Abstract
Xanthomonas axonopodis pv. citri is a phytopathogen bacterium that causes severe citrus canker disease. Similar to other phytopathogens, after infection by this bacterium, plants trigger a defense mechanism that produces reactive oxygen species. Ferredoxin-NADP+ reductases (FNRs) are redox flavoenzymes that participate in several metabolic functions, including the response to reactive oxygen species. Xanthomonas axonopodis pv. citri has a gene (fpr) that encodes for a FNR (Xac-FNR) that belongs to the subclass I bacterial FNRs. The aim of this work was to search for the physiological role of this enzyme and to characterize its structural and functional properties. The functionality of Xac-FNR was tested by cross-complementation of a FNR knockout Escherichia coli strain, which exhibit high susceptibility to agents that produce an abnormal accumulation of •O2 -. Xac-FNR was able to substitute for the FNR in E. coli in its antioxidant role. The expression of fpr in X. axonopodis pv. citri was assessed using semiquantitative RT-PCR and Western blot analysis. A 2.2-fold induction was observed in the presence of the superoxide-generating agents methyl viologen and 2,3-dimethoxy-1,4-naphthoquinone. Structural and functional studies showed that Xac-FNR displayed different functional features from other subclass I bacterial FNRs. Our analyses suggest that these differences may be due to the unusual carboxy-terminal region. We propose a further classification of subclass I bacterial FNRs, which is useful to determine the nature of their ferredoxin redox partners. Using sequence analysis, we identified a ferredoxin (XAC1762) as a potential substrate of Xac-FNR. The purified ferredoxin protein displayed the typical broad UV-visible spectrum of [4Fe-4S] clusters and was able to function as substrate of Xac-FNR in the cytochrome c reductase activity. Our results suggest that Xac-FNR is involved in the oxidative stress response of Xanthomonas axonopodis pv. citri and performs its biological function most likely through the interaction with ferredoxin XAC1762.
Introduction
Xanthomonas axonopodis pv. citri is a Gram-negative obligate aerobic bacterium that is responsible for severe citrus canker disease, which affects most commercial citrus cultivars. The disease appears as raised necrotic corky lesions on the leaves, stems and fruits, which reduces the fruit quality and quantity. The pathogen enters host plant tissues through the stomata or tissue wounds, and the infection is visualized as circular spots on the surface of the leaves. Subsequently, the bacteria colonize the apoplast and cause the leaf epidermis to break due to cell hyperplasia [1], [2].
In response to pathogens, the plant metabolism changes to produce reactive oxygen species, including superoxide radicals (•O2 -), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH) which kill the infectious agent [3]. Therefore, pathogens need to prevent and overcome oxidative stress in order to establish and maintain infections [4]. Different studies have demonstrated the protective role that catalases and peroxidases perform in Xanthomonas spp during oxidative stress developed by the plant's defence mechanisms [5], [6]. However, in other Gram-negative bacteria, such as Escherichia coli and Pseudomonas putida, alternative mechanisms of response to oxidative stress have been reported where ferredoxin-NADP+ reductase (FNR) performs an important function [7]–[12].
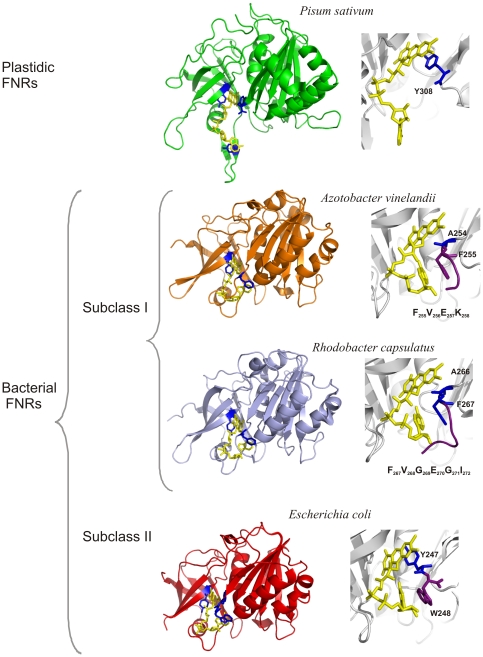
FNR is a flavoenzyme that is distributed in a large range of organisms. It participates in metabolic processes as dissimilar as photosynthesis [13], nitrogen assimilation [14], [15] and fatty acid desaturation [16]. The FAD prosthetic group enables these enzymes to catalyze electron transfer from obligate two-electron carriers, such as NADP(H), to one-electron proteins [17], such as ferredoxin, flavodoxin [18] or hemoxygenase [19]. In photosyntetic tissues and organisms, the reaction is directed to NADP+ reduction to produce NADPH; however, in non-photosynthetic tissues or organisms, the reaction is mainly displaced towards the oxidation of NADPH in order to produce low-potential electron donors that will be used in different metabolic functions [17], [20]. FNRs are grouped into two classes according to their structural and phylogenetic features [21], [22]: a plastidic class, which is found in photosynthetic tissues and displays high catalytic efficiency, and a bacterial class, which has a low catalytic efficiency (Figure 1). The bacterial class is further subdivided into two subclasses: subclass I, which has a structural prototype that is similar to the FNR from Azotobacter vinelandii, and subclass II, which is represented by the FNR from Escherichia coli [21]. As shown in Figure 1, the main differences between subclasses I and II bacterial FNRs are located in the carboxy-terminal region where E. coli FNR (Ec-FNR) has a tyrosine that faces the isoalloxazine of FAD, and subclass I enzymes have an alanine in the equivalent position and a longer carboxy-terminal extension [21].
Figure 1. Classification of plant-type FNRs according to structural features.
Plant-type FNRs are classified as plastidic and bacterial FNRs [21]. Structures of representative prototypes of the plastidic and the bacterial groups are shown. Bacterial-type FNRs are subdivided into two subclasses, subclass I and subclass II. FNRs from Azobacter vinelandii and Rhodobacter capsulatus belong to subclass I; however, they differ in length and sequence of the carboxy-terminal region upstream of the alanine that faces the isoalloxazine of FAD. A view of the environments of the different prosthetic groups and the sequences of the carboxy-terminal extensions are shown to the right of each enzyme structure. FNRs from Pisum sativum (1qg0), E. coli (1fdr), A. vinelandii (1a8p) and R. capsulatus (2bgj) were used as model proteins. Figures were generated using PyMol. Available: http://pymol.sourceforge.net/.
In a previous study, we identified a FNR in X. axonopodis pv. citri (Xac-FNR) that has all the structural and functional features of a typical subclass I bacterial FNR [23]. The aim of this work was to search for the physiological role of the Xac-FNR and its natural substrate in X. axonopodis pv. citri and to investigate its participation in the bacterial oxidative stress response. Furthermore, we performed a structural and functional characterization of Xac-FNR and analyzed the obtained data in the context of other plant-type FNRs.
Results and Discussion
Complementation of an E. coli fpr-null mutant with Xac-FNR
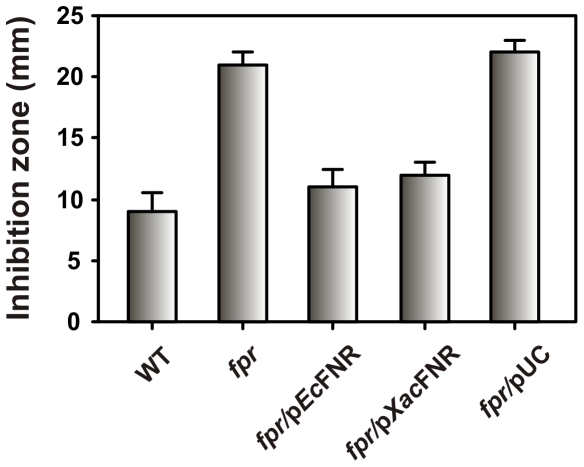
The functionality of Xac-FNR was initially tested by cross-complementation of an E. coli fpr mutant (the fpr gene encodes Ec-FNR). The E. coli fpr RR6A strain exhibited a high susceptibility to the bactericidal effects of methyl viologen (MV) [9]. These bacteria displayed a lower growth rate compared to wild-type cells when exposed to oxidants due to the abnormal accumulation of •O2 - in the cytosol [9]. The Xac-FNR coding sequence was amplified and cloned into the pUC119 vector, and the resulting plasmid (pUC/XacFNR) was transformed into E. coli RR6A. The resistance to MV of the resulting strain was evaluated using the inhibition zone assay. As shown in Figure 2, the Xac-FNR enzyme was able to restore the E. coli mutant to similar levels of resistance as the wild-type strain. This result indicates that Xac-FNR is able to substitute Ec-FNR in its antioxidant role. Attempts to construct a X. axonopodis pv. citri fpr-knockout strain were unsuccessful. This observation may result from the protective function of Xac-FNR or from another role of the enzyme yet to be uncovered. In general, the impossibility to recover knockout strains of a gene in bacteria suggests its participation in essential housekeeping metabolic steps. This issue needs to be further investigated.
