Abstract
The assembly mechanism for the human immunodeficiency virus type-1 (HIV) synaptic complex (SC) capable of concerted integration is unknown. Molecular and structural studies have established that the HIV SC and prototype foamy virus (PFV) intasome contain a tetramer of integrase (IN) that catalyzes concerted integration. HIV IN purified in the presence of 1 mM EDTA and 10 mM MgSO4 was predominately a monomer. IN efficiently promoted concerted integration of micromolar concentrations of 3′ OH recessed and blunt ended U5 long terminal repeat (LTR) oligonucleotides (ODN) substrates (19 bp to 42 bp) into circular target DNA. Varying HIV IN to U5 DNA showed that a IN dimer:DNA end molar ratio of 1 was optimal for concerted integration. Integration activities decreased with increasing length of the ODN, starting from the recessed 18/20 bp or 19/21 bp set to the 31/33 bp and 40/42 bp set. Under these conditions, the average fidelity for the HIV 5 bp host site duplication with recessed and blunt ended substrates was 56%. Modifications of U5 LTR sequences beyond 21 bp from the terminus on longer DNA (1.6 kb) did not alter the ~32 bp DNaseI protective footprint suggesting, viral sequences beyond 21 bp were not essential for IN binding. The results suggest IN binds differentially to an 18/20 bp than to an 40/42 bp ODN substrate for concerted integration. The HIV IN monomer may be a suitable candidate to attempt crystallization of an IN-DNA complex in the absence or presence of strand transfer inhibitors.
After retrovirus infection of cells, the reverse transcriptase produces a linear viral DNA genome which results in the formation of a cytoplasmic preintegration complex (PIC)(1–4). The biochemical mechanisms by which integrase (IN) first catalyzes the 3′ OH processing of two nucleotides from the viral DNA blunt-ends in the cytoplasmic PIC and the subsequent concerted integration of the recessed ends into the cell DNA have been elucidated (5).
Efforts to evaluate the structure and function of nucleoprotein complexes assembled in vitro that have properties of the PIC have been undertaken. The human immunodeficiency virus type-1 (HIV) synaptic complex (SC)(6–8) and the prototype foamy virus (PFV) intasome (9, 10) have demonstrable capability to catalyze both the 3′ OH processing and concerted integration reactions. Both complexes possess a tetramer of IN (6, 8, 9) that produces their respective HIV 5 bp (11, 12) and PFV 4 bp (13) host site duplications. Activity of the HIV SC is effectively inhibited by clinically relevant strand transfer inhibitors (STI) at low physiologically relevant nM concentrations (7, 14). The three-dimensional structure of the PFV intasome formed with 3′ OH recessed 17/19 bp oligonucleotides (ODN)(9) and the intasome in the presence of target DNA (15) have revealed numerous mechanistic insights into the protein-protein and protein-DNA interactions necessary for binding and catalysis. Modeling of the HIV SC or intasome (16) suggests that the interactions of the HIV IN within this nucleoprotein complex may be similar to the PFV intasome.
The structure of the HIV SC has not been determined. A mystery has existed for 20 years why recombinant HIV IN was not capable of efficiently using ODN for concerted integration, a necessary requirement for atomic resolution studies. In this report, we show that our preparation of HIV IN, which is purified in the presence of 1 mM EDTA and 10 mM MgSO4, exists as a monomer and exhibits some of the properties previously described for Zn++-deficient HIV IN in solution (17). We demonstrated that this preparation can use U5 long terminal repeat (LTR) ODN (20 bp to 42 bp) for highly efficient concerted integration activity where both IN and ODN are at near equal micromolar concentrations (Figure 1). The ODN substrates are designated by the length of their non-cleaved strand and either blunt (B) or 3′ OH recessed ended (R). The observed concerted integration activity of the 20R and 21R substrates used with HIV IN was similar to PFV IN monomer activity with 19R (9, 13). The relative concerted integration activities of HIV IN decreased with increasing length of the ODN from the 20R or 21R set to the 33R and 42R set. Using a longer DNA substrate (1.6 kb) containing varying lengths of U5 sequences, we determined that modifications of the sequences just beyond 21 bp in the U5 LTR did not alter the concerted integration activity or the ~32 bp DNaseI protective footprint previously observed (8, 14, 18), suggesting protection of these internal sequences by IN is not dependent on viral sequences. In summary, the results suggests that HIV IN appears to possess different DNA binding modes on the U5 LTR for concerted integration depending upon the length of the DNA substrate.
Figure 1.
The HIV IN monomer forms a tetramer in solution and on DNA. Purified IN monomers form tetramers upon preincubation with Zn++ (top). In the presence of viral DNA, IN forms the synaptic complex (SC) containing a tetramer. Upon addition of circular target DNA, the SC produces the linear concerted integration product as well as products resulting from multiple concerted events when ODN substrates are used.
