Abstract
The protein kinase CK2 (formerly casein kinase II) is thought to be involved in light-regulated gene expression in plants because of its ability to phosphorylate transcription factors that bind to the promoter regions of light-regulated genes in vitro. To address this possibility in vivo and to learn more about the potential physiological roles of CK2 in plants, we transformed Arabidopsis with an antisense construct of the CK2 α-subunit gene and investigated both morphological and molecular phenotypes. Antisense transformants had a smaller adult leaf size and showed increased expression of chs in darkness and of cab and rbcS after red-light treatment. The latter molecular phenotype implied that CK2 might serve as one of several negative and quantitative effectors in light-regulated gene expression. The possible mechanism of CK2 action and its involvement in the phytochrome signal transduction pathway are discussed.
One of the most important mechanisms in signal transduction is protein phosphorylation. This reaction is mediated by several groups of protein kinases in the cell and controls cellular processes that determine the state and rate of cell growth, metabolism, and differentiation (Krebs and Beavo, 1979; Trewavas and Gilroy, 1991; Hunter, 1995; Stone and Walker, 1995). Light signals that induce changes in growth and development also stimulate phosphorylation in plant cells (Datta et al., 1985; Otto and Schäfer, 1988; Reymond et al., 1992; Biermann et al., 1994; Fallon and Trewavas, 1994; Tong et al., 1996).
CK2 (formerly casein kinase II) is one of the major multifunctional protein kinases in cells. It recognizes Ser/Thr residues situated in an acidic environment in the substrate (X-S/T-X-X-E/D) (Pinna, 1990; Litchfield and Lüscher, 1993; Allende and Allende, 1995). This enzyme occurs ubiquitously and is essential for survival (Padmanabha et al., 1990). It is involved in the control of DNA replication and transcription, RNA processing and translation, cell metabolism, and motility of cells (Litchfield and Lüscher, 1993). The activity of CK2 is not directly affected by any known secondary messenger, but it can be stimulated or inhibited by extracellular signals (Tuazon and Traugh, 1991). To understand the regulation and functional significance of CK2, it is necessary to identify downstream and/or upstream components of the signaling pathways involving this kinase.
CK2 is composed of two α-subunits (α and α′) and two autophosphorylated β-subunits (Tuazon and Traugh, 1991). Their primary and quaternary structures are highly conserved and are readily evident in the structure of CK2 genes from a variety of organisms (Wirkner et al., 1992). All CK2s have several biochemical characteristics in common: a high sensitivity to polyanions such as heparin, the ability to use GTP as well as ATP as phosphoryl donors, a preference for acidic substrates such as phosvitin and casein, and being stimulated by polyamines (for review, see Pinna, 1990).
Several laboratories are trying to identify the signal transduction pathways that are initiated by light. One of the key issues in this field is to find the physiological significance of phosphorylation/dephosphorylation events in the light-triggered signal transduction pathway. In this context, CK2s are of particular interest, not only because of increasing recognition of the major role of protein phosphorylation in regulating plant metabolism but also because these kinases have been specifically implicated in the phosphorylation of trans-acting factors that regulate gene expression (Roux, 1994).
Phosphorylation of the transcription factors AT-1 and ATBP-1 by CK2 was thought to inhibit their binding to AT-rich regions because this binding was decreased in nuclear extracts treated with Mg2+ and ATP or GTP (Datta and Cashmore, 1989; Tjaden and Coruzzi, 1994; for review, see Terzaghi and Cashmore, 1995). In another study, dephosphorylation reduced G-box binding by GBFs, and this binding could be restored by phosphorylation by CK2 (Klimczak et al., 1992). This result was confirmed by in vitro studies using recombinant GBF1 and CK2 reconstituted from recombinant plant catalytic subunits and regulatory subunits (Klimczak et al., 1995) and by inhibitor studies using protein kinases and phosphatase inhibitors that affected the activity of cytosolic GBFs in parsley (Harter et al., 1994). However, there is no in vivo evidence, to our knowledge, that the phosphorylation of transcription factors affects the expression of light-regulated genes.
Other evidence for phosphorylation of a transcription factor by CK2 can be found in maize seed development (Ciceri et al., 1997). A basic Leu-zipper transcriptional activator, O2 (Opaque 2), regulates the expression of a storage protein zein that is the major product during endosperm development. This transcription factor can be phosphorylated by CK2, and its dephosphorylated and hypophosphorylated forms are able to bind to the zein promoter. The activity of the hyperphosphorylated form of O2 is increased during the night and decreased during the day, indicating that light may regulate the phosphorylating activity of CK2.
This report investigates the regulation and function of CK2 in plants, with a focus on further clarifying its role in gene regulation by light. As a step toward accomplishing this goal, we transformed Arabidopsis with an antisense construct of one of the CK2 α-subunit genes, ATHCK2A1. This allowed physiological and phenotypic comparisons between the transformants and wild-type plants and provided further evidence for the involvement of CK2 in the regulation of light-modulated genes.
MATERIALS AND METHODS
Arabidopsis Strain and Growth Conditions
Arabidopsis ecotype Wassilewskija was used for transformation and for the physiological investigations carried out on independent mutant lines. Arabidopsis seeds were surface sterilized in 70% ethanol for 30 s, transferred to 20% bleach solution for 15 min, and washed five times with sterile water. Seeds were sown on germination medium (Valvekens et al., 1988) and vernalized for 2 to 3 d at 4°C in foil-wrapped plates. They were then irradiated with white light for 2 h and transferred to darkness for germination at 25°C. For phenotypic investigation in soil, sterilized seeds were sown in a 10-cm pot containing soil (Sunshine mix no. 1, Sun Gro Horticulture, Bellevue, WA) purchased from a local market.
Construction of Recombinant ATHCK2A1 Antisense Binary Vector
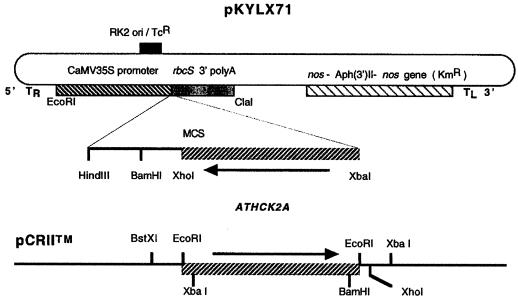
An ATHCK2A1 antisense insert was PCR amplified using a sense primer (5′-CATGCATCTAGAATGTCGAAAGCTCGTGTTTA- 3′; the XbaI site is underlined) and an antisense primer (5′-ACTAGGGATCCGCCGCAGTTAATCTGTCTTG-3′) to span the coding region of the cDNA. The template, ATHCK2A1 in pZL, was obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus; clone identification no. 20C12T7) and was identical to the gene that was cloned by Mizoguchi et al. (1993). The amplified product was gel-purified and subcloned into a TA-cloning vector (pCRII, Invitrogen, Carlsbad, CA). The orientation of the subcloned PCR product was checked by restriction analysis using XbaI and XhoI. The appropriate recombinant plasmid was cut with XbaI and XhoI, and the ATHCK2A1 fragment was ligated into XbaI and XhoI cut in pKYLX71. pKYLX71 is a derivative of pKYLX7 (Schardl et al., 1987). This cloning results in an antisense orientation of ATHCK2A1 under the transcriptional regulation of the cauliflower mosaic virus 35S promoter in a T-DNA-based plant-transformation vector. The construct of the fusion vector is diagrammed in Figure 1. This plasmid and a vector control (the plasmid minus the ATHCK2A1) were transformed into Agrobacterium tumefaciens by electroporation (Bio-Rad), and the positive clones were screened by kanamycin (100 mg/mL) resistance. The presence of the insert was verified by PCR (data not shown).
Figure 1.