Figure 2. Complementation of the E. coli fpr-null mutant with Xac-FNR.
The E. coli fpr strain (RR6A) was transformed with pEcFNR that contained the endogenous fpr gene, pXacFNR that contained the Xac fpr gene, or pUC119. The susceptibility of E. coli strains to MV toxicity was evaluated using the disk diffusion assay. The diameters of the inhibition zones were measured after 24 h of incubation. Bars indicate mean ± standard deviation of three independent experiments.
The role of FNRs in oxidative stress protection is not completely understood. One of the possible targets of superoxide toxicity are the metal-dependent hydro-lyases that contain solvent-exposed [4Fe–4S]2+ clusters [24]. Recovery of these metal clusters requires reduction which is thought to be done by ferredoxin [25]. Thus, during oxidative stress, induction of Xac-FNR might be important for providing reduced ferredoxin. Another unwanted situation is the build-up of NADPH levels, which may favor the propagation of active oxygen species through the reduction of Fe3+ [9]. It is likely that FNR acts through NADPH oxidation using any electron acceptor that is available and maintains NADPH at tolerable levels during oxidative stress conditions.
Expression analysis of Xac-FNR under oxidative stress conditions
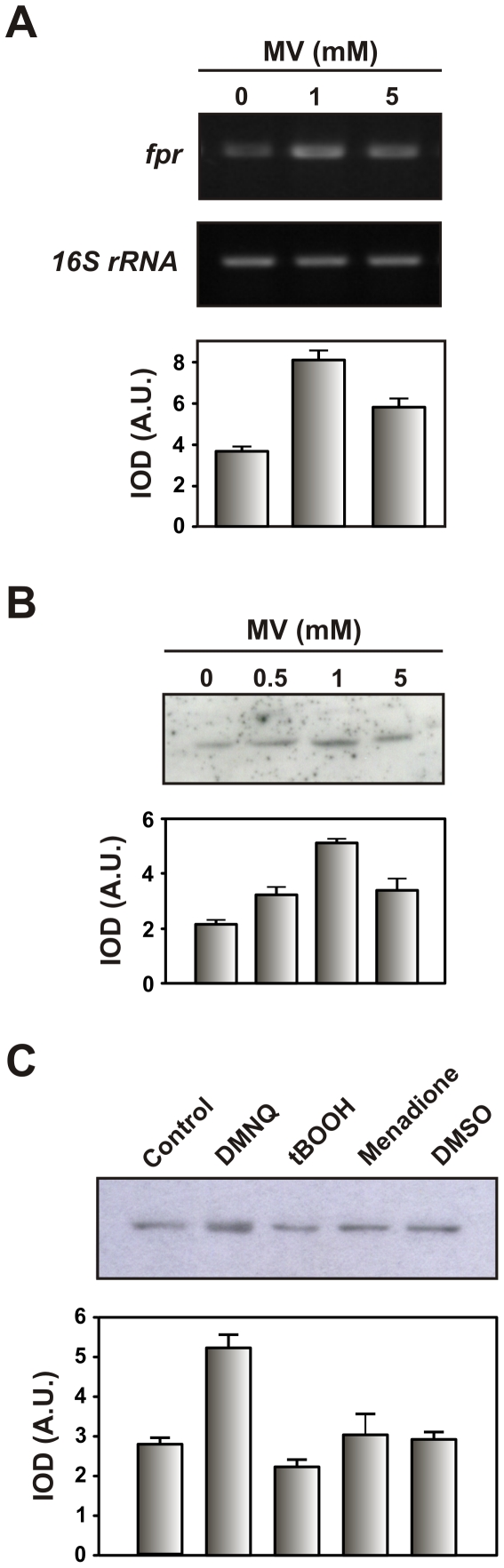
In order to investigate the involvement of Xac-FNR in the oxidative stress response in X. axonopodis pv. citri, fpr expression at the mRNA level was assessed using semiquantitative RT-PCR analysis. As shown in Figure 3A, expression of fpr was detected in normal growth conditions and exhibited a 2.2-fold induction in the presence of 1 mM MV. However, exposure to 5 mM MV led to less of an increased induction of the gene (1.5-fold). Analysis of the Xac-FNR protein abundance by Western blot analysis was highly correlated with the expression pattern obtained by RT-PCR (Figure 3B).
Figure 3. Expression analysis of Xac-FNR in response to oxidative treatments.
(A) Amplified products of the Xac fpr gene by semiquantitative RT-PCR using RNA preparations from Xanthomonas axonopodis pv. citri cultures grown in SB medium to the early exponential phase and exposed to the indicated concentrations of MV for 15 min. 16S rRNA was used as a loading control and for the quantitation of the total RNA in the RT-PCR experiments. (B) Xac-FNR accumulation after MV-dependent induction. Cleared extracts correspond to 25 µg of total soluble protein. Samples were analyzed by SDS-PAGE and immunoblot analysis using specific antisera. (C) Effect of 2,3-dimetoxy-1,4-naphthoquinone (DMNQ) 500 µM, tert-butyl hydroperoxide (tBOOH) 500 µM, menadione 100 µM and dimethyl sulfoxide (DMSO) on Xac-FNR protein expression. Cleared extracts correspond to 30 µg of total soluble protein and were analyzed by Western blot. The graphs below the gels in (A), (B) and (C) show the expression profiles that were obtained by densitometric quantification of the band intensities. Experiments were performed in triplicate with similar results, and the error bars indicate ±1 standard deviation of the mean (IOD, integrated optical density; A.U., arbitrary units).
Xac-FNR was also induced by exposure to the superoxide-generating agent 2,3-dimethoxy-1,4-naphthoquinone (DMNQ, 500 µM) to the same extent as MV 1 mM (Figure 3C). Induction of the fpr gene by superoxide-generating agents was previously reported in E. coli as a member of the SoxRS regulon [26] and in Pseudomonas putida under the control of FinR, a redox-sensing transcriptional regulator [27]. In E. coli the SoxRS regulon is also activated by hydrogen peroxide [28]. In X. axonopodis pv. citri SoxR was identified [29] but there is no evidence concerning the inclusion of Xac-FNR in this regulon. Nevertheless, the induction of fpr that was observed after exposure to superoxide-generating agents in X. axonopodis pv. citri suggests that FNR could serve a protective role against oxidative stress in this bacterium.
Structural and kinetic analyses
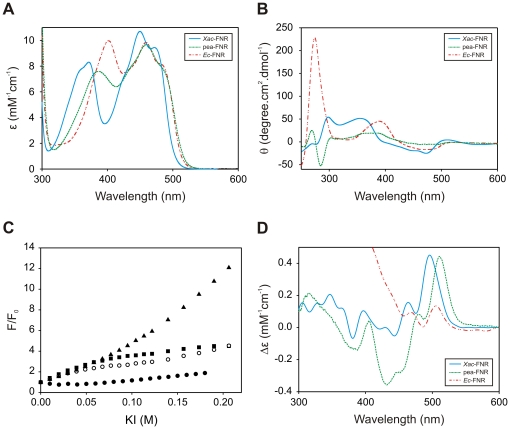
The UV-visible absorption and CD spectra were collected in order to obtain information about the FAD isoalloxazine environment. Figure 4A shows the UV-visible spectrum of Xac-FNR, and the representative spectra of plastidic and subclass II bacterial FNRs (pea and E. coli, respectively) for comparison. The UV-visible spectrum of Xac-FNR showed the typical pattern observed for subclass I bacterial FNRs with maxima at 450 nm and 372 nm [30]. Maxima for Xac-FNR were detected at lower wavelengths with respect to pea-FNR and Ec-FNR. In plastidic and subclass II FNRs, a tyrosine is stacked on the re-face of the FAD isoalloxazine, which stabilizes the prosthetic group through an aromatic interaction. The Xac-FNR contains an alanine that faces the isoalloxazine. Thus, the absence of this stabilizing aromatic interaction may increase the energetic levels of the isoalloxazine electronic transitions and result in the observed blue-shift spectral change [31].
