A truncated heterodimer of human PSPC1–NONO protein, a paraspeckle-specific complex, has been crystallized and the diffraction data collected to a resolution of 1.9 Å.
Keywords: paraspeckles, PSPC1–NONO heterodimer, RNA-recognition motifs, DBHS-family proteins
Abstract
The paraspeckle component 1 (PSPC1) and non-POU-domain-containing octamer-binding protein (NONO) heterodimer is an essential structural component of paraspeckles, ribonucleoprotein bodies found in the interchromatin space of mammalian cell nuclei. PSPC1 and NONO both belong to the Drosophila behaviour and human splicing (DBHS) protein family, which has been implicated in many aspects of RNA processing. A heterodimer of the core DBHS conserved region of PSPC1 and NONO comprising two tandemly arranged RNA-recognition motifs (RRMs), a NONA/paraspeckle (NOPS) domain and part of a predicted coiled-coil domain has been crystallized in space group C2, with unit-cell parameters a = 90.90, b = 67.18, c = 94.08 Å, β = 99.96°. The crystal contained one heterodimer in the asymmetric unit and diffracted to 1.9 Å resolution using synchrotron radiation.
1. Introduction
Eukaryotic gene expression with its messenger RNA (mRNA) metabolism is subject to diverse post-transcriptional control (Moore, 2005 ▶). An indicator of the multiplicity of this control is the multitude of proteins with RNA-binding capabilities. Beside RNA-binding motifs such as the Pumilio homology domain in PUF proteins (Wang et al., 2002 ▶), zinc-binding domains (D’Souza & Summers, 2004 ▶), K-homology (KH) domains (Grishin, 2001 ▶) and arginine–glycine–glycine (RGG) boxes (Godin & Varani, 2007 ▶), RNA-recognition motifs (RRMs) are abundantly represented in eukaryotic proteins, with 0.5–1% of human genes containing at least one RRM (Venter et al., 2001 ▶). The majority of RRMs have a length of 80–90 residues with a ‘sandwich’ structure consisting of four-stranded antiparallel β-sheets packed against two α-helices (Cléry et al., 2008 ▶). One RRM can bind between two and eight nucleotides (Calero et al., 2002 ▶). The maximum nucleotide length and affinity can be altered depending on the C- and N-terminal extensions and the number of RRMs in the structure (Maris et al., 2005 ▶).
This work focuses on a family of multifunctional nuclear RRM-containing proteins named DBHS (Drosophila behaviour and human splicing) proteins. DBHS proteins have a conserved domain ensemble in common: two RRMs followed by a NONA/paraspeckle (NOPS) domain and a coiled-coil domain. The DBHS domain is highly conserved (Bond & Fox, 2009 ▶). For example, the three mammalian DBHS proteins PSPC1 (also called PSP1), NONO (p54nrb) and splicing factor proline/glutamine-rich protein (SFPQ; also called PSF) show over 70% sequence identity within this region. Furthermore, this domain is crucial for the homodimerization and heterodimerization of the proteins with each other (Myojin et al., 2004 ▶; Fox et al., 2005 ▶). DBHS proteins are involved in several stages of mRNA metabolism: transcription initiation (Dong et al., 1993 ▶; Yang et al., 1993 ▶, 1997 ▶), co-activation (Kuwahara et al., 2006 ▶; Dong et al., 2005 ▶), constitutive and alternative splicing (Patton et al., 1993 ▶; Peng et al., 2002 ▶; Kameoka et al., 2004 ▶; Ito et al., 2008 ▶), transcriptional termination (Kaneko et al., 2007 ▶) and nuclear retention of A-to-I hyperedited RNA (Zhang & Carmichael, 2001 ▶; Chen & Carmichael, 2009 ▶; Prasanth et al., 2005 ▶).