Structure of the ATHCK2A antisense chimeric gene used to transform Arabidopsis. The PCR product amplified from the coding region of the ATHCK2A cDNA was inserted into pCRII (lower diagram). The 5′ primer had an XbaI site for further cloning. The recombinant pCRII vector was cut with XbaI and XhoI, and the insert was ligated into a similarly cut binary vector, pKYLX71 (upper diagram). This process gave the antisense orientation of ATHCK2A under the control of the cauliflower mosaic virus (CaMV) 35S promoter. MCS, Multiple cloning sites; nos, nopaline synthase gene; KmR, kanamycin-resistance gene; TcR, tetracyclin-resistance gene; RK2 ori, bacterial replication origin; TR and TL, right and left border of T-DNA, respectively.
Transformation of Arabidopsis
A. tumefaciens-mediated transformation was performed as described by Valvekens et al. (1988). Segregation of the T-DNA was checked by sowing T2 seeds on germination medium plates containing 50 mg/mL kanamycin. At this stage homozygous lines were identified. T3 seeds from the homozygous lines were harvested and used in the experiments.
Isolation of Genomic DNA and Southern-Blot Hybridization
Total Arabidopsis DNA was isolated as described by Dellaporta (1994). Isolated genomic DNA was cut with HindIII, loaded on a 0.8% (w/v) agarose gel, treated with 0.25 m HCl, and transferred to a Zeta-Probe membrane (Bio-Rad). After the blotted membrane was treated with a prehybridization solution (0.5 m sodium phosphate, pH 7.2, and 7% SDS), the blot was hybridized with radiolabeled cauliflower mosaic virus 35S promoter, which was excised from pBI221 (Clontech, Palo Alto, CA) for 18 h at 65°C. After hybridization the membrane was washed with 40 mm sodium phosphate buffer, pH 7.2, containing 5% SDS, and then washed with the same phosphate buffer containing 1% SDS at 65°C for 30 min. After the membrane was washed, it was exposed to radiographic film (Kodak) at −70°C.
Isolation of RNA and Northern-Blot Hybridization
Total RNA from 2 to 4 g of frozen, 5-d-old, etiolated and/or red-light-treated wild-type and transformed seedlings was extracted according to the method of Ausubel et al. (1987) with slight modification. Tissues were ground in TLE buffer (0.18 m Tris, pH 8.2, 0.09 m LiCl, and 4.5 mm EDTA) containing 1% SDS and an equal amount of TLE-saturated phenol at 65°C. After one extraction with phenol, one with phenol:chloroform (1:1, v/v), and one with chloroform, RNA was precipitated with ethanol and 0.1 volume of 3.3 m sodium acetate, pH 5.5, at −20°C overnight. RNA was pelleted by centrifugation at 18,000 rpm at 4°C, the pellet was resuspended in 500 μL of diethyl pyrocarbonate-water, and DNA and carbohydrate contamination were removed by precipitating the RNA in 2 m LiCl (final concentration) at −20°C overnight. After it was centrifuged at 18,000 rpm for 15 min, the RNA was washed with 80% ethanol and finally resuspended in diethyl pyrocarbonate-water at a concentration of about 4 mg/mL.
RNA (20 μg) was denatured in 50% formamide, 5.9% formaldehyde, and Mops buffer (20 mm Mops, pH 7.0, 10 mm sodium acetate, and 3 mm EDTA) at 65°C for 10 min and fractionated on a 1.2% agarose gel containing 2.2 m formaldehyde in Mops buffer. To ensure that equal amounts of RNA were loaded in each well, ethidium bromide was included in the RNA-loading buffer (50% glycerol, 1 mm EDTA, pH 8.0, 0.25% bromphenol blue, and 0.25% xylene cyanol FF) or hybridized with radiolabeled rDNA probes after transfer. After checking for equal loading, the RNA was transferred to a Zeta-Probe membrane (Bio-Rad) in 10× SSC (1.5 m NaCl and 0.15 m sodium citrate). The bound RNA was cross-linked using a Stratalinker (Stratagene).
The RNA-bound membrane was hybridized with the 32P-labeled coding region of each cDNA. Labeling was performed by a random-primed method using a DNA-labeling kit (DECAprime, Ambion, Austin, TX) and [α-32P]dATP.
After a 2-h prehybridization (0.5 m Na2HPO4, pH 7.2, and 7% SDS), hybridization with labeled probe was carried out for 18 h at 60°C in a buffer identical to the prehybridization solution. After hybridization, washes were in 25 mm Na2HPO4, pH 7.2, containing 5% SDS at 60°C for 1 h, and then in the same phosphate buffer including 1% SDS at 60°C for 1 h. The washed membrane was exposed to radiographic film at −70°C with intensifying screens. The developed film was scanned into NIH Image software (National Institutes of Health, Bethesda, MD), and the density of the bands was measured.
For northern-blot analyses using riboprobes, the probes were synthesized by PCR. For the CK2 α-subunit, a 928-bp riboprobe was synthesized using a sense primer (5′-TAATACGACTCACTATAGGGATGTCGAAAGCTCGTGTTTA-3′; the T7 promoter site is underlined) and an antisense primer (5′-ATTGAATTTAGGTGACACTATAGGCAGTTAATCTGTCTTG-3′; the SP6 promoter site is underlined), the region of which is the same as that of the antisense construct. T7 DNA polymerase was used for making antisense-strand-specific CK2α riboprobe, and SP6 DNA polymerase was used for synthesizing sense-strand-specific CK2α riboprobe. For cdc2, a 511-bp riboprobe (from Glu-41 to Gln-213) was made using a sense primer (5′-AAGGTGTTCCTAGCACAGCAA-3′, EGVPSTA) and an antisense primer (5′-GAATCCATTGAATTTAGGTGACACTATAGGATCAATCTCGGAGTCTCC-3′, GDSEIDQ; the SP6 promoter region is underlined). Riboprobes were synthesized using a DNA probe synthesis and removal kit (Strip-EZ, Ambion) using T7 or SP6 polymerase and [α-32P]UTP (800 mCi/mL).
Northern-blot transfer was performed as described above, and the hybridization was done following the Zeta-Probe membrane manufacturer's manual. In this experiment, 50% formamide was included in the prehybridization solution, and prehybridization was performed at 60°C for 18 h. Washing was performed using 2× SSC, 0.5× SSC including 0.1% SDS at room temperature and 0.1× SSC including 0.1% SDS at room temperature for CK2 and at 60°C for cdc2. After it was washed, the blot was exposed to a phosphor imager (Molecular Dynamics, Sunnyvale, CA) screen for 1 week.
Dot-Blot Analysis
cDNA inserts of ATHCK2A1 (SalI/XbaI cut, 1.2 kb), CK2B1 (SalI/XbaI cut, 1.1 kb), cdc2a (EcoRI cut, 1.4 kb), clone 2F1T7P (SalI/XbaI cut, 0.7 kb), and clone CD3-18 (EcoRI cut, 0.63 kb) were loaded onto a Zeta-Probe membrane using a dot-blotting apparatus (Bio-Dot, Bio-Rad). The dotted membrane was prehybridized with prehybridization solution (0.5 m NaH2PO4, pH 7.2, 7% SDS, and 50% formamide) and probed with radiolabeled ATHCK2A1 cDNA for 18 h. The probe was synthesized using a random-primed labeling kit (DECAprime, Ambion). After hybridization, the membrane was washed (40 mm NaH2PO4, pH 7.2, including 5% SDS, and 40 mm NaH2PO4, pH 7.2, including 1% SDS, subsequently) at 65°C for 15 min for each wash.
Probes Used for Northern- and Dot-Blot Analyses
The following expressed-sequence-tag clones were obtained from the Arabidopsis Biological Resource Center: ATHCK2A1 (stock no. 20C12T7; accession no. T04132), cab (stock no. 35G10T7; accession no. T04281), chs (stock no. YAP097T3; accession no. T04269), rbcS (stock no. 88G20T7; accession no. T20583), CKB1 (accession no. L22563; Collinge and Walker, 1994), CDC2A (stock no. CD3-83; accession no. M59198; Ferreira et al., 1991), Arabidopsis protein kinase homolog (stock no. 2F1T7P; accession no. T04181), plant-specific protein kinase (stock no. CD3-18; partial sequence of clone AtKin7; accession no. U79744), and rDNA repeat (stock no. CD3-197; clone identification no. JHD2-15A).