Figure 4. Spectroscopic analyses of Xac-FNR and comparison with the pea and E. coli enzymes.
(A) UV-visible spectra displayed by the different plant-type FNRs. (B) Near-UV and visible CD spectra of the enzymes. (C) FAD solvent accessibility studied by quenching with KI. Xac-FNR (○), pea-FNR (▪), Ec-FNR (•), free FAD (▴). (D) Differential UV-visible spectra elicited by the interaction between the enzymes and NADP+. The spectra were obtained by the subtraction of the spectra of FNR in the presence of 0.3 mM NADP+ and the free enzyme. Different colors were employed for each FNR variant: cyan, Xac-FNR; green, pea-FNR; and red, Ec-FNR.
The CD spectra of Xac-FNR displayed the same spectral trend that was observed for subclass I bacterial FNR from Rhodobacter capsulatus (Figure 4B) [32] except in the near UV region. Previous studies have demonstrated that this CD spectral region is susceptible to the polarity of the solvent [33]. The maximum observed at 270 nm in the R. capsulatus FNR CD spectrum was not detected in Xac-FNR. Therefore, in spite of the high structural homology between these enzymes, there are differences in the isoalloxazine environment between both members of the subclass I FNR. These differences may be related to the lengths of the carboxy-terminal region, which is shorter in Xac-FNR compared to R. capsulatus FNR [34] (Figure 1). The shortened carboxy-terminus may allow for the FAD isoalloxazine to be more accessible to the solvent.
Measurement of FNR fluorescence quenching by titration with a dynamic quencher can be used to analyze the accessibility of FAD [35], [36]. Using this experimental approach, we detected that the FAD isoalloxazine was more exposed to the solvent in Xac-FNR and in pea-FNR than in Ec-FNR (Figure 4C).
It has been previously observed that the intensity of the peak at 515 nm in the differential UV-visible spectra of FNR elicited by NADP+ was proportional to the extent of NADP+ nicotinamide stacking on the isoalloxazine [37]. The intensity of the signal obtained with Xac-FNR was similar to the plastidic FNR, and it was higher than the signal obtained with Ec-FNR (Figure 4D) and R. capsulatus FNR [38]. The K d value for the Xac-FNR-NADP+ complex was lower than those reported for R. capsulatus FNR (9.7 µM vs. 222 µM, Table 1, Figure S1 and [38]) and similar to that of the plastidic type FNRs (Table 1 and [20]). Consequently, in Xac-FNR the productive binding of NADP(H) was improved compared to other bacterial subclass I enzymes. These results indicate that in Xac-FNR when the NADP is bound to the enzyme, the catalytic competent conformation of the nucleotide is enhanced, resulting in a more efficient enzyme.
Table 1. Kinetic parametersa of NADPH and NADH diaphorase reactions that were catalyzed by Xac-FNR, pea-FNR and Ec-FNR, and the dissociation constants for the different complexes with NADP+ b.
| NADPH | NADH | |||||||
| FNR | K m (µM) | k cat (s−1) | k cat/K m (µM−1s−1) | K d (µM) | K m (mM) | k cat (s−1) | k cat/K m (µM−1s−1) | NADPH/NADH specificity |
| Xac-FNR | 10.8±0.5 | 121.9 ±− 1.8 | 11.3 | 9.7±0.4 | 3.3±1.4 | 3.2±1.3 | 0.001 | 1600 |
| pea-FNR | 15.3±4.3 c | 374.3±18 c | 24.5 c | 10.9±3.1c | 14.3±3.9 c | 7.0±2.1 c | 0.0005 c | 49060 c |
| Ec-FNR | 8.3±1.3 c | 38.2±3.5 c | 4.6 c | 5.9±0.6 c | <0.05 d | Nd e | Nd e | Nd e |
Each parameter value represents the average of three independent determinations. A description of the calculation methods that were employed is reported in the Materials and Methods. The original data are depicted in Figure S2.
Potassium ferricyanide reduction was assessed using the diaphorase assay of Zanetti [41] in 50 mM Tris-HCl (pH 8.0) using NADPH or NADH as the substrate.
Values of parameters for pea-FNR and Ec-FNR were obtained from a reference [49].
An estimate of the limit of the determination based on the tested sensitivity of the method.
Not determined.
FNRs display strong preference for NADP(H) and are very poor NAD(H) oxidoreductases (Table 1, Figure S2 and [20]). In contrast, various redox compounds, including complexed metals and aromatic molecules, can operate as mono and bi-electronic acceptors in vitro, in the so-called diaphorase reaction [39]. Xac-FNR showed higher NADPH-diaphorase activity compared to R. capsulatus FNR (121.9 s-1 vs. 7.2 s−1, Table 1 and [32]) and Ec-FNR (Table 1). In addition, Xac-FNR showed higher NADH-diaphorase activity than Ec-FNR (Table 1). An increase in the interaction between the nicotinamide and the isoalloxazine has been postulated to be the cause for the decrease in the discrimination of substrate in FNR proteins [37]. Therefore, our results indicate that in Xac-FNR a greater interaction between the isoalloxazine and NADP(H) nicotinamide occurs when compared to other FNR proteins from bacterial subclass I. This increased interaction may have some functional relevance.
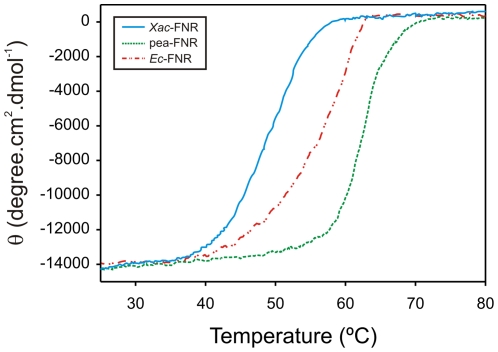
Profile of thermal denaturation
Figure 5 shows the thermal unfolding curves for the different FNR variants, where Xac-FNR has the lowest thermal stability. However, the low stability does not impede Xac-FNR to perform its biological function during the life cycle of the bacteria. The optimal growth temperature of X. axonopodis pv. citri is 28°C [5], which is lower than the melting temperature of Xac-FNR (Table 2 and Figure S3). The absence of an aromatic residue that stacks against the FAD isoalloxazine may contribute to the decreased stability of Xac-FNR. Site-directed mutagenesis studies have revealed that the replacement of the carboxy-terminal tyrosine in pea-FNR to a serine induced a 2.6 kcal/mol destabilization [40]. The lack of an aromatic residue that faces the FAD isoalloxazine in Xac-FNR allows for an improved interaction of the nicotinamide portion of NADP(H) with the prosthetic group; however, it could be the cause of the low thermal stability of the protein.
Figure 5. Thermal stability of the different plant-type FNRs according to the folding-unfolding transitions that were observed for the enzymes.
Cyan, Xac-FNR; green, pea-FNR; and red, Ec-FNR.