It has previously been reported that the interaction between PSPC1 and NONO is crucial for the integrity and function of paraspeckles, subnuclear bodies that have been implicated in the nuclear retention of hyperedited mRNA (Fox et al., 2005 ▶; Sasaki et al., 2009 ▶). To gain further insight into the interaction and the function of PSPC1 and NONO in paraspeckles at the molecular level, we here report the production, crystallization and preliminary X-ray diffraction analysis of a truncated heterodimer of human PSPC1–NONO protein.
2. Materials and methods
2.1. Cloning and expression
The conserved regions of PSPC1 (residues 61–320 of UniProt entry PSPC1_HUMAN) and NONO (residues 53–312 of UniProt entry NONO_HUMAN) were amplified by PCR and subcloned into the first and second multiple cloning sites of pETDuet-1 (Novagen), respectively. A tobacco etch virus (TEV) protease cleavage site encoded in the 5′ primer was introduced between the hexahistidine tag (His6) and the N-terminus of PSPC1 to facilitate removal at a later stage of purification.
pETDuet-1-His6PSPC1-NONO was transformed into E. coli Rosetta 2 (DE3) (Novagen). 500 ml LB growth medium supplemented with 100 µg ml−1 ampicillin and 50 µg ml−1 chloramphenicol was inoculated with a bacterial pre-culture at 310 K. When the culture reached an optical density (600 nm) of 0.5–0.8, it was induced with 0.5 mM IPTG and incubated with shaking overnight at 298 K.
To produce selenomethionine-substituted PSPC1–NONO protein, a 50 ml overnight pre-culture was pelleted and then resuspended in 500 ml M9 medium supplemented with 100 µg ml−1 ampicillin, 50 µg ml−1 chloramphenicol and an additional 0.3% glucose and incubated until the optical density (600 nm) reached 0.6. The methionine-biosynthesis pathway was inhibited by the addition of 100 µg ml−1 threonine/lysine/phenylalanine and 50 µg ml−1 valine/leucine/proline (modified from Doublié, 2007 ▶). Following 30 min incubation, protein production was induced with 1 mM IPTG and by the addition of 25 mg selenomethionine and 25 ml DMEM medium-M (Thermo Scientific). The culture was incubated for 42 h at 298 K before being harvested by centrifugation.
2.2. Purification
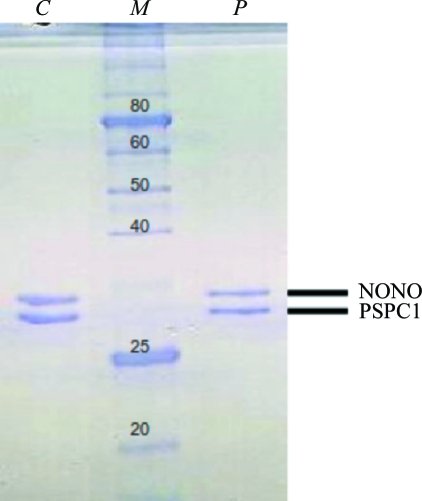
The bacterial pellet was resuspended in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM imidazole and 10%(v/v) glycerol and lysed using an Emulsiflex C5 high-pressure homogeniser (Avestin). The lysate was loaded onto a 5 ml HisTrap column (GE Healthcare) and washed and nickel-affinity chromatography was performed (BioLogic DuoFlow, Bio-Rad) with a 50–500 mM imidazole gradient to elute the native or the selenomethionine-substituted His6PSPC1–NONO complex. The peak fraction was subjected to buffer exchange [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10%(v/v) glycerol] using a PD-10 desalting column (GE Healthcare) and the tag was then removed by incubation with His6-tagged TEV protease overnight at 277 K. PSPC1–NONO protein was separated from uncleaved protein, His6 tag and His6-TEV protease via filtration over a His-Trap column. The flowthrough containing the cleaved protein was collected, concentrated, loaded onto a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare) and developed with 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10%(v/v) glycerol. Peak fractions were collected and the purity of the protein sample was confirmed by SDS–PAGE stained with Coomassie Brilliant Blue (Fig. 1 ▶).