Measurement of Leaf Area and Length of Hypocotyls and Roots
Round juvenile leaves in the rosette and cauline early-adult leaves (both diagrammatically illustrated in Fig. 5) from 21-d-old light-grown plants were measured. Leaves were taken from plants, attached to white cardboard with transparent tape, and scanned for computer analysis using Adobe Photoshop (version 3.0, Adobe Systems, Mountain View, CA) and NIH Image software to calculate the leaf area from a trace of perimeters. Each measurement involved an average of at least 15 plants. The statistical significance of differences was determined by the Student's t test.
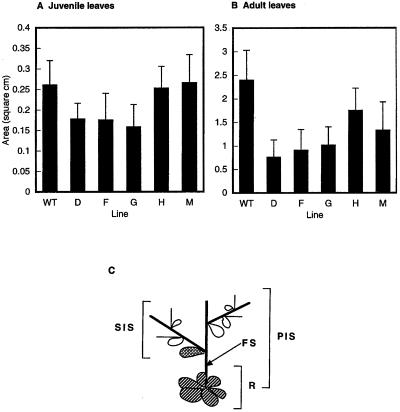
Figure 5.
Leaf size in Arabidopsis ATHCK2A1 antisense transformants. A, Measurement of juvenile leaves of the wild-type (WT) and antisense transformants (D, F, G, H, and M). B, Measurement of adult leaves of the wild type and antisense transformants taken from 21-d-old plants grown under continuous white light. Data are means of more than 15 plants of each line. Error bars show +sd of measurements. C, Schematic representation of the lower part of an Arabidopsis plant. A cauline leaf (cross-hatched) between the floral stem (FS) and the secondary inflorescence stem (SIS) was taken as an adult leaf. Juvenile leaves (striped) were taken from the rosette (R). PIS, Primary inflorescence stem. The leaves from plants transformed with the plasmid vector only were indistinguishable from wild-type leaves with regard to size.
To measure the length of the hypocotyl and root, 5-d-old etiolated seedlings were used. These seedlings were also treated with red light to test for phytochrome effects. For the hypocotyl-elongation experiment, Suc was deleted from the germination medium to maximize the effect of red light. Hypocotyl length was measured with a metric ruler. At least 40 seedlings were measured for each data point.
Light Treatment
Five-day-old etiolated seedlings were irradiated with red light (1 μmol m−2 s−1) for 15 min. For the red/far-red-light reversibility test, the same intensity of red light was given for 5 min, and the far-red fluence was 7 μmol m−2 s−1 for 1 min. After light was given, seedlings were returned to the dark and harvested at various times for other experiments. Seedlings were quickly frozen with liquid nitrogen in total darkness to eliminate the possibility of the green-light effect and were then stored at −70°C. For raising plants under white light, four fluorescent lightbulbs were used at a total intensity of 150 μE m−2 s−1.
CK2 Activity Assay
Crude extract for the assay was prepared by homogenizing 21-d-old green plants in a homogenization buffer (40 mm Hepes, pH 7.4, 15 mm MgCl2, 1 mm EDTA, 10 mm β-mercaptoethanol, 1 mm PMSF, 30 mm p-nitrophenyl phosphate, and 40 mm β-glycerophosphate). The homogenate was centrifuged at 8000 rpm for 15 min. Protein concentration in the resulting supernatant was measured in triplicate with a protein assay reagent (Bio-Rad) using BSA as a standard.
The CK2 activity assay was carried out according to the method of Litchfield et al. (1990). Crude extract (10 μL) was incubated at room temperature or 37°C for 10 min with 0.73 mm peptide substrate RRREEETEEE (Promega) in 20 μL of kinase buffer (100 mm Tris-HCl, pH 7.6, 20 mm MgCl2, 150 mm NaCl, 5 mm NaF, and 0.1 mm [γ-32P]GTP; 550 cpm/pmol). After the reaction, the reaction mixture was spotted onto a P81 membrane (Whatman), washed with 75 mm phosphoric acid five times, and dried in the air. The dried membranes were put into scintillation fluid, and their bound radioactivity was measured with a liquid-scintillation counter.
Western-Blot Analysis
Protein samples were the same as those used for the protein kinase assay. Thirty micrograms of total crude extract was loaded onto a 12% SDS-PAGE gel and electrophoresed. After electrophoresis, fractionated proteins were transferred to a nitrocellulose membrane. The membrane was blocked with 3% Blotto in PBS for 2 h at room temperature and probed with 1:200 rabbit anti-human cdc2 kinase polyclonal antibody (Upstate Biotechnology, Lake Placid, NY) directed to the PSTAIRE region overnight at 4°C. After the blot was probed with the primary antibody, it was washed with 3% Blotto in PBS three times for 10 min each and probed with horseradish peroxidase-conjugated goat anti-rabbit IgG (Kirkegaard and Perry Laboratory, Gaithersburg, MD; 1:2000) for 2 h at room temperature. After the blot was washed two times with PBS and two times with PBS plus 0.05% Tween 20 for 15 min each, chemiluminescence of the signal was developed using a kit (ECL, Amersham) and the blot was exposed to radiographic film.
RESULTS
Antisense Transformation of the CK2 α-Subunit Gene in Arabidopsis
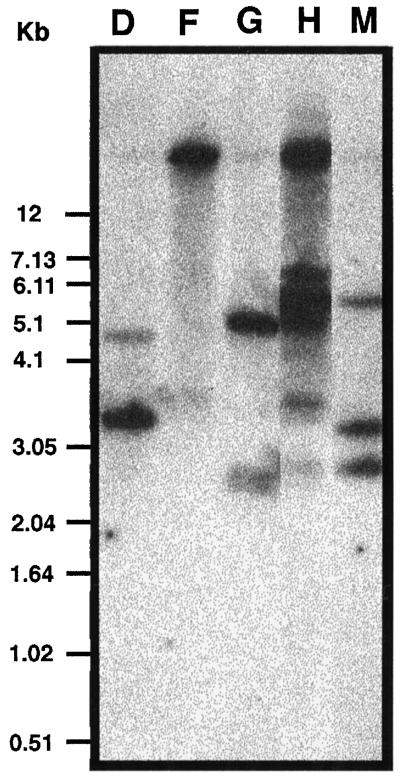
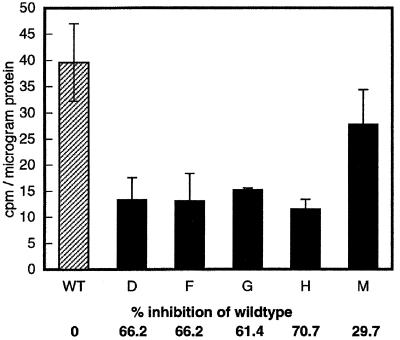
Fourteen CK2α antisense transgenic lines were produced. Southern-blot hybridization revealed that 5 of the 14 lines had independent insertions (Fig. 2). Both 21-d-old light-grown plants and etiolated 5-d-old seedlings showed overexpression of the antisense CK2α RNA as measured by northern-blot analysis using an antisense-specific riboprobe (data not shown). To assess the effect of the antisense expression, we performed a CK2 kinase assay using a CK2-specific peptide substrate and radiolabeled GTP to maximize the specificity of the kinase reaction. Except for line M, all of the antisense transgenic lines showed more than 60% inhibition of kinase activity compared with the wild type (Fig. 3). None of the transgenic lines showed complete loss of activity.
Figure 2.
Southern-blot hybridization of genomic DNA from antisense ATHCK2A1 transformants. Genomic DNA was digested with HindIII, separated by electrophoresis, transferred to a nylon membrane, and hybridized with a 32P-labeled cauliflower mosaic virus 35S promoter region cut from pBI121 (Clontech). D, F, G, H, and M, Five different antisense transformants.
