to control expression levels of several related genes at the same time. While we cannot rule that PSF functions indirectly to control the expression of H2AE, H3F and H4H, given that PSF has been shown to alter 3′ end processing and stability of canonical poly-adenylated mRNAs [14; 15], a direct role for this protein in histone processing and stability seems reasonable. Indeed, many of the factors involved in the processing and stability of the replication-dependent histone genes also have known activities in the stability and processing of poly-adenylated transcripts [4]. Moreover, we observe no significant cell-cycle alterations or defects in the PSF-deficient thymocytes which might indirectly influence histone mRNA expression. Finally, a recent analysis of RNAs that co-precipitate with PSF identified several histone mRNAs [16] suggesting that PSF may bind directly to these mRNAs, although an indirect interaction remains possible. Although determining the precise mechanism by which PSF may control the expression of replication-dependent histones is beyond the scope of this present study, this is clearly a potentially exciting aspect of PSF and histone activity that merits future investigation.
Supplementary Material
Highlights.
- The multifunctional RNA processing factor PSF is essential for cell viability
- T cell-specific knock down of PSF causes loss of thymic cellularity via apoptosis
- PSF-depleted thymocytes have a specific reduction in a subset of histone mRNAs
- Reduced expression of histone cluster 1 H2AE specifically induces apoptosis
Acknowledgements
We thank Robert Hammer, Mercy Gohil and Gary Koretzky for assistance with the generation and maintenance of the mouse lines. The mouse work done under protocol #2008-0216 from UT Southwestern Medical Center and #802852 from the University of Pennsylvania. This work was supported by R01 GM067719 and start-up funds from both UT Southwestern Medical Center and the University of Pennsylvania to K.W.L. F.H. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 2.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–98. [PubMed] [Google Scholar]
- 3.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biol Chem. 2008;389:333–43. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 4.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–54. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–14. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 6.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236:1347–57. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 7.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40:126–37. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao MK, Wilkinson MF. Tissue-specific and cell type-specific RNA interference in vivo. Nat Protoc. 2006;1:1494–501. doi: 10.1038/nprot.2006.260. [DOI] [PubMed] [Google Scholar]
- 9.Lynch KW, Weiss A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–84. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyd F, ten Dam G, Moroy T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat Immunol. 2006;7:859–67. doi: 10.1038/ni1361. [DOI] [PubMed] [Google Scholar]
- 12.Mallory MJ, Jackson J, Weber B, Chi A, Heyd F, Lynch KW. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol Cell Biol. 2011;31:2184–95. doi: 10.1128/MCB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. Rna. 2008;14:2038–49. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3' processing and transcription termination. Genes Dev. 2007;21:1779–89. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR. Rna. 2007;13:1103–15. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa A, Kotani H, Toda Y, Mazan-Mamczarz K, Mueller EC, Otto A, Disch L, Norman M, Ramdasi RM, Keshtgar M, Gorospe M, Fujita Y. Novel roles of hakai in cell proliferation and oncogenesis. Mol Biol Cell. 2009;20:3533–42. doi: 10.1091/mbc.E08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.