Table 2. Kinetic parameters a for cytochrome c reductase with pea ferredoxin, E. coli flavodoxin, and E. coli ferredoxin, and the melting temperatures of thermal unfolding transitions for Xac-FNR, pea-FNR and Ec-FNR b.
| pea ferredoxin | E. coli ferredoxin | E. coli flavodoxin | ||||||||
| FNR | Km (µM) | kcat (s−1) | kcat/Km (µM−1s−1) | Km (µM) | kcat (s−1) | kcat/Km (µM−1s−1) | Km (µM) | kcat (s−1) | kcat/Km (µM−1s−1) | TM (°C) c |
| Xac-FNR | <0.1 d | <0.1 d | Nd e | <0.1 d | <0.1 d | Nd e | <0.1 d | <0.1 d | Nd e | 48.1±0.8 |
| pea-FNR | 2.2±0.2 f | 75.0±0.5 f | 34.1 f | Nd e | Nd e | Nd e | Nd e | Nd e | Nd e | 61.5±0.5 |
| Ec-FNR | 1.4±0.1 f | 22.8±0.2 f | 16.3 f | 0.074±0.023 f | 12.3±1.2 f | 164.9 f | 2.3±0.2 f | 8.9±1.0 f | 3.9 f | 57.5±0.7 |
Each value represents the average of three independent determinations. A description of the calculation methods that were employed and the activity determinations are reported in the Materials and Methods. The original data are depicted in Figure S3.
Cytochrome c reduction was determined at 550 nm (ε550 = 19 mM−1 cm−1) as described in the Materials and Methods.
TM is the temperature of the midpoint of the thermal denaturation transition and was determined as described in the Materials and Methods.
An estimate of the limit of the determination based on the tested sensitivity of the method.
Not determined.
Values of parameters for pea-FNR and Ec-FNR were obtained from a reference [49].
Analysis of the redox partner of Xac-FNR
Ferredoxins and flavodoxins are considered the main redox-partners of FNRs [18], [20]. The electron transfer from NADPH to ferredoxin catalyzed by FNRs can be followed using cytochrome c as final electron acceptor in a coupled assay known as cytochrome c reductase activity [41]. Reduction of cytochrome c shows a strict requirement for ferredoxin. The reaction is most often described as consisting of two hemi-reactions: FNR-catalyzed reduction of ferredoxin by NADPH, and the subsequent reoxidation of the iron-sulfur protein by cytochrome c. Xac-FNR was not able to reduce ferredoxin or flavodoxin from E. coli or pea ferredoxin. Expected activity values were obtained in parallel experiments with the plastidic and E. coli enzymes (Table 2).
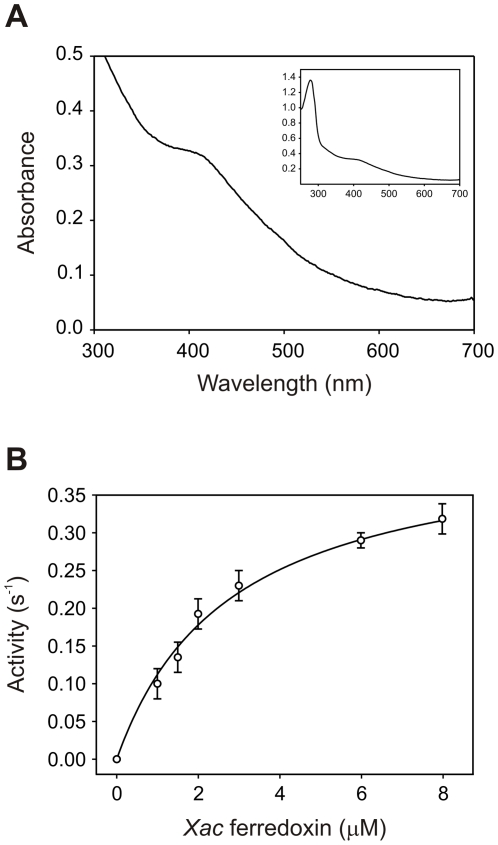
The analysis of the X. axonopodis pv. citri genome showed the existence of five putative ferredoxins and one flavodoxin [29]. Interestingly, the ferredoxin XAC1762 showed 68% identity and 80% similarity to ferredoxin I from A. vinelandii, which has been demonstrated to interact productively with FNR in this bacterium [42]. Taking into account the high similarity between Xac-FNR and the FNR from A. vinelandii (Figure 6A and Figure S4), we postulated ferredoxin XAC1762 as a potential redox partner of Xac-FNR. To test this hypothesis, we cloned, expressed and purified the ferredoxin coded by the XAC1762 sequence. The purified protein displayed a typical broad UV-visible spectrum with a band at 407 nm, which is indicative of [4Fe-4S] or [3Fe-4S] ferredoxins [43] (Figure 7A). The NADPH-cytochrome c reductase activity of Xac-FNR with different amounts of XAC1762 ferredoxin was measured under argon (Figure 7B) and a K m value of 2.8 µM and a k cat of 0.42 s−1 were obtained. These results indicate that XAC1762 ferredoxin is one of the possible redox partners of Xac-FNR. The activity observed with Xac-FNR and the ferredoxin XAC1762 is lower than the corresponding value obtained for the E. coli couple, although Xac-FNR displayed higher diaphorase activity.
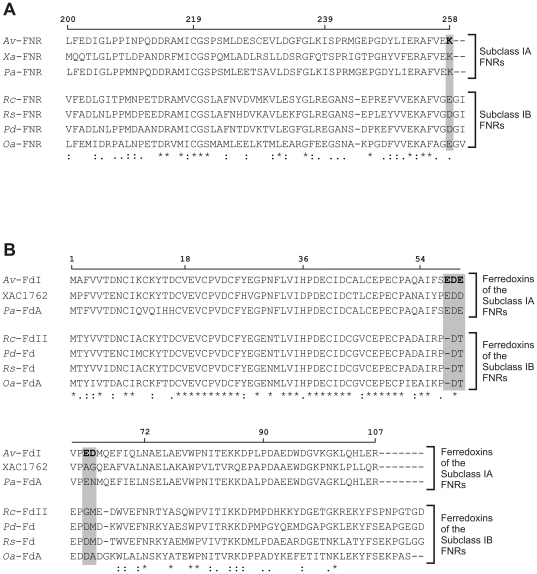
Figure 6. Analysis of Xac-FNR's redox partner by homology sequence.
(A) Alignment of primary structures of subclass I bacterial FNRs from A. vinelandii (Av-FNR, gb: YP_002800963.1), X. axonopodis pv. citri (Xa-FNR, gb: NP_641792.1), Pseudomonas aeruginosa (Pa-FNR, gb: YP_001347117.1), R. capsulatus (Rc-FNR, gb: ADE85336.1), Rhodobacter sphaeroides (Rs-FNR, gb: YP_002524612.1), Paracoccus denitrificans (Pd-FNR, gb: ABL68770.1) and Oceanicaulis alexandrii (Oa-FNR, gb: ZP_00952506.1). Sequence regions from amino acid 200 to the carboxy-terminus are shown. In bold is the amino acid from Av-FNR that is involved in the interaction with ferredoxin I, as was previously reported [42]. (B) Alignment of ferredoxin I from A. vinelandii (Av-FdI, gb: AAA22125.1) and ferredoxins from X. axonopodis pv. citri (XAC1762, gb: NP_642090.1), P. aeruginosa (ferredoxin A, Pa-FdA, gb: AAF89693.1), R. capsulatus (ferredoxin II, Rc-FdII, gb: YP_003578927.1), P. denitrificans (Pd-Fd, gb: ABL69923.1), Rhodobacter sp. (Rs-Fd, gb: ZP_05844833.1) and O. alexandrii (ferredoxin A, Oa-FdA, gb: ZP_00953239.1). In bold is the peptide involved in the interaction with Av-FNR identified by cross-linking experiments, as was previously reported [42]. Potential residues of Av-FdI that interact with Lys258 of Av-FNR are shaded in gray. Numbers over the sequences correspond to the A. vinelandii proteins. The alignments were performed using ClustalX 2.0.11.
Figure 7. Characterization of ferredoxin XAC1762.
(A) UV-visible spectrum displayed by ferredoxin XAC1762. (B) Kinetics of the cytochrome c reductase reaction of Xac-FNR with ferredoxin XAC1762 as substrate.