Figure 3.
Results of CK2 activity test. The phosphotransferase activity of the wild-type (WT) and antisense transformant lines (D, F, G, H, and M) was measured using a synthetic peptide RRREEETEEE (0.73 mm) and radiolabeled GTP as substrates. Two independent assays were performed for each line and each kinase assay was carried out with triplicate samples. The data shown are calculated average values ± sd.
Transformants Express Wild-Type Levels of cdc2 mRNA and Protein
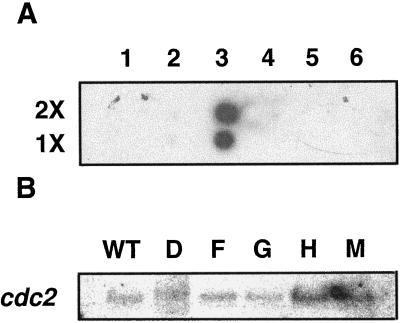
The results shown in Figure 4 address the question of the specificity of the antisense RNA of CK2α and whether antisense transformation results in the suppression of the expression of a closely related protein kinase. The CK2 α-subunit gene did not bind to cDNA inserts of other kinases (Fig. 4A). The endogenous level of mRNA for cdc2, a protein kinase very closely related to CK2 (Hanks and Hunter, 1995), was not significantly changed in the transformants compared with the wild type (Fig. 4B). More importantly, when the same protein samples used in the CK2 activity assay were assayed by immunoblot using antihuman CDC2 kinase (a polyclonal antibody raised against the PSTAIRE region), there was no significant change in the amount of signal between the wild type and the transformants (data not shown).
Figure 4.
Dot-blot analysis of ATHCKA1 binding to cDNA inserts of other protein kinases (A) and northern-blot analyses of cdc2 in wild type (WT) and the transformants (D, F, G, H, and M) (B). A, Either 100 ng (1×) or 200 ng (2×) of the 1.2-kb ATHCK2A1 cDNA insert (column 3, second lane) was loaded, and the amount of other DNAs (the number of molecules) was adjusted according to their size. As a negative control, the cDNA insert of CK2B1 and the plasmid 100 ng of pUC18 was loaded. Lane 1, CK2B1 insert (1.1 kb, SalI/XbaI cut); lane 2, pUC18; lane 3, CK2A1 insert (1.2 kb, SalI/XbaI cut); lane 4, cdc2a (1.4 kb, EcoRI cut); lane 5, clone 2F1T7P Arabidopsis protein kinase homolog (0.7 kb, SalI/XbaI cut); and lane 6, plant-specific protein-kinase clone CD-18 (0.63 kb, EcoRI cut). B, Northern-blot analyses of the expression of cdc2 kinase in the wild type and the transformants. Poly(A+) RNA was isolated from the total RNA extracted from 21-d-old light-grown plants and 0.5 μg was loaded in each lane. Riboprobes were synthesized as described in Methods. The probed blot was stripped off using a kit and reprobed.
Antisense Effect Seen in the Adult Leaves of Transformants
In a previous report by Mizoguchi et al. (1993), two Arabidopsis CK2 α-subunit genes were cloned and their expression was analyzed. Because the two genes are more than 97% identical, we reasoned that antisense RNA spanning the entire cDNA of ATHCK2A1 might effectively inhibit the expression of both CK2 α-subunit genes. Because the CK2 α-subunit was highly expressed in flower, root, and leaf tissues (Mizoguchi et al., 1993), we investigated their phenotype.
When the wild type and the transformants were grown under continuous white light for 21 d, flowers showed no significant differences between the transgenic lines and the wild type, although the bolting time was slightly slower in the antisense transformants than in the wild type (data not shown). When we investigated the root growth of 5-d-old etiolated seedlings, we saw no significant difference in root length between the wild type and the antisense transformants.
There was a significant difference in the size of adult leaves (P < 0.05), but there was no difference in the size of juvenile leaves in the rosette (Fig. 5, A and B). We found no difference in the leaf shapes.
Effect of Red Light on Hypocotyl Elongation of Etiolated Seedlings
There was no significant difference between the wild type and the antisense transformants in hypocotyl elongation either in darkness or after red-light treatment. The degree of inhibition by red light was 50% in both the wild type and the antisense transformants (data not shown).
Differential Effect of Antisense Transformation on the Expression of Light-Regulated Genes
CK2 can phosphorylate Arabidopsis GBF and AT-1, and this phosphorylation affects their binding to their promoter sequences (Klimczak et al., 1992, 1995). The antisense approach enabled us to test for differences in the expression of light-regulated genes in the transformants, because the G-box and the AT-1 box are present in the promoter regions of light-regulated genes. If CK2 interacts with the GBF or the AT-1-box-binding factor in vivo, then antisense inhibition of ATHCK2A1 might affect phytochrome-regulated gene expression. To address this possibility, we investigated the effect of red light on cab, chs, and rbcS gene expression in the wild type and in the antisense trans-formants.
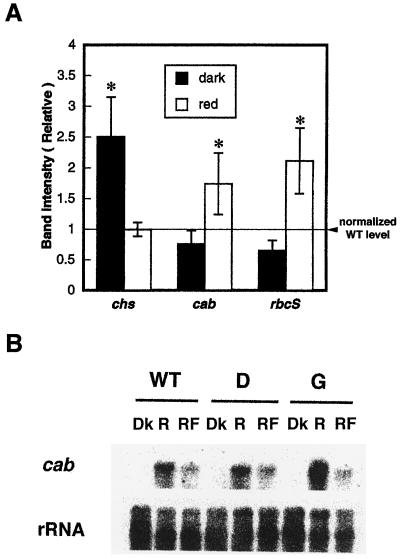
Figure 6A shows the relative mRNA abundance of all three genes tested in wild-type plants and in antisense transformants. The mean value of the band intensity of transformants was normalized to that of the wild type, vwith the band intensity of the wild type taken as 1 for the dark control and red-light treatment. The expression of all three light-regulated genes was increased by red light, both in the wild type and in the transformant lines (data not shown). However, the transformants showed two gene-expression changes: chs expression was significantly (P < 0.05 by Student's t test) increased in the dark control of the transformants, and after red-light treatment a significantly increased amount of cab and rbcS gene expression was seen in transformants over that of the wild-type control. The wild type and the transformants did not differ in their expression of chs after red-light treatment. The qualitative differences shown as averages in Figure 6A were also seen in each individual transformant line (data not shown).
Figure 6.
Effect of red light on cab, chs, and rbcS gene expression in wild-type and antisense lines. A, Quantitation and normalization of cab, chs, and rbcS gene-expression levels. Density of the bands was measured using NIH Image software and normalized to the intensity of rRNA bands. The expression ratio of each transformant line was calculated with reference to the band intensity of the wild type for each light condition after normalizing for differences in RNA loading, and the mean value of the band intensity of transformants was then calculated. The dotted line shows the value of the wild-type control (1) for dark and red-light treatments. Asterisks indicate that the mean value of gene expression is statistically different (P < 0.05) from that of the wild-type control. B, Red/far-red-light reversibility test of cab expression in the wild type and in the transformants. Five-day-old dark-grown seedlings were frozen at time 0 in the dark (Dk) or were treated with 5 min of red light (R) (1 μmol m−2 s−1) or 5 min of red light plus 1 min of far-red light (RF) (7 μmol m−2 s−1) at time 0 and returned to the dark. After 4 h light-treated seedlings were harvested and transferred to liquid N2 in the dark. The two durations of red light given (15 and 5 min) were both above the level needed to fully induce the response (Karlin-Neumann et al., 1988).