EXPERIMENTAL PROCEDURES
HIV 1 IN Purification
Bacterial recombinant wt HIV and F185K IN (pNY strain) were expressed in E. coli BL21(DE3) and purified to near homogeneity as described (11, 19). In the Heparin-HiTrap purification step, the buffer contained 50 mM HEPES (pH 6.8), 10 mM MgSO4, 1 mM EDTA, 3 mM DTT, and 10% glycerol (HEPES buffer). IN was eluted with a linear NaCl gradient in the HEPES buffer. The HEPES buffer was used for storage of IN at approximately 0.75 M NaCl. This purified IN was used for analysis and strand transfer assays unless otherwise indicated. HIV IN was free of DNA endonuclease activities using supercoiled DNA as substrate. RSV IN was purified in the presence of EDTA similar to a method described earlier (20).
Analysis by Size Exclusion Chromatography
IN was concentrated to 0.5 to 1.1 mg/ml using an Amicon Ultra-4 (30K) ultrafiltration device at 2°C. There was essentially no aggregation of the IN as judged by light scattering. IN was analyzed by size exclusion chromatography (SEC) on a Superdex 200 10/300 GL column (GE Healthcare) at 4°C. HIV IN was incubated on ice for 30 min without or with 50 μM ZnCl2 in 250 μl of the above HEPES buffer. IN was loaded (250 μl loop, auto injector) to the column attached to a BioLogic DuoFlow system. The elution was performed at 0.5 ml/min with 20 mM HEPES (pH 6.8), 500 mM NaCl, 10 mM MgSO4, 0.2 mM EDTA, 3 mM DTT, and 5% glycerol, without and with Zn++ at 25 μM. Elution of IN was monitored at 280 nm. Six molecular mass protein standards (17– 670 kDa)(BioRad) with added apoferritin and bovine serum albumin (Sigma) were used for column calibration. Molecular mass (M) was determined from a plot of log M versus Ve/V0 (where Ve is the elution volume of the particular protein and V0 is the void volume (8 ml)).
Viral DNA Substrates
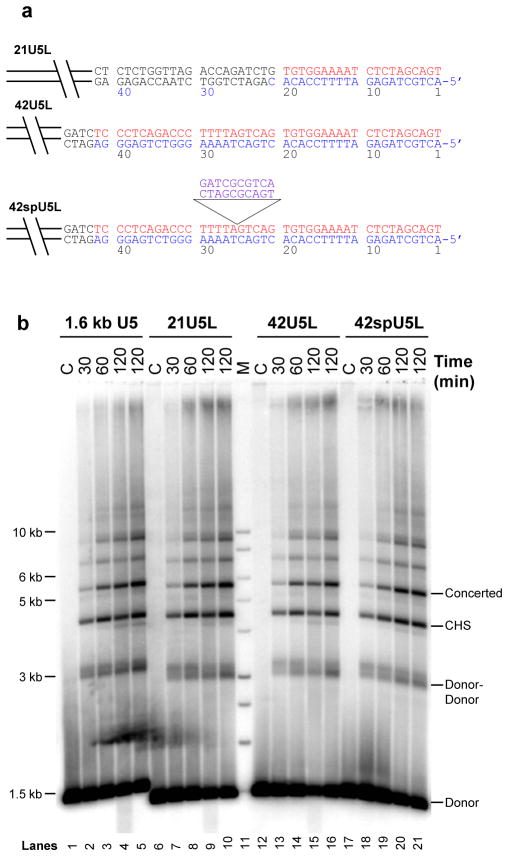
The ODN substrates containing U5 LTR sequences were prepared by annealing the respective length of cleaved and non-cleaved strand. All the ODN were synthesized and PAGE purified by Integrated DNA Technologies (IDT). A 1.6 kb blunt-ended U5 DNA substrate containing 168 bp of U5 sequences was prepared as described (21). To prepare the 1.5 kb DNA substrate containing varying lengths of U5 LTR sequences, the ODN containing U5 LTR sequences with a BglII site at non-LTR terminus (Fig. 4a) were synthesized, annealed and ligated to a pUCLTRKanII digested with BglII-ScaI. pUCLTRKanII was constructed by cloning the SalI (blunt-ended) kanamycin cassette from pUC4K (GE Healthcare) into the blunt-ended BspHI-SalI fragment of pUCLTR. pUCLTR contained the blunt-ended KpnI-PstI possessing U5-U3 HIV LTR from pU3U5 (22) ligated to blunt-ended SspI-AflIII fragment of pUC19. The 1.5 kb DNA ScaI-HindIII fragment containing 21 bp, 42 bp and 42 bp U5 sequences interrupted by a 10 bp spacer at position 25 are referred to as 21U5L, 42U5L, and 42spU5L, respectively (Fig. 4a).
Figure 4.