Crosslinking experiments between FNR and ferredoxin I from A. vinelandii were employed to determine the important residues for complex formation [42]. Lys258 from FNR and an acidic patch formed by Glu57-Asp58-Glu59/Glu62-Asp63 of the above mentioned ferredoxin were identified. Xac-FNR and ferredoxin XAC1762 from X. axonopodis pv. citri contain homologous residues at equivalent positions (see Figure 6A and B). Thus, it may be suggested that Xac-FNR and ferredoxin XAC1762 from X. axonopodis pv. citri contain the structural features necessary for this interaction. Lys258 is not conserved in all subclass I bacterial FNRs and is replaced in some of them by a glutamate in the equivalent position as occurs in R. capsulatus [21] (Figure 6A). Consequently, it can be suggested that the FNRs may have evolved in order to acquire structural features on the carboxy-terminal region that allow for the modulation of the specificity of the interaction with their redox partners. We suggest that subclass I bacterial FNRs be further subdivided into two new groups: subclass IA, which the prototype would be the FNR from A. vinelandii; and subclass IB, which the representative member would be the FNR from R. capsulatus. The main differences between both subclasses are located at the carboxy-terminal region. While enzymes from subclass IA have Lys258 (numbering for A. vinelandii), the subclass IB FNR proteins contain a glutamate or an aspartate at the equivalent position and a longer carboxy-terminal region (Figure 1 and Figure 6A). We searched for all proteins with carboxy-terminal regions that were similar to FNR from R. capsulatus in the Data Bank. We found that the FNRs from Rhodobacter sp., Paracoccus denitrificans and Oceanicaulis alexandrii meet these criteria (Figure 6A). Consequently, subclass IA is defined by the carboxy terminal sequence VEK and the subclass IB by the sequence (V/A)G(E/D)G(I/V).We analyzed the ferredoxins that may function as substrates for these enzymes. In all cases, the ferredoxins that might act as redox partners of subclass IA FNRs contain the Glu-Asp-Glu triad and the acidic patches while those of subclass IB FNRs displayed conserved Asp-Thr-Glu and basic amino acids at positions 56 and 73/74 (Figure 6B).
In our work, we demonstrated that Xac-FNR was regulated by the accumulation of reactive oxygen species, and that this protein is able to substitute the endogenous E. coli FNR in its antioxidant role. Purified ferredoxin XAC1762 was shown to be one of the possible substrates of Xac-FNR. Reduction of this ferredoxin by Xac-FNR may contribute to the oxidative stress response in X axonopodis pv. citri, by promoting a decrease in the intracellular NADPH levels. Structural and functional analyses of Xac-FNR suggests that the bacterial subclass I can be further classified into subgroups IA and IB. Subclass IA bacterial FNRs (to which Xac-FNR belongs) may interact with ferredoxins similar to ferredoxin I of A. vinelandii and ferredoxin XAC1762 of X. axonopodis pv. citri.
Materials and Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are described in Table 3. X. axonopodis pv. citri cells were grown aerobically in Silva Buddenhagen (SB) medium (5 g l−1 sucrose, 5 g l−1 yeast extract, 5 g l−1 peptone, and 1 g l−1 glutamic acid at pH 7.0) at 28°C with shaking at 200 rpm. E. coli strains were grown at 37°C in Luria-Bertani (LB) or M9 minimal media that was supplemented with 0.2% (w/v) glucose [44]. IPTG was added to a final concentration of 0.5 mM when the expression of plasmid-borne genes was desired. Antibiotics were added to the media at the following final concentrations: ampicillin (Ap), 25 µg ml−1 for X. axonopodis pv. citri and 100 µg ml−1 for E. coli; kanamycin (Km), 40 µg ml−1; tetracycline (Tc), 15 µg ml−1 and chloramphenicol (Cm), 30 µg ml−1. The X. axonopodis pv. citri strain Xcc99–1330 was kindly provided by Blanca I. Canteros (INTA; Bella Vista, Argentina).
Table 3. Bacterial strains and plasmids used in this study.
| Strain/plasmid | Genotype or relevant characteristics | Source/reference |
| Strains | ||
| Xanthomonas axonopodis pv. citri | ||
| Xcc99-1330 | Wild type, Apr | B. I. Canteros |
| Escherichia coli | ||
| JM109 | HsdR17 endA1 Recal thi gyrA96 relA1 recA1 supE44 λ-Δ (lac-proAB), [F', traD36, proA+B+, lacIqZΔM15] | [44] |
| GC4468 | F- Δlac U169 rpsL | [54] |
| RR6A | GC4468 fpr, Kmr | [9] |
| BL21(DE3)pLysS | F- ompT hsdS B (r- Bm- B) dcm gal (DE3) pLysS, Cmr | Novagen |
| C41(DE3) | F- ompT hsdS B (r- Bm- B) dcm gal (DE3) | [55] |
| Plasmids | ||
| pGEM-T Easy | Vector for cloning PCR products, Apr | Promega |
| pUC119 | pBR322 derivative, lacZ, Apr | [45] |
| pEE1010 | pUC18 carrying the E. coli fpr gene, Apr | [56] |
| pRKISC | pRK415 vector carrying an ISC operon, Tcr | [47] |
| pUC/XacFNR | pUC119 carrying the Xac fpr gene, Apr | This study |
| pET/XacFNR | pET28a carrying the Xac fpr gene, Kmr | This study |
| pET/XacFd | pET28a carrying the XAC1762 gene, Kmr | This study |
Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol; Tc, tetracycline.
Complementation of the E. coli fpr-null mutant with Xac-FNR
The coding sequence for the Xac-FNR was amplified by PCR using oligonucleotides FPR-F2 (5'-TTCCAAGCTTCATGTCTTCCGCTTTTGGCGC-3') and FPR-R (5'-TTCCGAATTCGCGCGTCACTTTTCGACGAA-3') as primers, and the X. axonopodis pv. citri genomic DNA was used as the template. The PCR product (806 bp) was cloned into the pGEM-T Easy plasmid (Promega), digested with HindIII and EcoRI, and ligated to compatible sites in pUC119 [45]. After sequencing the resulting plasmid, pUC/XacFNR was transformed into E. coli strain RR6A (fpr-null mutant) [9]. Expression of the recombinant protein in soluble cell extracts was verified by SDS-PAGE, and immunoblot analysis was performed with specific antisera.
Bacterial viability assay
Bacterial resistance to MV was evaluated by the disk diffusion method. Briefly, 100 µl of a bacterial suspension that contained ∼109 cells ml−1 was mixed with 3 ml 0.7% (w/v) molten agar at 42°C and was poured onto M9-agar plates supplemented with the corresponding antibiotics and 0.5 mM IPTG when required. After hardening, 5 µl of a 100 mM MV solution was added onto paper disks (5-mm diameter) placed on the agar surface. The zones of growth inhibition were measured after incubation for 24 h at 37°C.
Determination of Xac-FNR expression
X. axonopodis pv. citri overnight cultures were diluted into fresh SB medium with 2% inoculum. Bacterial suspensions were grown at 28°C to OD600 0.5–0.7 (exponential phase) and were incubated with the oxidative agents for 15 min. For cell extract preparation, the cultures were harvested by centrifugation at 10000 g for 10 min at 4°C. Bacteria were washed and resuspended in 500 µl of ice-cold potassium phosphate buffer (50 mM; pH 7.0) that contained 1 mM PMSF and were disrupted by intermittent sonication. The suspensions were clarified by centrifugation at 12000 g for 20 min at 4°C. Protein concentrations in the soluble cell extracts were determined using a dye-binding assay [46] that used bovine serum albumin as a standard. The soluble fractions were resolved by SDS-PAGE and transferred to nitrocellulose membranes, and FNR was detected with specific antisera using secondary antibodies that were conjugated to alkaline phosphatase. Immunoreactive bands were integrated using Gel-Pro Analyzer Software 3.1 (Media Cybernetics).