Reversibility by Far-Red Light of the Red-Light Effect in the Antisense Transformants
To rule out the possibility that the increase or decrease of light-regulated genes in the antisense transformants was caused by a secondary effect of reduced photosynthesis resulting from a defect in red-light-stimulated chloroplast development and chlorophyll production, the red/far-red reversibility of light-regulated genes was tested. To further minimize an effect of photosynthesis on the results of this experiment, the intensity of red light used (300 μmol m−2) was lower than that used for the experiments shown in Figure 6A (900–1000 μmol m−2). Two transgenic lines were selected as representative lines. As shown in Figure 6B, the reversibility of red-light-induced cab expression was observed in both transformants. The expression pattern of the red-light-induced expression of cab was not qualitatively different from the previous result (Fig. 6A). The ineffectiveness of far-red light in reversing the effects of red light on chs expression was seen in both wild-type plants and transformants (data not shown).
DISCUSSION
Specificity of Antisense RNA of CK2α to CK2 Activity Suppression
Because protein kinases have 11 homologous subdomains in the catalytic domain (Hanks and Hunter, 1995) and we used the 928-bp coding region of the CK2 α-subunit gene, we were concerned that the antisense RNA might bind to the mRNA of other kinases and inhibit their expression. This possibility was rendered less likely by showing that the CK2 α-subunit gene did not bind to cDNA inserts of other kinases (Fig. 4A). We also tested this possibility by assaying the level of mRNA and protein for cdc2 kinase, evolutionarily one of the closest of all of the protein kinases to that of the CK2 α-subunit (Hanks and Hunter, 1995). The fact that both the mRNA and protein levels for cdc2 in the transformants were indistinguishable from their levels in wild-type plants argues for the specificity of the antisense phenotypes observed, i.e. they are most plausibly attributed to suppression of CK2 activity rather than to any cosuppression of other related protein kinases.
Potential Involvement of CK2 in Leaf Growth
Leaf development is one of the most dramatic of the developmental changes that occur in the deetiolation processes of dicotyledonous plants and is the result of both cell expansion and cell division (Neff and van Volkenburgh, 1994). CK2 is known to be involved in cell division and differentiation in animal systems (Pinna, 1990; Ralph et al., 1990). Overexpression of CK2 in animal cells results in oncogenesis (Seldin and Leder, 1995), and CK2 is phosphorylated by cdc2 kinase (Mulner-Lorillon et al., 1990; Litchfield et al., 1992, 1995) and vice versa (Russo et al., 1992). Moreover, it is required for the transition of G0/G1, early G1, and G1/S phases in the cell (Pepperkok et al., 1994). Based on these findings in animals, one might predict that the severe reduction in CK2 activity in antisense transformants would affect leaf growth by reducing cell division and/or cell expansion in Arabidopsis. In comparing the CK2 antisense transformants with wild-type plants, we did not see any difference in leaf shape, but we found a significant difference in leaf size. This phenotype is similar to that of the dominant negative mutation of cdc2 kinase in tobacco (Hemerly et al., 1995).
Leaf tissue shows a high level of gene expression of the CK2 α-subunit gene (Mizoguchi et al., 1993), and here we show a high correlation between reduced kinase activity in the transformants and reduced leaf size. Both findings indicate an involvement of CK2 in leaf physiology. There have been recent reports stating that chloroplast ATP synthase and ribonucleoproteins are phosphorylated by CK2 in spinach chloroplasts in vivo and in vitro (Kanekatsu et al., 1995; Kanekatsu and Hiroshi, 1997), demonstrating the importance of CK2 in chloroplast function.
CK2 May Play a Role in the Repression of Light-Regulated Genes
Phytochrome is a major regulator of the deetiolation process in dicotyledonous seedlings. One of the key light-induced changes during deetiolation is an increase in the expression of a variety of genes (Furuya and Schäfer, 1996).
Gene expression of chs in the CK2α antisense transformants was derepressed in the dark and showed almost no difference after red-light treatment. On the other hand, amplified gene expression of cab and rbcS was seen after red-light treatment (Fig. 6). The fact that an increase in gene expression was seen in both dark and light conditions suggests that CK2 might act as a negative regulator of the expression of genes that are up-regulated by light.
Analysis of the changes in gene expression seen in the nonirradiated and the red-light-treated transformants suggests that CK2 may act differently for regulation of chs than for regulation of cab and rbcS expression. There are several reports that the phytochrome signal transduction pathways that lead to cab gene expression and chs gene expression are different (Neuhaus et al., 1993; Bowler et al., 1994a, 1994b). The pathway that leads to anthocyanin-biosynthesis gene expression is mediated by cGMP, and the other pathway, which affects cab, rbcS gene expression, and chloroplast development, requires calcium and calmodulin for its induction. PSI, Cyt b6f, and FNR (Fd NADP+ reductase) gene expression are induced by both pathways (for review, see Hiratsuka and Chua, 1997).
There are at least two possibilities to explain the changes in light-regulated gene expression in the transformants. One is the regulation of DNA-binding activity of the transcription factors by phosphorylation, and the other is the regulation of their nuclear localization by phosphorylation. The first possibility requires that CK2 primarily localizes in the nucleus and/or can be translocated into the nucleus in response to extracellular signals such as light. In animal systems CK2 is localized primarily in the nucleus (Krek et al., 1992; Gruppuso and Boylan, 1995; Penner et al., 1997), or it can be translocated into the nucleus by mitogenic signaling (Ahmed et al., 1993; Lorenz et al., 1993; Boulikas, 1996). Whether light affects CK2 localization in plants is not known.
CK2 is known to phosphorylate GBF1, and this factor is known to bind to the light-responsive elements of phytochrome-regulated genes such as cab and rbcS (Datta and Cashmore, 1989; Klimczak et al., 1992, 1995). The AT-1 box is on the promoter of numerous light-regulated promoters, including pea rbcS-3A and tobacco cab-E genes. The AT-1-box-binding factor is phosphorylated by CK2, and this phosphorylation decreases the AT-1-binding activity (Datta and Cashmore, 1989). However, high-mobility proteins also can bind to the AT-rich regions and can be phosphorylated by CK2 without this affecting their binding affinity to DNA (Klimczak and Cashmore, 1994).
G-box elements are present in the promoters of many genes that are induced by environmental factors such as light, auxin, hypoxia, ethylene, and drought (for review, see Daugherty et al., 1994; Menkens et al., 1995), and the role of these elements in mediating plant responsiveness to light has been reported previously (Block et al., 1990; Donald and Cashmore, 1990). cab, rbcS, and chs, which are up-regulated by phytochrome, have this element (Terzaghi and Cashmore, 1995). The presence of G-box-binding proteins that bind to the G-box in the promoters of chs, rbcS, and cab has been shown by in vitro and in vivo footprinting (Giuliano et al., 1988; Schulze-Lefert et al., 1989; Schindler and Cashmore, 1990; Manzara et al., 1993). One of the G-box-binding proteins that has been functionally identified is O2 in maize (Schmidt et al., 1992; Ueda et al., 1992; Izawa et al., 1993), and this protein is phosphorylated by CK2 in vitro (Ciceri et al., 1997). Phosphorylation of O2 is light dependent and under diurnal control (Ciceri et al., 1997). GBF1 of Arabidopsis can be phosphorylated by a nuclear extract from broccoli that has CK2-like activity, and its phosphorylation increases its G-box-binding activity (Klimczak et al., 1992). This finding has been confirmed in recombinant GBF1 and recombinant CK2 α- and β-subunits (Klimczak et al., 1995).
Results implicating CK2α in the regulation of transcription factors for light-regulated genes provide a framework for interpreting the transgenic results reported here. Because all three genes tested have a G-box element in their promoters, and because the G-box is commonly found within the 5′ upstream region of non-light-regulated genes, it may act as a quantitative element that modulates the expression level of light-responsive genes (Hiratsuka and Chua, 1997). The G-box element itself is not light specific, and an additional element is important for the appropriate response (Menkens et al., 1995).