Concerted integration activity of IN with a large size DNA containing varying lengths of U5 LTR sequences. (a) Construction of 1.5 kb DNA substrates containing varying lengths of U5 LTR sequences at the termini. ODN containing a BglII restriction site along with 21 bp (21U5L) and 42 bp (42U5L) of blunt ended U5 LTR sequences were cloned into a vector. A 10 bp spacer containing non-specific sequences inserted at nucleotide position 25 of U5 was also constructed (42spU5L). The clones were digested with restriction enzymes to yield a 1.5 kb U5 blunt-ended DNA substrate. The cleaved strand is indicated in red and the non-cleaved strand in blue while the spacer region is shown in purple. The Bglll restriction site and the vector sequence in 21U5L are in black. (b) Strand transfer activities of varying length U5 substrates. IN (20 nM) was preassembled with 0.5 nM of 5′-32P end-labeled 1.6 kb wt blunt-ended U5 DNA (lanes 1–5), 21U5L (lanes 6–10), 42U5L (lanes 12–16) and 42spU5L (lanes 17–21) for 15 min at 14°C. Upon addition of target DNA (1.5 nM), strand transfer was allowed to proceed for indicated time (marked on the top) at 37°C. Reactions were stopped with EDTA to a final concentration of 25 mM and samples were deproteinized. Strand transfer products were separated on 0.7% agarose gel, dried and analyzed by scanning on Typhoon Trio Variable Mode Imager. At 120 min, the percentage of donor incorporated into the concerted and CHS products was 8.6, 10.1, 9, 10 and 7.9, 9.5, 8.6, and 9.2 in lanes 5, 10, 16 and 21, respectively.
Concerted Integration Assay
Concerted integration assays with the 1.6 kb U5 blunt ended DNA substrates were performed as described (19). Briefly, IN was preassembled with DNA substrates in 20 mM HEPES (pH 7.0), 100 mM NaCl, 5 mM dithiothreitol, 10 mM MgCl2, 25 μM ZnCl2, and 10 % poly(ethylene glycol) 6000 at 14°C for 15 min. The assay with ODN substrates was slightly modified and supplemented with 10% dimethyl sulfoxide (DMSO). IN (0.2 to 3.0 μM as dimers) was preincubated with DNA substrates (0.2 to 4.0 μM) for 15 min at 14°C. Upon addition of target DNA (8 to 24 nM), the strand transfer was for 1 h at 37°C. Reactions were stopped with EDTA to a final concentration of 25 mM and samples were deproteinized with 0.5 % SDS, 1 mg/ml proteinase K for 1 h at 37°C. Strand transfer products were separated on 1.8% agarose gel, stained with SYBR Gold (Invitrogen) and identified by scanning on Typhoon Trio Variable Imager (GE Healthcare).
DNaseI Footprint Analysis of IN-DNA Complexes
DNaseI treatment of IN-DNA complexes and the isolation of the treated DNAs were performed as described earlier (8).
Sequencing of Concerted Integration Products
Concerted integration products obtained with ODN substrates were excised from the agarose gel and the DNA was purified by Qiaquick gel extraction kit (Qiagen). The integration products were amplified by PCR with a single primer (5′-GTCAGTGTGGAAAATCTCTAGC-3′) using Accuprime Taq DNA polymerase (Invitrogen). PCR products (~2.7 kb) were purified from agarose gels and cloned in pCRII-TOPO (Invitrogen). Recombinant clones were sequenced using SP6 primer and a custom primer (KKP-TA392R 5′-CTAGATGCATGCTCGAGCGGCC-3′) to analyze the LTR-target junction and host site duplications.
RESULTS
Different purification procedures for recombinant HIV IN have resulted in a variety of arrangements of IN subunits ranging from monomer, dimer, tetramer, octamer, and aggregates, depending on the concentration and other factors (17, 23–26). His-tagged HIV IN and IN with the tag removed by a protease usually form dimers and/or tetramers which are in equilibrium (27–30). It is not apparent what form of IN is required for efficient assembly of HIV SC. In contrast, PFV IN is predominately a monomer in solution but assembles into a tetramer in the presence of a 19R ODN (13).
Determination of the oligomeric form of HIV IN
We have observed that HIV IN purified in the presence of 1 mM EDTA and 10 mM MgSO4 in a HEPES buffer (pH 6.8)(11, 19) is a monomer in solution and is converted to a tetramer in the presence of Zn++ (Figure 2). IN was concentrated to 15 μM (as monomers) by an ultrafiltration device and applied to a Superdex 200 column. IN eluted predominately as a monomer (95%) with a small quantity of tetramer (3%) present (Figure 2a). A similar elution profile was obtained at 5.0 μM IN. At 26 μM, a similar elution profile for the IN monomer was observed with a slight increase in the quantity of tetramer. The calculated molecular weights of the monomer and tetramer were 35 and 131 kDa, respectively. Removal of MgSO4 in the buffer did not affect the elution profile of IN monomers. Preincubation of 15 μM IN in the presence of 50 μM ZnCl2 on ice for 30 min resulted in the almost exclusive conversion to a tetramer (Figure 2a). Minimal quantities of IN dimers were observed in the absence or presence of Zn++. The elution profiles of HIV IN F185K (23) and wt Rous sarcoma virus (RSV) IN (20) showed these proteins to be dimeric (Figure 2b,c, respectively). Preincubation of F185K IN with Zn++ resulted in a 65% conversion to tetramers but did not significantly affect the RSV IN dimer/tetramer equilibrium. In summary, HIV IN purified in the presence of 1 mM EDTA and 10 mM MgSO4 resulted in predominately monomers but were converted to tetramers in the presence of Zn++, confirming earlier studies of HIV IN (17). Analytical ultracentrifugation studies of HIV IN lacking Zn++ followed by reconstitution with Zn++ demonstrated a monomer-tetramer-octamer equilibrium without forming a dimer intermediate (17).