Figure 1.
SDS–PAGE of PSPC1–NONO heterodimer. 12% SDS–polyacrylamide gel stained with Coomassie Brilliant Blue (lane C, a dissolved crystal of PSPC1–NONO; lane M, molecular-weight markers labelled in kDa; lane P, purified PSPC1–NONO heterodimer). Theoretical masses: PSPC1, 29.9 kDa; NONO, 30.2 kDa.
2.3. Crystallization
Initial crystallization screening of the native complex [9.5 mg ml−1 in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10%(v/v) glycerol] was performed in 96-well sitting-drop format using a Phoenix liquid-handling robot (Art Robbins Scientific). A variety of commercially available crystallization screens were trialled, including Crystal Screen, Crystal Screen 2, Crystal Screen Lite, Index, PEG/Ion and Natrix (Hampton Research), Wizard Screens I, II and III (Emerald BioSystems) and Nextal Anions and Cations Suites (Qiagen). A typical drop of 300 nl in volume with three different protein:reservoir ratios (1:2, 1:1 and 2:1) was equilibrated against 80 µl reservoir solution at 293 K in the initial screening stage. Use of these drop ratios allows the efficient robotic setup of trays using a single protein stock, rather than the alternative of setting up 1:1 drops with diluted protein stocks.
Small single crystals appeared after 1 d in Index condition No. 71 [0.1 M Bis-Tris pH 6.5, 0.2 M NaCl, 25%(w/v) PEG 3350] with a 1:2 protein:reservoir ratio in the drop. Based on this result, an extensive optimization in 24-well hanging-drop format was carried out in approximately 1800 drops. Ultimately, diffraction-quality crystals (0.2 × 0.2 × 0.15 mm) were obtained by mixing 3 µl protein solution (10 mg ml−1) and 6 µl reservoir solution [0.1 M Bis-Tris pH 5.5, 0.5 M NaCl, 28%(w/v) PEG 3350] and equilibrating the drops against 1 ml reservoir solution at 293 K. Optimal crystallization conditions for the selenomethionine-substituted protein consisting of 4 µl protein solution (8 mg ml−1) and 6.4 µl reservoir solution [0.5 M NaCl, 0.1 M Bis-Tris pH 5.5, 18%(w/v) PEG 3350] led to smaller crystals (0.1 × 0.05 × 0.05 mm).
2.4. Data collection and preliminary X-ray diffraction analysis
Complete X-ray diffraction data were collected on beamline MX2 of the Australian Synchrotron with an ADSC Quantum 315r detector and cooling to 100 K. 360° of native data were collected at a wavelength of 1.077 Å in 0.5° oscillations with 0.5 s exposure per image and 95% attenuation of the beam.
The data were processed using XDS (Kabsch, 2010 ▶); the space group was evaluated with POINTLESS (Evans, 2006 ▶) and the data were merged and scaled with SCALA (Winn et al., 2011 ▶). The crystals belonged to space group C2, with unit-cell parameters a = 90.90, b = 67.18, c = 94.08 Å, β = 99.96°. For the best native data set, we chose a data cutoff at 1.90 Å on the basis that at this resolution the data have an average I/σ(I) of 2 and the otherwise excellent-looking Wilson plot begins to degrade beyond 1.89 Å.
Additionally, a full multiple anomalous dispersion (MAD) data set was collected. Selenomethionine-substituted PSPC1–NONO crystals were cryoprotected with Paratone oil (Hampton Research) and flash-cooled to 100 K. A similar data-collection protocol to that used for the native crystal was used, resulting in complete (360°) data sets at three wavelengths (high-energy remote, 0.95372 Å; inflection point, 0.97973 Å; peak, 0.97963 Å). Data-collection and processing statistics are listed in Table 1 ▶.
Table 1. Crystallographic data statistics for native and MAD data sets.