Interaction between the G-box element and the I-box element in the Arabidopsis rbcS-1A gene is important for light responsiveness (Donald and Cashmore, 1990). The affinity of GBF1 for the G-box element is determined by the flanking sequences (Amstrong et al., 1992; Williams et al., 1992; Foster et al., 1994; Menkens and Cashmore, 1994), and the G-box element does not work solely during inducible gene expression (Daugherty et al., 1994; Terzaghi and Cashmore, 1995). Therefore, there is a high probability that another protein factor that is involved in light-regulated gene expression might be regulated by CK2 phosphorylation and may repress the light-regulated gene expression when it is in a phosphorylated form. This phosphorylated factor should be dephosphorylated by a phosphatase with activity that is activated by red light (or phytochrome). This speculation is consistent with the finding that protein phosphatase activity is required for light-inducible gene expression in maize (Sheen, 1993). To test this idea, it would be necessary to verify whether GBF or AT-1 is hypophosphorylated in the transformants or to find the factor responsible for the repression of chs gene expression in the dark.
Another possibility to be considered is whether the phosphorylation of a transcription factor or a retention factor by CK2 changes the localization of these factors. Gene expression can be regulated by controlling the nucleo-cytoplasmic partitioning of transcription factors (Whiteside and Goodbourn, 1993), and this partitioning can be modulated by the phosphorylation of the amino acids neighboring the nuclear-localization signal (Jans, 1995). For example, the nuclear uptake of the simian virus 40 large T-antigen is enhanced by phosphorylation of a Ser residue by CK2 (Rihs et al., 1991), which increases the binding of this protein to importins (Hübner et al., 1997).
All four GBFs described in the literature thus far have putative CK2-phosphorylation sites, and a deletion construct of GBF-1 that is missing one of these sites has increased nuclear localization (Terzaghi et al., 1997). In parsley cell cultures, protein kinase inhibitors and phosphatase inhibitors change the activity of a cytosolic GBF whose transport to the nucleus is stimulated by red light (Harter et al., 1994). Therefore, it is reasonable to consider a potential role for CK2 in regulating the nucleo-cytoplasmic partitioning of GBFs and thereby influencing gene expression.
To interpret the gene-expression changes observed in transgenic plants, two things should be pointed out. First, the measured abundance of cab and rbcS must be a cumulative expression of several genes of a family, because we used homologous cDNA probes for this experiment. In chs the major band shown by northern-blot analysis must reflect the expression of a single gene, because it has been found that chs is a single-copy gene in Arabidopsis (Feinbaum and Ausubel, 1988). However, it has been shown that the Arabidopsis rbcS gene family consists of four genes (Krebbers et al., 1988), and five genes have been reported for chlorophyll a/b-binding proteins in Arabidopsis (Leutwiler et al., 1986; Zhang et al., 1991). Genes of both the cab and rbcS families show differential regulation by light (Karlin-Neumann et al., 1988; Dedonder et al., 1993), and our results do not provide information about a specific promoter that is affected by the antisense transformation of the CK2 α-subunit. Second, our results give the steady-state level of gene expression; therefore, we cannot exclude the possibility that the changes observed may be attributable to a posttranscriptional control. Gene expression can be affected at a posttranscriptional level by light (Ernst et al., 1988; Jenkins, 1991; Wanner and Gruissem, 1991).
Other light-regulated DNA-binding activities have been found in tobacco and Arabidopsis. 3AF5 and 3AF3 have been found in tobacco, and their binding sites are located at the 5′ and 3′ ends of box III of the promoter region of the rbcS-3A gene (Sarokin and Chua, 1992). Gel-shift assays showed that DNA-protein complexes moved faster in dark-adapted plants than those from light-grown plants and that the mobility of complexes from light-grown plants was affected by alkaline phosphatase treatment. This finding supports the notion that rbcS gene expression can be regulated by phosphorylation of transcription factors, but there is no evidence regarding which kinase is responsible for this phosphorylation.
In Arabidopsis, CCA-1, which binds to the promoter of the Lhcb*3 gene (formerly cab 140), has been cloned and antisense transformants have been produced (Wang et al., 1997). The antisense transformants have a reduction in red-light-stimulated cab gene expression. CCA-1 is a myb-related protein and has an acidic C-terminal region that has potential CK2-phosphorylation sites. CK2 is an acidophilic kinase (Pinna, 1997) and is known to phosphorylate myb proteins in animals (Lüscher et al., 1990).
HY5, which is a basic Leu zipper DNA-binding protein, has a putative CK2-phosphorylation site (Oyama et al., 1997). The Hy5 mutant has been genetically identified by its insensitivity to the inhibition of hypocotyl elongation by light (Koornneef et al., 1980). It has defects in light-induced chlorophyll accumulation, root development, and the expression of cab expression affecting the circadian control (Ang and Deng, 1994; Anderson et al., 1997; Oyama et al., 1997). It will be useful to determine whether HY5 and CCA-1 are phosphorylated by CK2 and, if so, how this affects the expression of light-regulated genes.
CK2 RNA Suppression Does Not Affect the Reversibility of cab Expression by Far-Red Light
Because the reversibility of cab expression was not changed in the transformants (Fig. 6B), it is unlikely that the altered effects of red light on cab expression seen in the transformants were caused primarily by some defect in photosynthesis. This result also implies that CK2 influences the phytochrome signal transduction pathway only indirectly and that CK2 is not one of the main components in the pathway controlling the red/far-red reversible induction of gene expression. Earlier reports of a phytochrome-associated kinase (Wong and Lagarias, 1989; Wong et al., 1989) indicated that it had none of the characteristics of CK2. More likely, CK2 influences phytochrome signaling only indirectly, e.g. by modulating the activity of one or more components in the transduction pathway leading to the regulation of gene expression. Consistent with this speculation, one of the substrates of CK2 is calmodulin, which influences the expression of light-regulated genes (Lam et al., 1989; Sacks et al., 1992).
Antisense Transformation Affects Two Red-Light Responses Differently
Although the effect of red light on hypocotyl elongation was unchanged in the transformants, the regulation of genes by phytochrome in these plants was clearly altered. This implies that a reduction in the levels of CK2α in Arabidopsis can lead to an alteration in the expression of phytochrome-regulated genes without having much influence on the signal transduction pathways that control the hypocotyl elongation response to light. There is no direct evidence that these phytochrome responses have different signal transduction pathways. Nonetheless, such a conclusion is consistent with the finding that two phytochrome-related mutants, hy3 and hy5, show normal induction of cab expression by a brief red-light treatment (Sun and Tobin, 1990; Chory, 1992; Ang and Deng, 1994; Pepper and Chory, 1997), whereas the inhibition of hypocotyl elongation of both mutants is insensitive to red light (Koornneef et al., 1980; Reed et al., 1994).
Abbreviation:
- GBF
G-box-binding factor
Footnotes
This work was supported by grants from the National Science Foundation (no. DCB-9106245) and from the Institute of Cellular and Molecular Biology at The University of Texas, Austin.