Figure 2.
HIV IN exists predominantly as a monomer. (a) SEC profile of wt HIV IN. HIV IN (15 μM, 115 μg) was predominantly a monomer with a minor quantity of tetramer (blue line). The IN monomer was converted to a tetramer after incubation in 50 μM ZnCl2 (red line). A minimal quantity of dimers was present in both IN samples. (b) SEC profile of HIV F185K IN without and with incubation in ZnCl2. F185K IN (18 μM) was exclusively a dimer (blue line). Upon incubation of F185K IN (16.5 μM) with 50 μM ZnCl2, dimers and tetramers were present. (c) SEC profile of wt RSV IN (286 residues) was unaffected by the ZnCl2. RSV IN (25 μM) eluted as a dimer/tetramer without (blue line) or after incubation in 50 μM ZnCl2 (red line).
HIV IN Monomer and Tetramer Equally Promote Concerted Integration
We examined whether the purified IN monomer and tetramer fractions were equally proficient in promoting the concerted integration reaction using a 1.6 kb blunt ended U5 DNA substrate (7). IN monomer and tetramer fractions were purified by Superdex 200 chromatography in the absence of ZnCl2. The integration reactions were performed with 1.6 kb blunt-ended U5 DNA and IN at 25 nM. Under these assay conditions in the presence of Zn++, both the purified IN monomer and tetramer equally promoted concerted and circular half-site (CHS) integration into supercoiled DNA target (Figure S1 of the Supporting Information).
Sub-Micromolar and Micromolar Concentrations of HIV IN Efficiently Produce Concerted Integration Products using ODN
In the past, it has not been possible to effectively promote concerted integration using ODN substrates by HIV IN. We investigated whether IN preparations which contained predominately monomers were capable of performing efficient concerted integration with ODN substrates. Varying quantities of IN and ODN of various sizes (20 bp to 42 bp) with blunt or 3′ OH recessed ends (Figure 3a) were preincubated at 14°C prior to strand transfer at 37°C for 1 h. The 20B and 20R substrates (Figure 3d) were the most efficient substrates for concerted integration in comparison to the 33B and 33R or the 42B and 42R substrates (Figure 3c and 3b, respectively). At a constant IN concentration (0.5 μM) with varying DNA, a calculated IN dimer:DNA end molar ratio near 1 was optimal for concerted integration with the both the 20B and 20R (Figure 3d, lanes 6 and 7; 15 and 16, respectively). The efficiency of the 20R (Figure 3d, lanes 11 and 12; and lanes 14–17) or the 21R substrates (Figure S2 of the Supporting Information) to promote concerted integration by IN monomers was sufficiently high to promote multiple concerted integrations into the same DNA target that resulted in smearing of the DNA immediately below the linear concerted integration product (Figure 1 and Figure 3). Similar efficiency was observed for PFV IN with the cognate 19R ODN substrate (9, 13). The efficiency of the 20B substrate for concerted integration is significantly less (Figure 3d, lanes 2–8) than the 20R because 3′ OH processing occurs slowly in the context of the HIV SC (6–8). At a highest IN dimer:DNA end molar ratio of 5, both the 20B and 20R (Figure 3d, lanes 2–4, lanes 11–13, respectively) reactions were inhibited as were the same reactions using the 33 bp and 42 bp ODN sets (Figure 3c,b). Finally, the quantities of the CHS products are less than the concerted products with the 20R substrate (Figure 3d)(Figure S3 of the Supporting Information). In summary, the 20R substrate was significantly better than the 20B for concerted integration.
Figure 3.
Concerted integration with ODN possessing 3′ OH recessed and blunt ended U5 LTR sequences. (a) Sequences of ODN containing U5 LTR sequences. The blunt-ended (designated B) counterparts of each recessed (designated R) ODN had the normal GT bases on the top cleaved strand. The 20R substrate had a cytosine at the nucleotide position 20 on the non-cleaved strand instead of adenine. (b, c, and d) Strand transfer reactions were performed with varying amounts of IN (0.2 μM to 1.0 μM as dimers) and 42B, 42R, 33B, 33R, 20B and 20R (0.2 μM to 0.8 μM). The ODN are marked on the top. The filled triangle shows increasing concentrations of IN with a fixed quantity of ODN (0.2 μM). The open triangle indicates a fixed concentration of IN (0.5 μM) with increasing quantities of ODN. In each separate panel, blunt ended ODN are on left and recessed ODN on the right. Strand transfer was for 1 h at 37°C. Concerted and circular half-site (CHS) products are marked on the right. The proficiency of producing multiple concerted products (marked on right) was highest with 20R. O.C. and S.C. are open circle and supercoiled target, respectively. Molecular weight markers are on the left and the donor ODN on the right.