Values in parentheses are for the highest resolution shell.
| Native | Inflection | Peak | Remote | |
|---|---|---|---|---|
| Wavelength (Å) | 1.07716 | 0.97973 | 0.97963 | 0.95372 |
| Space group | C2 | C2 | C2 | C2 |
| Unit-cell parameters (Å, °) | a = 90.90, b = 67.18, c = 94.08, β = 99.96 | a = 91.00, b = 67.28, c = 94.03, β = 100.37 | a = 91.28 b = 67.41, c = 94.28, β = 100.33 | a = 91.12, b = 67.36, c = 94.02, β = 100.47 |
| Resolution (Å) | 19.83–1.90 (2.00–1.90) | 19.36–2.53 (2.67–2.53) | 19.40–2.58 (2.72–2.56) | 19.37–2.50 (2.64–2.50) |
| No. of observed reflections | 326294 (47355) | 131355 (13149) | 130106 (15645) | 141572 (16338) |
| Data-collection range (°) | 360 | 360 | 360 | 360 |
| Oscillation per image (°) | 0.5 | 0.5 | 0.5 | 0.5 |
| Unique reflections | 43472 (6235) | 18667 (2623) | 17815 (2560) | 19471 (2786) |
| Completeness (%) | 98.8 (97.8) | 99.2 (96.5) | 99.7 (99.5) | 99.6 (98.9) |
| Average I/σ(I) | 16.3 (2.1) | 18.9 (2.0) | 18.4 (2.0) | 18.3 (2.2) |
| Multiplicity | 7.5 (7.6) | 7.0 (5.0) | 7.3 (6.1) | 7.3 (5.9) |
| Rmerge (%) | 9.7 (100.2) | 8.6 (68.3) | 7.9 (81.3) | 8.0 (76.6) |
| Rmeas (%) | 10.4 (109.3) | 9.2 (76.4) | 8.5 (88.9) | 8.6 (84.2) |
| Rp.i.m. (%) | 3.8 (39.5) | 3.4 (33.4) | 3.1 (35.5) | 3.2 (34.5) |
| Anomalous completeness (%) | 98.5 (93.0) | 99.5 (98.5) | 99.4 (97.5) | |
| Ranom (%) | 3.5 (39.7) | 5.0 (37.8) | 4.3 (38.2) |
3. Results and discussion
The full-length heterodimer of PSPC1–NONO exhibited very low expression and solubility in the E. coli expression system tested in our hands (data not shown). Thus, it was necessary to optimize the lengths and the termini of the heterodimer to make the proteins suitable for crystallographic studies. The construct described here (PSPC1 residues 61–320 and NONO 53–312) includes a partial DBHS domain: it contains the two RRM domains, the NOPS domain and part of the coiled-coil domain. Prediction programs [COILS (Lupas et al., 1991 ▶) and Paircoil (Berger et al., 1995 ▶)] indicate that the coiled-coil domains of PSPC1 and NONO extend a further 60 residues beyond the C-terminus of this construct.