LITERATURE CITED
- Ahmed K, Davis A, Hanten J, Lambert D, McIvor RS, Goueli SA. Cloning of cDNAs encoding the alpha and beta subunits of rat casein kinase 2 (CK-2): investigation of molecular regulation of CK-2 by androgens in rat ventral prostate. Cell Mol Biol Res. 1993;39:451–462. [PubMed] [Google Scholar]
- Allende JE, Allende CC. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- Amstrong GA, Weisshaar B, Hahlbrock K. Homodimeric and heterodimeric leucine zipper proteins and nuclear factors from parsley recognize diverse promoter elements with ACGT cores. Plant Cell. 1992;4:525–537. doi: 10.1105/tpc.4.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA. Attenuation of phytochrome A and B signalling pathways by the Arabidopsis circadian clock. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L-H, Deng X-W. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York
- Biermann BJ, Pao LI, Feldman LJ. Pr-specific phytochrome phosphorylation in vitro by a protein kinase present in anti-phytochrome maize immunoprecipitates. Plant Physiol. 1994;105:243–251. doi: 10.1104/pp.105.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Dangl JL, Hahlbrock K, Schulze-Lefert P. Functional borders, genetic fine structure, and distance requirements of cis-elements mediating light responsiveness of the parsley chalcone synthase promoter. Proc Natl Acad Sci USA. 1990;87:5387–5391. doi: 10.1073/pnas.87.14.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. Nuclear import of protein kinases and cyclins. J Cell Biochem. 1996;60:61–96. doi: 10.1002/(sici)1097-4644(19960101)60:1<61::aid-jcb10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994a;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Bowler C, Yamagata H, Neuhaus G, Chua N-H. Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev. 1994b;8:2188–2202. doi: 10.1101/gad.8.18.2188. [DOI] [PubMed] [Google Scholar]
- Chory J. A genetic model for light-regulated seedling development in Arabidopsis. Development. 1992;115:337–354. [Google Scholar]
- Ciceri P, Gianazza E, Lazzari B, Lippoli G, Ganga A, Hoschek G, Schmidt RJ, Viotti A. Phosphorylation of Opaque 2 changes diurnally and impacts its DNA binding activity. Plant Cell. 1997;9:97–108. doi: 10.1105/tpc.9.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge MA, Walker JC. Isolation of an Arabidopsis thaliana casein kinase II beta subunit by complementation in Saccharomyces cerevisiae. Plant Mol Biol. 1994;25:649–658. doi: 10.1007/BF00029603. [DOI] [PubMed] [Google Scholar]
- Datta N, Cashmore AR. Binding of a pea nuclear protein to promoters of certain photoregulated genes is modulated by phosphorylation. Plant Cell. 1989;1:1069–1077. doi: 10.1105/tpc.1.11.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, Chen YR, Roux SJ. Phytochrome and calcium stimulation of protein phosphorylation in isolated pea nuclei. Biochem Biophys Res Commun. 1985;128:1403–1408. doi: 10.1016/0006-291x(85)91096-4. [DOI] [PubMed] [Google Scholar]
- Daugherty CJ, Rooney MF, Paul A-L, de Vetten N, Vega-Palas MA, Lu G, Gurley WB, Ferl RJ. Environmental stress and gene regulation. In: Sommerville CR, Meyerowitz EM, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 769–806. [Google Scholar]
- Dedonder A, Rethy R, Frederiq H, van Montagu M, Krebbers E. Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol. 1993;101:801–808. doi: 10.1104/pp.101.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S. Plant DNA miniprep and microprep: versions 2.1–2.3. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 522–525. [Google Scholar]
- Donald RGK, Cashmore AR. Mutation in either G box or I box sequences profoundly affects expression from the Arabidopsis thaliana rbcS-1A promoter. EMBO J. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst D, Pfeiffer F, Eilfeld P, Rudiger W, Oesterhelt D (1988) Phytochrome regulation of mRNA levels of ribulose-1,5-bisphosphate carboxylase and light-harvesting chlorophyll a/b protein in etiolated rye seedlings (Secale cereale). In P von Wettstein, NH Chua, eds, Plant Molecular Biology. Plenum Press, New York, pp 679–688
- Fallon KM, Trewavas AJ. Phosphorylation of renatured protein from etiolated wheat leaf protoplast is modulated by blue and red light. Plant Physiol. 1994;105:253–258. doi: 10.1104/pp.105.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PCG, Hemerly AS, Villarroel R, Montagu MV, Inze D. The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell. 1991;3:531–540. doi: 10.1105/tpc.3.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N-H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Furuya M, Schäfer E. Photoperception and signaling of induction reactions by different phytochromes. Trends Plant Sci. 1996;9:301–307. [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruppuso PA, Boylan JM. Developmental changes in the activity and cellular localization of hepatic casein kinase II in rat. J Cell Biochem. 1995;58:65–72. doi: 10.1002/jcb.240580109. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Harter K, Kircher S, Frohnmeyer H, Krenz M, Nagy F, Schäfer E. Light-regulated modification and nuclear translocation of cytosolic G-box binding factors in parsley. Plant Cell. 1994;6:545–559. doi: 10.1105/tpc.6.4.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Montagu MV, Engler G, Inze D, Ferreira P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Chua N-H. Light-regulated transcription in higher plants. J Plant Physiol. 1997;110:131–139. doi: 10.1007/BF02506852. [DOI] [PubMed] [Google Scholar]
- Hübner S, Xiao C-Y, Jans DA. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua N-H. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jans DA. The regulation of protein transport to the nucleus by phosphorylation. Biochem J. 1995;311:705–716. doi: 10.1042/bj3110705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (1991) Photoregulation of plant gene expression in plants. In D Grierson, ed, Developmental Regulation of Plant Gene Expression. Blackie & Son, Glasgow, Scotland, pp 1–41
- Kanekatsu M, Ezumi A, Nakamura T, Ohtsuki K. Chloroplast ribonucleoproteins (RNPs) as phosphate acceptors for casein kinase II: purification by ssDNA-cellulose column chromatography. Plant Cell Physiol. 1995;36:1649–1656. [PubMed] [Google Scholar]
- Kanekatsu M, Hiroshi S. Phosphorylation of chloroplast ATP synthase by casein kinase II (CK-II) (abstract no. 700) Plant Physiol. 1997;114:S-148. [Google Scholar]
- Karlin-Neumann GA, Sun L, Tobin EM. Plant Physiol. 1988;88:1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Cashmore AR. Microheterogenous cytosolic high-mobility group proteins from broccoli co-purify with and are phosphorylated by casein kinase II. Plant Physiol. 1994;105:911–919. doi: 10.1104/pp.105.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Collinge MA, Farani D, Giuliano G, Walker JC, Cashmore AR. Reconstitution of Arabidopsis casein kinase II from recombinant subunits and phosphorylation of transcription factor GBF1. Plant Cell. 1995;7:105–115. doi: 10.1105/tpc.7.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AJ. DNA binding activity of the Arabidopsis G-box factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Roff E, Spruit C. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP. Four genes in 2 diverged subfamilies encode the ribulose 1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol. 1988;11:745–759. doi: 10.1007/BF00019515. [DOI] [PubMed] [Google Scholar]
- Krebs EG, Beavo JA. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Krek W, Maridor G, Nigg EA. Casein kinase II is a predominantly nuclear enzyme. J Cell Biol. 1992;116:43–55. doi: 10.1083/jcb.116.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Benedyk M, Chua N-H. Characterization of phytochrome-regulated gene expression in photoautotrophic cell suspension: possible role for calmodulin. Mol Cell Biol. 1989;9:4819–4823. doi: 10.1128/mcb.9.11.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutwiler LS, Meyerowitz EM, Tobin EM. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield DW, Bosc DG, Slominski E. The protein kinase from mitotic human cells that phosphorylates Ser-209 on the casein kinase II β subunit is p34cdc2. Biochim Biophys Acta. 1995;1269:69–78. doi: 10.1016/0167-4889(95)00100-7. [DOI] [PubMed] [Google Scholar]
- Litchfield DW, Lozeman FJ, Piening C, Sommercorn J, Takio K, Walsh KA, Krebs EG. Subunit structure of casein kinase II from bovine testis: demonstration that the α and α′ subunits are distinct polypeptides. J Biol Chem. 1990;265:7638–7644. [PubMed] [Google Scholar]
- Litchfield DW, Lüscher B (1993) Casein kinase II in signal transduction and cell cycle regulation. Mol Cell Biochem 127/128: 187–199 [DOI] [PubMed]