Similar results were obtained with the 42B and 42R and 33B and 33R substrates (Figure 3b,c, respectively) as observed with 20B and 20R substrates. Both the 42R and 33R substrates were significantly better than their blunt-ended counterparts. As noted above, the 42R and 33R group were less effective than the 20R (Figure 3d) and 21R (Figure S3 of the Supporting Information) group for concerted integration because the former group had significantly less multiple concerted integrations events that produced smearing below the linear product.
As shown previously, the purified HIV IN monomer and tetramer at 25 nM possessed equivalent concerted integration activities with the 1.6 kb U5 DNA substrate (Figure S1 of the Supporting Information). Likewise, tetrameric IN produced by Zn++ and monomeric IN possessed similar activities for concerted integration at 0.5 μM IN using 21B and 21R substrates (Figure S4 of the Supporting Information). At optimal IN to DNA end molar ratios, micromolar concentrations of the IN monomer and ODN were also efficient at promoting concerted integration (Figure S5 of the Supporting Information).
HIV 5 bp Host Site Duplications with ODN Substrates
Integration of HIV DNA into target DNA results in a 5 bp host site duplication (12, 31, 32). The concerted integration products derived from 42B, 33B, 20B, and 21R were purified and cloned. Restriction analysis of the transformants demonstrated ~87% of the DNA clones were of the expected size. Sequence analysis of 89 clones revealed an average fidelity of 56% for the 5 bp duplication with the rest of the clones containing various size deletions (Table 1). The highest fidelity rate was 61% using the 21R substrate. Fidelity rates obtained with larger size DNA substrates (>300 bp) using either 3′ OH recessed or blunt-ended DNA substrates (12, 33–35) were also similar to these current ODN results. As anticipated (14), MK-2048 (Figure S6 of the Supporting Information) and Raltegravir (RAL) effectively inhibited the production of the linear concerted and multiple concerted integration products obtained with ODN substrates.
Table 1.
Sequence analysis of HIV concerted integration products obtained with U5 LTR ODN substrates.
| Substrate | Clones analyzed | Correct duplication | Deletions
|
|||||
|---|---|---|---|---|---|---|---|---|
| Total | 1–20 | 21–50 | 51–100 | 100–500 | 500–2000 | |||
| 20B | 20 | 11 | 9 | 1 | 4 | 2 | 0 | 2 |
| 33B | 20 | 9 | 11 | 1 | 2 | 3 | 5 | 0 |
| 42B | 18 | 11 | 7 | 3 | 0 | 1 | 2 | 1 |
| 21R | 31 | 19 | 12 | 1 | 3 | 1 | 6 | 1 |
| Total | 89 | 50 | 39 | 6 | 9 | 6 | 13 | 3 |
In total, 89 clones were analyzed and 50 of them were found to have the HIV 5 bp host site duplication (~56% fidelity). Deletions were categorized on the basis of their length in bp.
Modification of LTR Sequences Upstream of 21 Nucleotides from the DNA Terminus does not Alter the ~32 bp DNaseI Protective Footprint by HIV IN
We had established that wt IN binds ~32 bp of U5 and U3 LTR sequences in HIV SC in the absence and presence of L-841,411 and RAL (8, 14), the strand transfer complex (8), and the IN-single DNA (ISD) complex formed in the presence of the above inhibitors (18). Using DNaseI footprint analysis, we determined whether modification of viral sequences upstream of 21 nucleotides from the terminus affected concerted integration activity and DNA binding interactions of IN. Blunt ended U5 ODN ranging in size from 21 bp (21U5L), 42 bp (42U5L) and modified 42 bp (42spU5L) were cloned (Figure 4a). The 42spU5L ODN contained a 10 bp spacer with non-specific sequences inserted between nucleotides 25 and 26 of U5. All three modified U5 DNA substrates (1.5 kb)(Figure 4b, lanes 6–21) including the U5 1.6 kb substrate containing contiguous 168 bp of U5 sequences (Figure 4b, lanes 1–5) equally promoted both concerted and CHS integration activities.
We determined whether the ~32 bp DNaseI protective footprint by IN observed with wt U5 sequences on the 1.5 kb substrates were modified by disruption of U5 sequences upstream of the 21th nucleotide (Figure 4a)(8, 14). IN-DNA complexes formed in the integration reaction were subjected to DNaseI protective footprint analysis prior to isolation of the concerted and CHS integration products on agarose gels. The wt U5 LTR sequences within these two products were protected from DNaseI digestion by IN up to ~32 bp (Figure 5a,b, left panel). IN binding also enhanced DNaseI cleavage near position 32G with both the concerted and CHS products (Figure 5a,b, lanes 7 and 8) in comparison to control digests of naked DNA (Figure 5a,b, lanes 5 and 6). Both the 21U5L and 42spU5L substrates also produced a similar ~32 bp DNaseI protective footprints (Figure 5a,b, right panel). The boundary for DNaseI protection by IN with both modified U5 substrates was clearly evident (Figure 5a,b, lanes 9 and 10) in comparison to control digests of naked DNA (Figure 5a,b, lanes 11 and 12). Enhanced DNaseI cleavages mapping at ~32 nucleotides were more evident with 42spU5L than 21U5L substrate. In summary, concerted integration and binding of IN to ~32 bp on U5 were independent of viral sequences upstream of 21 nucleotides.