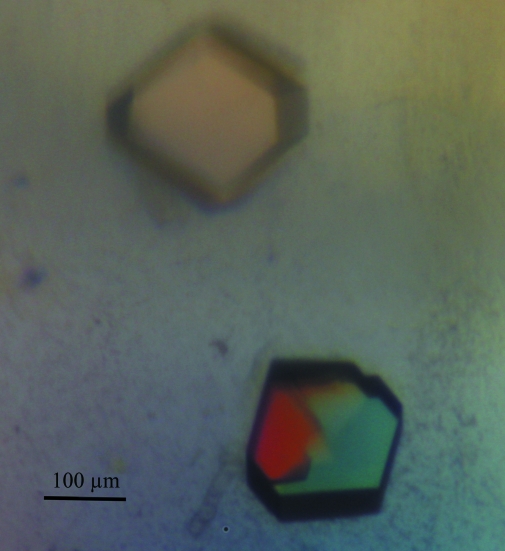
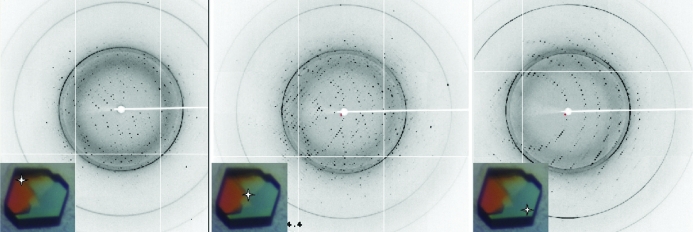
The crystals of the PSPC1–NONO heterodimer consistently appeared as symmetrical penetration twins (Fig. 2 ▶). The crystals proved to be highly sensitive to handling and exhaustive optimization of cryoprotection led to a condition consisting of brief (<2 s) washing in artificial mother liquor supplemented with 35%(v/v) ethylene glycol. Any prolonged incubation in the cryoprotectant caused the crystals to dissolve or crack. Thus, the very brief incubation in the cryoprotectant often resulted in incomplete cryoprotection and the presence of some ice rings in the diffraction images (Fig. 3 ▶).
Figure 2.
Crystals of the PSPC1–NONO heterodimer viewed under crossed polarizers. The different twin domains are clearly visible.
Figure 3.
X-ray diffraction patterns obtained from a PSPC1–NONO crystal. Different twin domains are isolated using the microfocus beam. The crystal shown is similar to that from which data were collected. Stars indicate how the crystal was centred to obtain the differing diffraction patterns. The more prominent ice rings visible in the diffraction pattern occur at 3.66, 2.25 and 1.93 Å.
The penetration-twinned nature of the crystals required careful attention to crystal centring during data collection. The use of a micro-focused beam (30 × 30 µm) made it possible to collect complete data from different parts of the crystals. Fig. 3 ▶ illustrates the effects of different centring on the resulting lattices of the twin domains. Clean single-crystal data were obtained by collecting data at a corner of the crystal. Solvent-content analysis indicated a predicted solvent content of 47% with a Matthews coefficient of 2.32 Å3 Da−1 (Matthews, 1968 ▶) for one PSPC1–NONO heterodimer in the asymmetric unit. The presence of both components in the crystals was confirmed by running the dissolved crystals on SDS–PAGE (Fig. 1 ▶). Structure solution and refinement are currently under way.
Acknowledgments
This work was funded by NHMRC Project Grant 588130 to CSB and AHF and an NHMRC Postdoctoral Training Fellowship to ML. Data collection was undertaken on the MX2 beamline at the Australian Synchrotron, Victoria, Australia. We are grateful to Tom Caradoc-Davies, Amanda Kirby and Rachel Williams for their assistance.