RNA extraction and semiquantitative reverse transcription PCR (RT-PCR)
The total RNA from X. axonopodis pv. citri cells was isolated using TRIzol® (Invitrogen) according to the manufacturer's instructions. After extraction, the RNA was treated with RNase-free DNase (Promega), and its integrity was determined by agarose gel electrophoresis. Semiquantitative analyses of fpr transcript levels were performed using a two-step RT-PCR approach that employed the primers fprRT-F (ATGTCTTCCGCTTTTGGCGC) and fprRT-R (CTGGGTGAGGATCACCTTGT). For cDNA synthesis, total RNA (1 µg) was added to 20 µl of a reverse transcription reaction that contained 4 µl 5× M-MLV buffer (Promega), 0.5 mM dNTP mixture, 0.5 µg gene-specific primer, and 200 U M-MLV reverse transcriptase (Promega), and the reaction was incubated for 60 min at 42°C. Reverse transcription was terminated by incubation at 94°C for 5 min. Control reactions, where RT was omitted, were performed in parallel for all the samples to rule out the possibility of amplification from contaminating DNA. PCR reactions were performed with 2 µl of cDNA template under the following conditions: 25 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min, and extension at 72°C for 1 min; and a final extension step at 72°C for 5 min. The number of cycles, which avoided reaching the plateau of PCR, was previously determined by taking samples at different cycle numbers during PCR amplification and analyzing the products obtained by agarose gel electrophoresis. As a control, a 217-bp fragment of 16S rRNA was amplified using primers 16S-F (TGGTAGTCCACGCCCTAAACG) and 16S-R (CTGGAAAGTTCCGTGGATGTC) and the same PCR conditions; however, only 1% of the cDNA synthesis reaction was used as the template due to the high abundance of 16S rRNA in the total RNA extracts. RT-PCR products were resolved on 1.5% (w/v) agarose gels, and the gels were densitometrically quantified using Gel-Pro Analyzer Software 3.1 (Media Cybernetics).
Preparation of recombinant proteins
Recombinant Xac-FNR was obtained by expression in E. coli. Briefly, a pET/XacFNR expression vector was constructed by inserting the coding sequence of Xac-FNR into the pET28a vector (Novagen). The coding sequence for Xac-FNR was amplified using PCR with the primers FPR-F (TATCTCTCCATATGTCTTCCGCTTTTGGCGC) and FPR-R (TTCCGAATTCGCGCGTCACTTTTCGACGAA), and the X. axonopodis pv. citri genomic DNA was used as the template. To facilitate the cloning process, the NdeI and EcoRI restriction sites were introduced in the primers FPR-F and FPR-R, respectively. The PCR product (806 bp) was cloned into the pGEM-T Easy plasmid (Promega), digested with NdeI and EcoRI and ligated into compatible sites in pET28a. The plasmid pET/XacFNR contained the entire Xac-FNR coding region fused in-frame to an N-terminal hexahistidine tag. For expression in E. coli BL21(DE3)pLysS cells, the cultures were grown at 37°C in LB medium supplemented with kanamycin and chloramphenicol for 3 h and were induced with 0.25 mM IPTG for 6 h at 20°C. Xac-FNR was purified by Ni-NTA affinity chromatography and dialyzed against 50 mM Tris-HCl buffer (pH 8.0) in the presence of 150 mM NaCl. The fusion protein was digested with thrombin, and the hexahistidine-tag was removed by another Ni-NTA affinity chromatography procedure.
Ferredoxin XAC1762 were overexpressed in E. coli C41 cells transformed with pET/XacFd and ISC operon expressing plasmid pRKISC [47], as a carboxy-terminal fusion with His6-tag and a TEV recognition site between the His6-tag and the last amino acid of the protein. The pET/XacFd was constructed by inserting the coding sequence for the ferredoxin XAC1762 into pET28a expression vector. This sequence was amplified by PCR using the primers XacFd-F (AAGGCCATGGCTTTTGTTGTCACCGAAAACTGC) and XacFd-R (TGGAAGCTTGCCCTGAAAATACAGGTTTTCGCGCTGCAGCAGCGGCAATTTGTTGGGCTTGCCATCCCATTCGGC) and the X. axonopodis pv. citri genomic DNA as a template. To facilitate cloning, the NcoI and HindIII restriction sites were introduced in the primers XacFd-F and XacFd-R, respectively. The PCR product (357 bp) was cloned into the pGEM-T Easy plasmid (Promega), digested with NcoI and HindIII and ligated into compatible sites in pET28a, rendering pET/XacFd plasmid. For functional expression, bacteria were grown at 37°C in LB medium supplemented with kanamycin and tetracycline for 3 h and then expression induced by the addition of 0.25 mM IPTG and supplemented with 2 mM ammonium ferric citrate. Then, the cultures were maintained during 16 h at 18°C with mild agitation. Xac ferredoxin was purified by Ni-NTA affinity chromatography and dialyzed against 50 mM Tris-HCl buffer (pH 8.0), 150 mM NaCl. The fusion protein was digested with TEV protease and the hexahistidine-tag was removed by another Ni-NTA affinity chromatography procedure.
Spectral Analyses
UV-visible absorption spectra were recorded on a Shimadzu UV-2450 spectrophotometer. CD spectra were obtained using a JASCO J-810 spectropolarimeter at 25°C. The spectra were recorded in 5.0 µM protein solutions in 0.1 cm path length cuvettes. Fluorescence spectra were monitored using a Varian (Palo Alto, CA) Cary Eclipse fluorescence spectrophotometer that was interfaced with a personal computer. The samples were filtered through G25 Sephadex spin columns that were equilibrated with 50 mM potassium phosphate (pH 8.0) before measurements were collected. The extinction coefficient of Xac-FNR was determined by releasing FAD from the protein by treatment with 0.2% (w/v) SDS and quantifying the flavin spectrophotometrically [48].
Enzymatic Assays
FNR-dependent NADPH-K3Fe(CN)6 diaphorase activity was determined using previously published methods [41]. The NADH-ferricyanide diaphorase activity was determined in 1 ml reaction medim that contained 50 mM Tris-HCl (pH 8.0), 1 mM potassium ferricyanide, and 1.25–2.5 µM FNR. The cytochrome c reductase activity of Xac-FNR, using either ferredoxin or flavodoxin, was assayed in reaction medium (1 ml) that contained 50 mM Tris-HCl (pH 8.0), 0.3 mM NADP+, 3 mM glucose 6-phosphate, 1 unit of glucose-6-phosphate dehydrogenase, and 50 µM cytochrome c [49]. After the addition of approximately 15–100 nM FNR, cytochrome c reduction was monitored spectrophotometrically by following absorbance changes at 550 nm (ε550 = 19 mM−1 cm−1). All kinetic experiments were performed at 30°C. In all cases, precautions were taken to ensure the linearity of the enzyme activity, and when appropriate, saturation of the Michaelis-Menten plots was verified.
Thermal unfolding transitions
Protein stock solutions were diluted to a final concentration of 0.5 µM in 50 mM potassium phosphate (pH 8.0). The CD signal was measured by excitation at 220vnm while the temperature of the sample was increased at a rate of 1°C min−1 (from 25 to 80°C). Thermal unfolding transitions were analyzed assuming a two-state approximation, which only the native and unfolded states were significantly populated. The TM was determined by fitting experimental data to the equation, ΔG(T) = ΔH(TM)+ΔCp(T−TM)−T(ΔH(TM)/TM+ΔCpln(T/TM)), as described elsewhere [49], [50].
Determination of dissociation constants of Xac-FNR complexed with NADP+ and protein substrates
The K d value of the complex between Xac-FNR and NADP+ was determined by difference absorption spectroscopy, which was previously described [51]. Briefly, 15 µM flavoprotein in 50 mM Tris-HCl (pH 8.0) was titrated at 25°C with NADP+. After each addition, the absorbance spectra (200–600 nm) were monitored. The difference spectra were calculated, and the absorbance differences at the stated wavelengths were plotted against the concentration of NADP+. The data were fitted to a theoretical equation for a 1∶1 complex. The sample had been previously filtered through a desalting column that had been equilibrated with 50 mM Tris-HCl (pH 8.0). To determine the K d values of the complex between Xac-FNR and pea ferredoxin, E. coli flavodoxin or E. coli ferredoxin, solutions that contained 3 µM enzyme in 50 mM Tris-HCl (pH 8.0) were titrated with the corresponding protein substrate. After each addition, fluorescence quenching at 340 nm (excitation at 270 nm) was determined. Controls were run in parallel to estimate the fluorescence contribution due to the addition of pea ferredoxin, E. coli flavodoxin or E. coli ferredoxin. The K d values were estimated by fitting the fluorescence data to a theoretical equation for a 1∶1 complex [49].