- Litchfield DW, Lüscher B, Lozeman FJ, Eisenman RN, Krebs EG. Phosphorylation of casein kinase II by p34cdc2 in vitro and at mitosis. J Biol Chem. 1992;267:13943–13951. [PubMed] [Google Scholar]
- Lorenz P, Pepperkok R, Ansorge W, Pyerin W. Cell biological studies with monoclonal and polyclonal antibodies against human casein kinase II subunit β demonstrate participation of the kinase in mitogenic signaling. J Biol Chem. 1993;268:2733–2739. [PubMed] [Google Scholar]
- Lüscher B, Chistenson E, Litchfield D, Krebs E, Eisenman R. Myb DNA binding inhibited by phosphorylation at a site deleted during oncogenic activation. Nature. 1990;344:517–522. doi: 10.1038/344517a0. [DOI] [PubMed] [Google Scholar]
- Manzara T, Carrasco P, Gruissem W. Developmental and organ-specific changes in DNA-protein interactions in the tomato rbcS1, rbcS2, and rbcS3A promoter regions. Plant Mol Biol. 1993;21:69–88. doi: 10.1007/BF00039619. [DOI] [PubMed] [Google Scholar]
- Menkens AE, Cashmore AR. Isolation and characterization of a forth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA. 1994;91:2522–2526. doi: 10.1073/pnas.91.7.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element bound by the GBF family of bZIP proteins. Trends Biochem. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Yamaguchi-Shinizaki K, Hayashida N, Kamada H, Shinozaki K. Plant Mol Biol. 1993;21:279–289. doi: 10.1007/BF00019944. [DOI] [PubMed] [Google Scholar]
- Mulner-Lorillon O, Cormier P, Labbe J-C, Doree M, Poulhe R, Osborne H, Belle R. M-phase-specific cdc2 protein kinase phosphorylates the beta subunit of casein kinase II and increases casein kinase II activity. Eur J Biochem. 1990;193:529–534. doi: 10.1111/j.1432-1033.1990.tb19368.x. [DOI] [PubMed] [Google Scholar]
- Neff MM, van Volkenburgh E. Light-stimulated cotyledon expansion in Arabidopsis seedling. The role of phytochrome B. Plant Physiol. 1994;104:1027–1032. doi: 10.1104/pp.104.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua N-H. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Otto V, Schäfer E. Rapid phytochrome-controlled protein phosphorylation and dephosphorylation in Avena sativa. Plant Cell Physiol. 1988;29:1115–1121. [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha R, Chen-Wu JL-P, Hanna DE, Glover CVC. Mol Cell Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner CG, Wang Z, Litchfield DW. Expression and localization of epitope-tagged protein kinase CK2. J Cell Biochem. 1997;64:525–537. [PubMed] [Google Scholar]
- Pepper AE, Chory J. Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Lorenz P, Ansorge W, Pyerin W. Casein kinase II is required for transition of G0/G1, early G1, and G1/S phases of the cell cycle. J Biol Chem. 1994;269:6986–6991. [PubMed] [Google Scholar]
- Pinna LA. Casein kinase 2: “eminence grise” in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- Pinna LA. Molecules in focus: protein kinase CK2. Int J Biochem Cell Biol. 1997;29:551–554. doi: 10.1016/s1357-2725(96)00142-2. [DOI] [PubMed] [Google Scholar]
- Ralph RK, Darkin-Rattray S, Schofield P. Growth-related protein kinases. BioEssays. 1990;12:121–124. doi: 10.1002/bies.950120305. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Short TW, Briggs WR, Poff KL (1992) Light-induced phosphorylation of a membrane protein plays an early role in signal transduction for phototropism in Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 4718–4721 [DOI] [PMC free article] [PubMed]
- Rihs H-P, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by a casein kinase II site flanking the nuclear localization sequence of the SV40 T antigen. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux SJ. Signal transduction in phytochrome responses. In: Kendrick RE, Krononberg GHM, editors. Photomorphogenesis in Plants, Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 187–209. [Google Scholar]
- Russo GL, Vandenberg MT, Yu IJ, Bae YS, Franza BRJ, Marshak DR. Casein kinase II phosphorylates p34cdc2 kinase in G1 phase of the HeLa cell division cycle. J Biol Chem. 1992;267:20317–20325. [PubMed] [Google Scholar]
- Sacks DB, Davis HW, Williams JP, Sheehaa EL, Garcia JGN, McDonald JM. Phosphorylation by casein kinase II alters the biological activity of calmodulin. Biochem J. 1992;283:21–24. doi: 10.1042/bj2830021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarokin LP, Chua N-H. Binding sites for two novel phosphoproteins, 3AF5 and 3AF3, are required for rbcS-3A expression. Plant Cell. 1992;4:473–483. doi: 10.1105/tpc.4.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Mildebrand DF, Hunt AG. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Schindler U, Cashmore AR. Photoregulated gene expression may involve ubiquitous DNA binding proteins. EMBO J. 1990;9:3415–3427. doi: 10.1002/j.1460-2075.1990.tb07549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific site in 22-kD zein genes. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Becker-Andre M, Schulz W, Hahlbrock K, Dangl JL. Functional architecture of the light-responsive chalcone synthase reporter from parsley. Plant Cell. 1989;1:707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin DC, Leder P. Casein kinase IIα transgene-induced murine lymphoma: relation to theileriosis in cattle. Science. 1995;267:894–897. doi: 10.1126/science.7846532. [DOI] [PubMed] [Google Scholar]
- Sheen J. Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO J. 1993;12:3497–3505. doi: 10.1002/j.1460-2075.1993.tb06024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Tobin EM. Photochem Photobiol. 1990;52:51–56. doi: 10.1111/j.1751-1097.1990.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Bertekap RLJ, Cashmore AR. Intracellular localization of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultured Arabidopsis and soybean cells. Plant J. 1997;11:967–982. doi: 10.1046/j.1365-313x.1997.11050967.x. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Tjaden G, Coruzzi GM. A novel AT-rich DNA binding protein that combines an HMG-like DNA binding domain with a putative transcription domain. Plant Cell. 1994;6:107–118. doi: 10.1105/tpc.6.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C-G, Kendrick RE, Roux SJ. Red-light-induced appearance of phosphotyrosine-like epitopes on nuclear proteins from pea (Pisum sativum) plumules. Photochem Photobiol. 1996;64:863–866. doi: 10.1111/j.1751-1097.1996.tb01848.x. [DOI] [PubMed] [Google Scholar]
- Trewavas A, Gilroy S. Signal transduction in plants cells. Trends Genet. 1991;7:356–361. doi: 10.1016/0168-9525(91)90255-o. [DOI] [PubMed] [Google Scholar]
- Tuazon PT, Traugh JA. Casein kinase I and II: multipotential serine protein kinases. Structure, function, and regulation. Adv Second Messenger Phosphoprotein Res. 1991;23:123–164. [PubMed] [Google Scholar]
- Ueda T, Waverczak W, Ward K, Sher N, Ketudat M, Schmidt RJ, Messing J. Mutations of 22- and 27-kD zein promoters affect transactivation by the Opaque-2 protein. Plant Cell. 1992;4:701–709. doi: 10.1105/tpc.4.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lijsebettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Gruissem W. Expression dynamics of the tomato rbcS gene family during development. Plant J. 1991;3:1289–1303. doi: 10.1105/tpc.3.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside ST, Goodbourn S. Signal transduction and nuclear targetting: regulation of transcription factor activity by subcellular localization. J Cell Sci. 1993;104:949–955. doi: 10.1242/jcs.104.4.949. [DOI] [PubMed] [Google Scholar]
- Williams ME, Foster R, Chua N-H. Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell. 1992;4:485–496. doi: 10.1105/tpc.4.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirkner U, Voss H, Lichter P, Weitz S, Ansorge W, Pyerin W. Human casein kinase II subunit α: sequence of a processed (pseudo) gene and its localization on chromosome 11. Biochim Biophys Acta. 1992;1131:220–222. doi: 10.1016/0167-4781(92)90083-c. [DOI] [PubMed] [Google Scholar]
- Wong YS, Lagarias JC. Affinity labeling of Avena phytochrome with ATP analogs. Proc Natl Acad Sci USA. 1989;86:3469–3473. doi: 10.1073/pnas.86.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YS, McMichael JRW, Lagarias JC. Properties of a polycation-stimulated protein kinase associated with purified Avena phytochrome. Plant Physiol. 1989;91:709–718. doi: 10.1104/pp.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hanley S, Goodman HM. Isolation, characterization, and chromosomal location of a new cab gene from Arabidopsis thaliana. Plant Physiol. 1991;96:1387–1388. doi: 10.1104/pp.96.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]