Figure 5.
HIV IN binding to the U5 LTR ends is independent of internal sequences beyond 21 nucleotides from the terminus. Panel (a) shows the DNaseI protective footprint located on the concerted and CHS products of the wt blunt ended U5 LTR sequences and 21U5L. Lanes 1–8 contain the products derived from wt blunt-ended U5 DNA. Lane 1 contains the input DNA, lanes 2– 4 contain the Maxam-Gilbert chemical sequence marker prepared from the input DNA; lanes 5 and 6 contain input DNA digested with DNaseI for 2 and 3 min, respectively; lanes 7 and 8 contain DNaseI treated samples producing concerted and CHS integration products, respectively. Lanes 9–16 contain the products derived from 21U5L. Lanes 9 and 10 contain the DNaseI treated samples producing CHS and concerted products, respectively. Lanes 11 and 12 contain the input DNA digested with DNaseI for 3 and 2 min, respectively. Lanes 13–15 contain the Maxam-Gilbert chemical sequence marker for 21U5L. Lane 16 contains the untreated input DNA. Panel (b) shows the DNaseI protective footprint by IN on 1.6 kb DNA wt U5 LTR sequences and 1.5 kb DNA 42spU5L with the 10 bp spacer. Lanes 1–8 contains the same samples as described in panel A. Lanes 9–13 contain the products derived from 42spU5L. Lanes 9 and 10 contain the DNaseI treated samples producing CHS and concerted integration products, respectively. Lanes 11 and 12 contain the input DNA digested with DNaseI for 3 and 2 min, respectively. Lane 13 contains the untreated input DNA.
DISSCUSSION
We employed several new strategies to address a long standing mystery why HIV IN was not capable of efficiently producing concerted integration products using ODN substrates. First, we verified that IN purified in the presence of 1 mM EDTA and 10 mM MgSO4 was predominantly monomeric (17) and highly soluble, at least up to ~ 1 mg/ml at 0.5 M NaCl. Secondly, these preparations efficiently promoted concerted integration with ODN substrates (18 bp to 42 bp), a prerequisite for crystallization attempts of HIV SC without and with strand transfer inhibitors. The 20R and 21R ODN substrates were better substrates for promoting concerted integration than the larger size 33R and 42R substrates. The blunt ended ODN counterparts were less efficient for concerted integration than the recessed ODN. With the 1.6 kb DNA substrate, IN bound to sequences beyond the 21 nucleotide of U5 and as we showed here, this binding is nonspecific. The binding extended to ~32 bp in length that resulted in DNaseI enhanced cleavages at this boundary suggesting distortion of the DNA by IN.
Why is HIV IN predominately a monomer upon purification in the presence of 1 mM EDTA and 10 mM MgSO4? We established that IN was a monomer by SEC at pH 6.8 (Figure 2a) similar to a previous report (17). In the presence of 50 μM ZnCl2, monomers are converted to tetramers (Fig. 2). Some Zn++ remains with IN purified in the presence of EDTA (0.12 equivalent of Zn++ per IN monomer) or with Zn++ during purification (0.67 equivalent per monomer)(17). This metal ion binds at the HH-CC motif in the N-terminus domain of IN (36). Further studies suggested that Zn++ and Mg++ are critical for enzymatic activities and IN multimerization properties (17, 28, 37). As previously stated, analytical ultracentrifugation studies of HIV IN lacking Zn++ followed by reconstitution with Zn++ demonstrated a monomer-tetramer-octamer equilibrium without forming a dimer intermediate (17). Our SEC studies support such a model for HIV IN in solution purified in the presence of EDTA (Figure 2a). In contrast, both the HIV F185K and RSV IN were purified in the presence of EDTA but formed mostly dimers (Figure 2b,c).
The assembly mechanisms of HIV IN onto two viral DNA ends to promote 3′ OH processing and concerted integration are unknown. The formation of an active IN-DNA complex is directly dependent on the correct positioning of IN on viral DNA ends and, the dimers which are formed on viral DNA ends rather than on nonspecific DNA ends are not functionally similar (26, 38, 39). Lesbats et al. (38) concluded that active dimers can be formed from monomers only after oligomerization of IN on a U5 21 bp ODN however, no concerted integration was reported with this ODN substrate. The exact reasons for the inability of IN monomers to catalyze concerted integration using ODN substrates is unknown but may be due to the presence of the nonionic detergent CHAPS during purification of IN or NP40 in the reaction mixture (28, 38, 40). However, with a longer viral DNA substrate (300 bp) and HIV IN purified with CHAPS and stored in the presence of ZnSO4 (50 μM), Lesbats et al. (38) demonstrated that purified IN containing monomers, dimers, and tetramers was capable of promoting concerted integration. Our HIV IN purified without detergents contain predominately monomers and was capable of efficient concerted integration with ODN substrates (Fig. 3 and Supporting Information Fig. S2 to S5) and 1.6 kb DNA substrates (7, 8, 14).