Determination of parameters
All experimental data were fit to theoretical curves using SigmaPlot (Systat Software Inc., Point Richmond, CA, USA).
Solvent accessibility of the FAD
Quenching of flavin fluorescence by iodide was used to investigate the relative accessibility of FAD in the FNR variants [52], [53]. The emission fluorescence at 525 nm (λ of emission 450 nm) of a 2 ml sample of FNR in Tris-HCl (pH 8.0) was determined during the titration of KI in cuvettes with a 1-cm path-length at 25°C. The samples were previously filtered through a sephadex G25 column that was equilibrated with 50 mM Tris-HCl (pH 8.0) to separate the free FAD.
Supporting Information
Determination of the dissociation constants for the FNR-NADP+ complexes. The absorbance changes elicited by NADP+ on each enzyme in the 490–510 nm region were used to calculate the K d values by fitting the data to a theoretical equation for a 1∶1 complex. Xac-FNR (○), pea-FNR (▪), Ec-FNR (•).
(TIF)
Determination of the kinetic parameters of the diaphorase reaction for the different enzymes. Kinetics of the ferricyanide reduction by Xac-FNR (○), pea-FNR (▪) and Ec-FNR (•) using NADPH (A) and NADH (B) as substrates.
(TIF)
Kinetics of cytochrome c reductase reactions of the different FNR enzymes. Reduction of cytochrome c by pea-FNR (▪) and Ec-FNR (•) using pea ferredoxin (A), E. coli ferredoxin (B) and E. coli flavodoxin (C) as substrates.
(TIF)
Alignment of the complete sequences of subclass I bacterial FNRs. Sequences of FNRs from A. vinelandii (Av-FNR), X. axonopodis pv. citri (Xa-FNR), Pseudomonas aeruginosa (Pa-FNR), R. capsulatus (Rc-FNR), Rhodobacter sphaeroides (Rs-FNR), Paracoccus denitrificans (Pd-FNR) and Oceanicaulis alexandrii (Oa-FNR) were analyzed.
(TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants PICT-2007 01–00645, Agencia Nacional de Promoción Científica y Tecnológica to EAC, Ministerio de Ciencia, Tecnología e Innovación Productiva, Argentina (www.agencia.mincyt.gov.ar); PIP 252 to EAC and 112–2009–01–00873 to EGO from Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina (www.conicet.gov.ar); BIO187 to EAC and BIO162 to EGO from the University of Rosario, Argentina (www.unr.edu.ar). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Mol Plant Pathol. 2003;4:141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 2.Graham JH, Gottwald TR, Cubero J, Achor DS. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol Plant Pathol. 2004;5:1–15. doi: 10.1046/j.1364-3703.2004.00197.x. [DOI] [PubMed] [Google Scholar]
- 3.Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green J, Paget MS. Bacterial redox sensors. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 5.Tondo ML, Petrocelli S, Ottado J, Orellano EG. The monofunctional catalase KatE of Xanthomonas axonopodis pv. citri is required for full virulence in citrus plants. PLoS One. 2010;5:e10803. doi: 10.1371/journal.pone.0010803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jittawuttipoka T, Buranajitpakorn S, Vattanaviboon P, Mongkolsuk S. The catalase-peroxidase KatG is required for virulence of Xanthomonas campestris pv. campestris in a host plant by providing protection against low levels of H2O2. J Bacteriol. 2009;191:7372–7377. doi: 10.1128/JB.00788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianchi V, Haggard-Ljungquist E, Pontis E, Reichard P. Interruption of the ferredoxin (flavodoxin) NADP+ oxidoreductase gene of Escherichia coli does not affect anaerobic growth but increases sensitivity to paraquat. J Bacteriol. 1995;177:4528–4531. doi: 10.1128/jb.177.15.4528-4531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krapp AR, Rodriguez RE, Poli HO, Paladini DH, Palatnik JF, et al. The flavoenzyme ferredoxin (flavodoxin)-NADP(H) reductase modulates NADP(H) homeostasis during the soxRS response of Escherichia coli. J Bacteriol. 2002;184:1474–1480. doi: 10.1128/JB.184.5.1474-1480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeom J, Jeon CO, Madsen EL, Park W. Ferredoxin-NADP+ reductase from Pseudomonas putida functions as a ferric reductase. J Bacteriol. 2009;191:1472–1479. doi: 10.1128/JB.01473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park W, Pena-Llopis S, Lee Y, Demple B. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem Biophys Res Commun. 2006;341:51–56. doi: 10.1016/j.bbrc.2005.12.142. [DOI] [PubMed] [Google Scholar]
- 13.Shin M, Arnon DI. Enzymatic mechanisms of pyridine nucleotide reduction in chloroplast. J Biol Chem. 1965;240:1405–1411. [PubMed] [Google Scholar]
- 14.Ritchie SW, Redinbaugh MG, Shiraishi N, Vrba JM, Campbell WH. Identification of a maize root transcript expressed in the primary response to nitrate: characterization of a cDNA with homology to ferredoxin-NADP+ oxidoreductase. Plant Mol Biol. 1994;26:679–690. doi: 10.1007/BF00013753. [DOI] [PubMed] [Google Scholar]
- 15.Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, et al. Differential interaction of maize root ferredoxin: NADP(+) oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 2000;123:1037–1045. doi: 10.1104/pp.123.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrillo N, Vallejos RH. Ferredoxin-NADP+ oxidoreductase. In: Barber J, editor. Topics in Photosynthesis. Amsterdam-New York-Oxford: Elsevier; 1987. pp. 527–560. [Google Scholar]
- 17.Arakaki AK, Ceccarelli EA, Carrillo N. Plant-type ferredoxin-NADP+ reductases: a basal structural framework and a multiplicity of functions. FASEB J. 1997;11:133–140. doi: 10.1096/fasebj.11.2.9039955. [DOI] [PubMed] [Google Scholar]
- 18.Medina M, Gomez-Moreno C. Interaction of ferredoxin-NADP(+) reductase with its substrates: optimal interaction for efficient electron transfer. Photosynth Res. 2004;79:113–131. doi: 10.1023/B:PRES.0000015386.67746.2c. [DOI] [PubMed] [Google Scholar]
- 19.Wang A, Zeng Y, Han H, Weeratunga S, Morgan BN, et al. Biochemical and structural characterization of Pseudomonas aeruginosa Bfd and FPR: ferredoxin NADP+ reductase and not ferredoxin is the redox partner of heme oxygenase under iron-starvation conditions. Biochemistry. 2007;46:12198–12211. doi: 10.1021/bi7013135. [DOI] [PubMed] [Google Scholar]
- 20.Carrillo N, Ceccarelli EA. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur J Biochem. 2003;270:1900–1915. doi: 10.1046/j.1432-1033.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 21.Ceccarelli EA, Arakaki AK, Cortez N, Carrillo N. Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim Biophys Acta. 2004;1698:155–165. doi: 10.1016/j.bbapap.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Milani M, Balconi E, Aliverti A, Mastrangelo E, Seeber F, et al. Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: functional and crystallographic analysis. J Mol Biol. 2007;367:501–513. doi: 10.1016/j.jmb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Tondo ML, Ottado J, Orellano EG. Expression, purification and characterization of the ferredoxin-NADP(H) reductase from the phytopatyhogen Xanthomonas axonopodis pv. citri. In: Frago S, Gomez-Moreno C, Medina M, editors. Flavins and Flavoproteins. Zaragoza: Prensas Universitarias de Zaragoza; 2008. pp. 255–259. [Google Scholar]
