Abstract
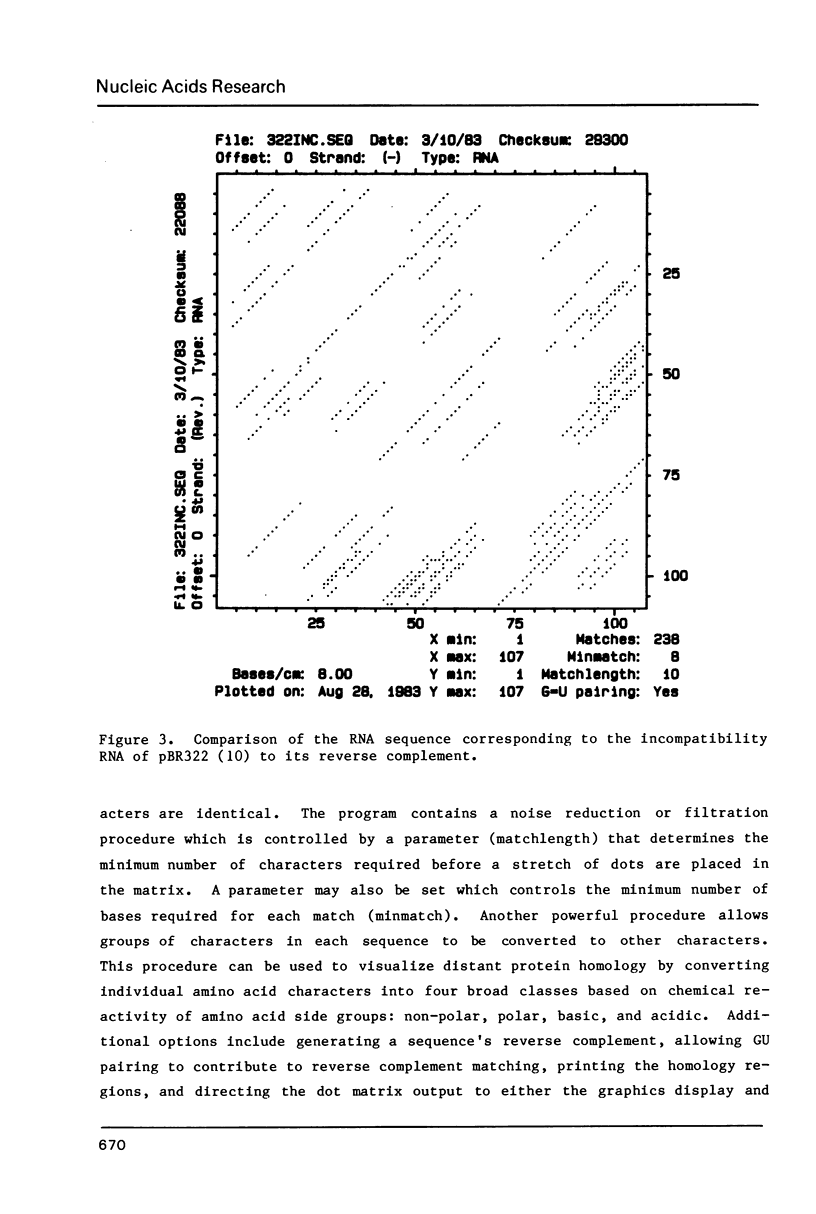
A group of interactive computer programs have been developed which aid in the collection and graphical analysis of nucleotide and protein sequence data. The programs perform the following basic functions: a) enter, edit, list, and rearrange sequence data; b) permit automatic entry of nucleotide sequence data directly from an autoradiograph into the computer; c) search for restriction sites or other specified patterns and plot a linear or circular restriction map, or print their locations; d) plot base composition; e) analyze homology between sequences by plotting a two-dimensional graphic matrix; and f) aid in plotting predicted secondary structures of RNA molecules.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Funnell B. E., Inman R. B. Comparison of partial denaturation maps with the known sequence of simian virus 40 and phi X174 replicative form DNA. J Mol Biol. 1979 Jun 25;131(2):331–340. doi: 10.1016/0022-2836(79)90079-2. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Rice P., Roberts R. J. A semi-automated method for the reading of nucleic acid sequencing gels. Nucleic Acids Res. 1982 Jan 11;10(1):103–114. doi: 10.1093/nar/10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A. A program for reading DNA sequence gels using a small computer equipped with a graphics tablet. Nucleic Acids Res. 1982 Jan 11;10(1):27–30. doi: 10.1093/nar/10.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Frelinger J. G., Fisher D., Hunkapiller T., Pereira D., Weissman S. M., Uehara H., Nathenson S., Hood L. Three cDNA clones encoding mouse transplantation antigens: homology to immunoglobulin genes. Cell. 1981 Apr;24(1):125–134. doi: 10.1016/0092-8674(81)90508-0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