The oligomeric state of HIV IN without DNA under the reaction conditions for concerted integration was determined by BS3 cross-linking. In presence of Zn++, multimeric forms of IN (dimers, trimers, and tetramers) were observed (Figure S7 of the Supporting Information, lanes 2 and 3), besides the monomers. The proportion of multimeric forms of IN was much higher under the reaction conditions (Figure S7 of the Supporting Information, lanes 6 and 7) when the IN was pre-incubated with only ZnCl2 to form IN tetramers (Fig 2a). Under the reactions conditions for concerted integration and in the absence of Zn++, only the monomer and dimer species of IN were observed suggesting that Zn++ was essential for multimerization of IN (Figure S7 of the Supporting Information, lanes 4 and 5). Additionally, IN was unable to perform concerted integration in the absence of Zn++ (data not shown). Similar pattern of multimeric forms of IN upon DSS or BS3 cross-linking of IN in the absence of DNA substrate were observed under reaction conditions containing Zn++ (6, 16). These results suggested that in the presence of Zn++, IN gets converted to multimeric forms of IN, which, in the presence of viral LTR substrate were capable of efficient concerted integration (Figure S4 of the Supporting Information). Dimers and tetramers of IN were also observed within the SC and strand transfer complex (8) and in stable synaptic complex (6).
We showed that our HIV IN preparations preferred the 20R and 21R substrates over the larger size 33R and 42R substrates for concerted integration (Figure 3)(Figure S2 and S3 of the Supporting Information). The data suggested that IN may be binding to the shorter ODN substrates differently than the longer ODN substrates. Previous reports have suggested that the length of ODN also affected 3′ OH processing activity, the single insertion of a viral DNA end into a target, and the cooperation and the stability of HIV IN subunits associated with the ODN (17, 41–43). The preference of all of the recessed ODN over their blunt ended counterpart substrates for concerted integration (Figure 3) suggests that 3′ OH processing is slow for concerted integration as shown previously in the context of the HIV SC using a 1.6 kb U5 substrate (6–8). In summary, the apparent reason why the 33R and 42R substrates are less efficient than the 20R and 21R substrates could be linked to binding of IN to sequences upstream of the 21th nucleotide of U5 (Figure 5), as shown by DNaseI protective footprint analysis (8, 14).
HIV IN catalyzes concerted integration with longer size DNA substrates (>300 bp) without or with the assistance of the lens epithelium-derived growth factor (LEDGF)(11, 21, 27, 33, 34). Previous attempts to promote HIV concerted integration using 32 bp U5 ODN substrates (blunt or recessed ended) were successful with wt IN however, activity only occurred in the presence of LEDGF (27, 44, 45). The apparent need for LEDGF for concerted integration activity with ODN may be linked to the multimeric state of IN in these studies. HIV IN in solution existed in a dimer-tetramer equilibrium (27) and 3′ OH processing was independent of LEDGF (44). Our HIV IN purified in the presence of EDTA was predominately a monomer with minimal quantities of dimers or tetramers, except in the presence of Zn++ when the majority was tetramers (Figure 2a). This IN preparation, or a Zn++-induced tetramer of this preparation, performed efficient concerted integration with ODN in the absence of LEDGF (Figure 3) and possessed efficient 3′ OH processing activity (18). The above data suggests that an HIV IN monomer-tetramer equilibrium (Figure 2a)(17) has an unknown advantage over a dimer-tetramer (27) equilibrium for concerted integration using ODN substrates.
HIV IN promotes concerted integration with 20R and 21R substrates (Figure 3)(Figure S2 and S3 of the Supporting Information) similar to the monomeric PFV IN with its cognate 19R substrate (9, 13). Therefore, both IN proteins are fully capable of concerted integration by binding to either ODN or 1.6 kb DNA substrates (11, 13). What accounts for the extended binding by HIV IN to ~32 bp on U5 (Figure 5) as well as U3 (8, 14, 18)? Does the extended protection by HIV IN on large size U5 substrates (Figure 5) suggest different DNA binding modes in the presence of different size DNA substrates? Perhaps the binding of the PFV IN monomer to its cognate 19R DNA substrate offers a possible explanation (9, 30). The PFV intasome crystal structures obtained with the 19R substrate clearly demonstrates that the asymmetric unit contains a dimer of IN bound to a single DNA molecule. The resolved inner PFV monomer only contacts the DNA for catalysis while the outer monomer forms the CCD:CCD bridge between the two monomers bound per viral DNA end. The functions of the unresolved N-terminus and C-terminus domains of the outer monomer were suggested to provide structural support for the intasome (30, 46). Genetic and molecular evidence suggests that the unresolved HIV C-terminus or “tail” region (residues 270 to 288) appear to play a role during the 3′ OH processing reaction and/or facilitate the assembly of the intasome (44, 47). Whether the extended protection of U5 beyond the 21th nucleotide involves the “tail” region of HIV IN or the unresolved domains is unknown. Atomic resolution studies of HIV IN with 20R and 33R will be necessary to resolve a functional role for the extended interactions of HIV IN with its different size substrates.