References
- Berger, B., Wilson, D. B., Wolf, E., Tonchev, T., Milla, M. & Kim, P. S. (1995). Proc. Natl Acad. Sci. USA, 92, 8259–8263. [DOI] [PMC free article] [PubMed]
- Bond, C. S. & Fox, A. H. (2009). J. Cell Biol. 186, 637–644. [DOI] [PMC free article] [PubMed]
- Calero, G., Wilson, K. F., Ly, T., Rios-Steiner, J. L., Clardy, J. C. & Cerione, R. A. (2002). Nature Struct. Biol. 9, 912–917. [DOI] [PubMed]
- Chen, L.-L. & Carmichael, G. G. (2009). Mol. Cell, 35, 467–478. [DOI] [PMC free article] [PubMed]
- Cléry, A., Blatter, M. & Allain, F. H. (2008). Curr. Opin. Struct. Biol. 18, 290–298. [DOI] [PubMed]
- Dong, B., Horowitz, D. S., Kobayashi, R. & Krainer, A. R. (1993). Nucleic Acids Res. 21, 4085–4092. [DOI] [PMC free article] [PubMed]
- Dong, X., Shylnova, O., Challis, J. R. & Lye, S. J. (2005). J. Biol. Chem. 280, 13329–13340. [DOI] [PubMed]
- Doublié, S. (2007). Methods Mol. Biol. 363, 91–108. [DOI] [PubMed]
- D’Souza, V. & Summers, M. F. (2004). Nature (London), 431, 586–590. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fox, A. H., Bond, C. S. & Lamond, A. I. (2005). Mol. Biol. Cell, 16, 5304–5315. [DOI] [PMC free article] [PubMed]
- Godin, K. S. & Varani, G. (2007). RNA Biol. 4, 69–75. [DOI] [PubMed]
- Grishin, N. V. (2001). Nucleic Acids Res. 29, 638–643. [DOI] [PMC free article] [PubMed]
- Ito, T., Watanabe, H., Yamamichi, N., Kondo, S., Tando, T., Haraguchi, T., Mizutani, T., Sakurai, K., Fujita, S., Izumi, T., Isobe, T. & Iba, H. (2008). Biochem. J. 411, 201–209. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kameoka, S., Duque, P. & Konarska, M. M. (2004). EMBO J. 23, 1782–1791. [DOI] [PMC free article] [PubMed]
- Kaneko, S., Rozenblatt-Rosen, O., Meyerson, M. & Manley, J. L. (2007). Genes Dev. 21, 1779–1789. [DOI] [PMC free article] [PubMed]
- Kuwahara, S., Ikei, A., Taguchi, Y., Tabuchi, Y., Fujimoto, N., Obinata, M., Uesugi, S. & Kurihara, Y. (2006). Biol. Reprod. 75, 352–359. [DOI] [PubMed]
- Lupas, A., Van Dyke, M. & Stock, J. (1991). Science, 252, 1162–1164. [DOI] [PubMed]
- Maris, C., Dominguez, C. & Allain, F. H. (2005). FEBS J. 272, 2118–2131. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Moore, M. J. (2005). Science, 309, 1514–1518. [DOI] [PubMed]
- Myojin, R., Kuwahara, S., Yasaki, T., Matsunaga, T., Sakurai, T., Kimura, M., Uesugi, S. & Kurihara, Y. (2004). Biol. Reprod. 71, 926–932. [DOI] [PubMed]
- Patton, J. G., Porro, E. B., Galceran, J., Tempst, P. & Nadal-Ginard, B. (1993). Genes Dev. 7, 393–406. [DOI] [PubMed]
- Peng, R., Dye, B. T., Pérez, I., Barnard, D. C., Thompson, A. B. & Patton, J. G. (2002). RNA, 8, 1334–1347. [DOI] [PMC free article] [PubMed]
- Prasanth, K. V., Prasanth, S. G., Xuan, Z., Hearn, S., Freier, S. M., Bennett, C. F., Zhang, M. Q. & Spector, D. L. (2005). Cell, 123, 249–263. [DOI] [PubMed]
- Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T. & Hirose, T. (2009). Proc. Natl Acad. Sci. USA, 106, 2525–2530. [DOI] [PMC free article] [PubMed]
- Venter, J. C. et al. (2001). Science, 291, 1304–1351. [DOI] [PubMed]
- Wang, X., McLachlan, J., Zamore, P. D. & Hall, T. M. (2002). Cell, 110, 501–512. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, Y.-S., Hanke, J. H., Carayannopoulos, L., Craft, C. M., Capra, J. D. & Tucker, P. W. (1993). Mol. Cell. Biol. 13, 5593–5603. [DOI] [PMC free article] [PubMed]
- Yang, Y.-S., Yang, M.-C., Tucker, P. W. & Capra, J. D. (1997). Nucleic Acids Res. 25, 2284–2292. [DOI] [PMC free article] [PubMed]
- Zhang, Z. & Carmichael, G. G. (2001). Cell, 106, 465–475. [DOI] [PubMed]