- 24.Giro M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152:1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 25.Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem. 2004;279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 26.Liochev SI, Hausladen A, Beyer WF, Jr, Fridovich I. NADPH: ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci U S A. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Pena-Llopis S, Kang YS, Shin HD, Demple B, et al. Expression analysis of the fpr (ferredoxin-NADP+ reductase) gene in Pseudomonas putida KT2440. Biochem Biophys Res Commun. 2006;339:1246–1254. doi: 10.1016/j.bbrc.2005.11.135. [DOI] [PubMed] [Google Scholar]
- 28.Manchado M, Michan C, Pueyo C. Hydrogen peroxide activates the SoxRS regulon in vivo. J Bacteriol. 2000;182:6842–6844. doi: 10.1128/jb.182.23.6842-6844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 30.Isas JM, Burgess BK. Purification and characterization of a NADP+/NADPH-specific flavoprotein that is overexpressed in FdI- strains of Azotobacter vinelandii. J Biol Chem. 1994;269:19404–19409. [PubMed] [Google Scholar]
- 31.Orellano EG, Calcaterra NB, Carrillo N, Ceccarelli EA. Probing the role of the carboxyl-terminal region of ferredoxin-NADP+ reductase by site-directed mutagenesis and deletion analysis. J Biol Chem. 1993;268:19267–19273. [PubMed] [Google Scholar]
- 32.Bittel C, Tabares LC, Armesto M, Carrillo N, Cortez N. The oxidant-responsive diaphorase of Rhodobacter capsulatus is a ferredoxin (flavodoxin)-NADP(H) reductase. FEBS Lett. 2003;553:408–412. doi: 10.1016/s0014-5793(03)01075-5. [DOI] [PubMed] [Google Scholar]
- 33.Edmondson DE, Tollin G. Circular dichroism studies of the flavin chromophore and of the relation between redox properties and flavin environment in oxidases and dehydrogenases. Biochemistry. 1971;10:113–124. doi: 10.1021/bi00777a018. [DOI] [PubMed] [Google Scholar]
- 34.Nogues I, Perez-Dorado I, Frago S, Bittel C, Mayhew SG, et al. The ferredoxin-NADP(H) reductase from Rhodobacter capsulatus: molecular structure and catalytic mechanism. Biochemistry. 2005;44:11730–11740. doi: 10.1021/bi0508183. [DOI] [PubMed] [Google Scholar]
- 35.Khan KK, Mazumdar S, Modi S, Sutcliffe M, Roberts GC, et al. Steady-state and picosecond-time-resolved fluorescence studies on the recombinant heme domain of Bacillus megaterium cytochrome P-450. Eur J Biochem. 1997;244:361–370. doi: 10.1111/j.1432-1033.1997.00361.x. [DOI] [PubMed] [Google Scholar]
- 36.Paladini DH, Musumeci MA, Carrillo N, Ceccarelli EA. Induced fit and equilibrium dynamics for high catalytic efficiency in ferredoxin-NADP(H) reductases. Biochemistry. 2009;48:5760–5768. doi: 10.1021/bi9004232. [DOI] [PubMed] [Google Scholar]
- 37.Piubelli L, Aliverti A, Arakaki AK, Carrillo N, Ceccarelli EA, et al. Competition between C-terminal tyrosine and nicotinamide modulates pyridine nucleotide affinity and specificity in plant ferredoxin-NADP(+) reductase. J Biol Chem. 2000;275:10472–10476. doi: 10.1074/jbc.275.14.10472. [DOI] [PubMed] [Google Scholar]
- 38.Bortolotti A, Perez-Dorado I, Goni G, Medina M, Hermoso JA, et al. Coenzyme binding and hydride transfer in Rhodobacter capsulatus ferredoxin/flavodoxin NADP(H) oxidoreductase. Biochim Biophys Acta. 2009;1794:199–210. doi: 10.1016/j.bbapap.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Avron M, Jagendorf AT. A TPNH diaphorase from chloroplasts. Arch Biochem Biophys. 1956;65:475–490. doi: 10.1016/0003-9861(56)90207-7. [DOI] [PubMed] [Google Scholar]
- 40.Calcaterra NB, Pico GA, Orellano EG, Ottado J, Carrillo N, et al. Contribution of the FAD binding site residue tyrosine 308 to the stability of pea ferredoxin-NADP+ oxidoreductase. Biochemistry. 1995;34:12842–12848. doi: 10.1021/bi00039a045. [DOI] [PubMed] [Google Scholar]
- 41.Zanetti G. A lysyl residue at the NADP binding site of ferredoxin-NADP reductase. Biochim Biophys Acta. 1976;445:14–24. doi: 10.1016/0005-2744(76)90157-1. [DOI] [PubMed] [Google Scholar]
- 42.Jung YS, Roberts VA, Stout CD, Burgess BK. Complex formation between Azotobacter vinelandii ferredoxin I and its physiological electron donor NADPH-ferredoxin reductase. J Biol Chem. 1999;274:2978–2987. doi: 10.1074/jbc.274.5.2978. [DOI] [PubMed] [Google Scholar]
- 43.Fukuyama K. Ferredoxins containing one [4Fe-4S] center. In: Messerschmidt A, Huber R, Poulos TL, Wieghardt K, editors. Handbook of Metalloproteins. New York: John Wiley & Sons; 2006. pp. 543–552. [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. New York: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: A laboratory manual. [Google Scholar]
- 45.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 46.Sedmak JJ, Grossberg SE. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977;79:544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura M, Saeki K, Takahashi Y. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hs cA-fdx-ORF3 gene cluster. J Biochem. 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 48.Aliverti A, Hagen WR, Zanetti G. Direct electrochemistry and EPR spectroscopy of spinach ferredoxin mutants with modified electron transfer properties. FEBS Lett. 1995;368:220–224. doi: 10.1016/0014-5793(95)00648-s. [DOI] [PubMed] [Google Scholar]
- 49.Musumeci MA, Botti H, Buschiazzo A, Ceccarelli EA. Swapping FAD binding motifs between plastidic and bacterial ferredoxin-NADP(H) reductases. Biochemistry. 2011;50:2111–2122. doi: 10.1021/bi101772a. [DOI] [PubMed] [Google Scholar]
- 50.Nascimento AS, Catalano-Dupuy DL, Bernardes A, de Oliveira NM, Santos MA, et al. Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+. BMC Struct Biol. 2007;7:69. doi: 10.1186/1472-6807-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catalano-Dupuy DL, Orecchia M, Rial DV, Ceccarelli EA. Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a carboxyl-terminal artificial metal binding site. Biochemistry. 2006;45:13899–13909. doi: 10.1021/bi061152v. [DOI] [PubMed] [Google Scholar]
- 52.Bastiaens PI, van Hoek A, van Berkel WJ, de Kok A, Visser AJ. Molecular relaxation spectroscopy of flavin adenine dinucleotide in wild type and mutant lipoamide dehydrogenase from Azotobacter vinelandii. Biochemistry. 1992;31:7061–7068. doi: 10.1021/bi00146a006. [DOI] [PubMed] [Google Scholar]
- 53.Centeno F, Gutierrez-Merino C. Location of functional centers in the microsomal cytochrome P450 system. Biochemistry. 1992;31:8473–8481. doi: 10.1021/bi00151a013. [DOI] [PubMed] [Google Scholar]
- 54.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 56.Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, et al. Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J Bacteriol. 1993;175:1590–1595. doi: 10.1128/jb.175.6.1590-1595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of the dissociation constants for the FNR-NADP+ complexes. The absorbance changes elicited by NADP+ on each enzyme in the 490–510 nm region were used to calculate the K d values by fitting the data to a theoretical equation for a 1∶1 complex. Xac-FNR (○), pea-FNR (▪), Ec-FNR (•).
(TIF)
Determination of the kinetic parameters of the diaphorase reaction for the different enzymes. Kinetics of the ferricyanide reduction by Xac-FNR (○), pea-FNR (▪) and Ec-FNR (•) using NADPH (A) and NADH (B) as substrates.
(TIF)
Kinetics of cytochrome c reductase reactions of the different FNR enzymes. Reduction of cytochrome c by pea-FNR (▪) and Ec-FNR (•) using pea ferredoxin (A), E. coli ferredoxin (B) and E. coli flavodoxin (C) as substrates.
(TIF)
Alignment of the complete sequences of subclass I bacterial FNRs. Sequences of FNRs from A. vinelandii (Av-FNR), X. axonopodis pv. citri (Xa-FNR), Pseudomonas aeruginosa (Pa-FNR), R. capsulatus (Rc-FNR), Rhodobacter sphaeroides (Rs-FNR), Paracoccus denitrificans (Pd-FNR) and Oceanicaulis alexandrii (Oa-FNR) were analyzed.
(TIF)