Supplementary Material
Acknowledgments
We thank Dr. Hideki Aihara (University of Minnesota) for reading the manuscript and providing his critical comments.
ABBREVIATIONS
- HIV
human immunodeficiency virus type-1
- IN
integrase
- ODN
oligonucleotides
- PIC
preintegration complex
- SC
synaptic complex
- PFV
prototype foamy virus
- LTR
long terminal repeat
- B
blunt
- R
recessed
- wt
wild type
- PEG
polyethylene glycol
- SEC
size exclusion chromatography
- IDT
Integrated DNA technologies
- DMSO
dimethyl sulfoxide
- RSV
Rous sarcoma virus
- RAL
raltegravir
- CHS
circular half site
Footnotes
This work was supported in part by National Institute of Allergy and Infectious Diseases grant R21AI081629, Merck & Co. Inc Investigator Initiated Program no. 37072, and by Saint Louis University
ASSOCIATED CONTENT
Supporting Information. Supporting figures (Figures S1–S7). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 3.Brown HE, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 5.Craigie R. Retroviral DNA Integration. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 613–630. [Google Scholar]
- 6.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of human immunodeficiency virus type-1 concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J Virol. 2007;81:12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A. 2010;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha S, Grandgenett D. Recombinant HIV-1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegraton complexes from virus-infected cells. J Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodarzi G, Chiu R, Brackmann K, Kohn K, Pommier Y, Grandgenett DP. Host site selection for concerted integration by human immunodeficiency virus type-1 virions in vitro. Virology. 1997;231:210–217. doi: 10.1006/viro.1997.8558. [DOI] [PubMed] [Google Scholar]
- 13.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey KK, Bera S, Vora AC, Grandgenett DP. Physical trapping of HIV-1 synaptic complex by different structural classes of integrase strand transfer inhibitors. Biochemistry. 2010;49:8376–8387. doi: 10.1021/bi100514s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 18.Bera S, Pandey KK, Vora AC, Grandgenett DP. HIV-1 integrase strand transfer inhibitors stabilize an integrase-single blunt-ended DNA complex. J Mol Biol. 2011;410:831–846. doi: 10.1016/j.jmb.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandgenett DP, Bera S, Pandey KK, Vora AC, Zahm J, Sinha S. Biochemical and biophysical analyses of concerted (U5/U3) integration. Methods. 2009;47:229–236. doi: 10.1016/j.ymeth.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCord M, Stahl SJ, Mueser TC, Hyde CC, Vora AC, Grandgenett DP. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Expr Purif. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- 21.Pandey KK, Sinha S, Grandgenett DP. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J Virol. 2007;81:3969–3979. doi: 10.1128/JVI.02322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 24.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 27.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Brochon JC. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- 29.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21:249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci U S A. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 33.Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci U S A. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lesbats P, Metifiot M, Calmels C, Baranova S, Nevinsky G, Andreola ML, Parissi V. In vitro initial attachment of HIV-1 integrase to viral ends: control of the DNA specific interaction by the oligomerization state. Nucleic Acids Res. 2008;36:7043–7058. doi: 10.1093/nar/gkn796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baranova S, Tuzikov FV, Zakharova OD, Tuzikova NA, Calmels C, Litvak S, Tarrago-Litvak L, Parissi V, Nevinsky GA. Small-angle X-ray characterization of the nucleoprotein complexes resulting from DNA-induced oligomerization of HIV-1 integrase. Nucleic Acids Res. 2007;35:975–987. doi: 10.1093/nar/gkl1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochon JC, LeCam E, Coulaud D, Auclair C, Mouscadet JF. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 41.Lee SP, Censullo ML, Kim HG, Han MK. Substrate-length-dependent activities of human immunodeficiency virus type 1 integrase in vitro: differential DNA binding affinities associated with different lengths of substrates. Biochemistry. 1995;34:10215–10223. doi: 10.1021/bi00032a015. [DOI] [PubMed] [Google Scholar]
- 42.Pemberton IK, Buc H, Buckle M. Displacement of viral DNA termini from stable HIV-1 integrase nucleoprotein complexes induced by secondary DNA-binding interactions. Biochemistry. 1998;37:2682–2690. doi: 10.1021/bi971893j. [DOI] [PubMed] [Google Scholar]
- 43.Pemberton IK, Buckle M, Buc H. The metal ion-induced cooperative binding of HIV-1 integrase to DNA exhibits a marked preference for Mn(II) rather than Mg(II) J Biol Chem. 1996;271:1498–1506. doi: 10.1074/jbc.271.3.1498. [DOI] [PubMed] [Google Scholar]
- 44.Dar MJ, Monel B, Krishnan L, Shun MC, Di Nunzio F, Helland DE, Engelman A. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology. 2009;6:94. doi: 10.1186/1742-4690-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohammed KD, Topper MB, Muesing MA. Sequential deletion of the integrase (Gag-Pol) carboxyl-terminus reveals distinct phenotypic classes of defective HIV-1. J Virol. 2011;85:4654–4666. doi: 10.1128/JVI.02374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.