Abstract
Aquatic hypoxia caused by organic pollution and eutrophication is a pressing worldwide water pollution problem. Better methods for monitoring oxygen levels are needed to assist efforts to maintain and protect the health of natural aquatic environments. In this project, we used a Japanese ricefish (medaka, Oryzias latipes) 8K oligonucleotide array as a platform to identify potential hypoxic biomarkers in different organs (fin, gill, liver and brain) upon exposure to hypoxia. The microarray results were validated by qRT-PCR employing a subset of candidate biomarkers. Interestingly, the largest number and most significant of hypoxia responding array features were detected in hypoxia exposed fin tissues. We identified 173 array features that exhibited a significant response (over 2 fold change in expression) upon exposure to hypoxic conditions and validated a subset of these by quantitative RT-PCR. These gene targets were subjected to annotation and gene ontology mining. Positively identifiable gene targets that may be useful for development of a rapid and accurate biomarker test using fin clips are discussed in relation to previous reports on hypoxia responsive genes.
Keywords: gene expression, hypoxia, medaka, fish, microarray, biomarker
1. Introduction
Hypoxic conditions occur when the dissolved oxygen content in water drops below 2 mg/L (Diaz 2001). Hypoxic “dead zones” in large estuary and marine systems are a worldwide problem and generally occur at river outflows due to organic run-off and corresponding eutrophication (Diaz and Rosenberg, 1995; Li and Brouwer, 2007, 2009a, 2009b; Mason et al., 1998; Wu, 2002; Zhang et al., 2009). Therefore, assays for monitoring oxygen levels and the biological impacts of hypoxia in aquatic systems are important to our understanding of natural aquatic environments. Unfortunately, frequent physical monitoring of oxygen levels over large regions and long periods of time are impractical and such measurements do not address the biological impact of these events. During the past decade, interest in the identification and utilization of biomarkers for assessment of environmental events has increased (Wu et al., 2003; Shang and Wu, 2004; Held et al., 2004; Nikinmaa and Rees, 2005). Biomarkers are defined as measurable biochemical, cellular, and/or physiological changes in a biological system caused by perturbations in the environmental conditions (such as hypoxia or toxicant exposure). Biomarkers may serve as stress indices with the assumption that low stress conditions may cause biochemical responses within individual organisms before prior to those effects being readily observed at higher levels of biological organization (McLoughlin et al., 2000; Schlenk, 1999). In this sense, the use of appropriate biomarkers as environmental indicators may alleviate difficulties in the accurate assessment of hypoxic episodes in aquatic systems.
Fish are one of the most important economic groups in aquatic systems, which make them valuable subjects of hypoxic effect studies. Among extreme the fishes, several species have evolved diverse mechanisms to modulate gene expression patterns in response to various levels and time periods of hypoxic stress. Thus, identification of quantitative changes in fish molecular genetic expression patterns brought about by hypoxia exposure is expected to produce biomarkers that represent both the duration and amplitude of hypoxic stress.
Recent progress in profiling molecular genetic modulation of gene expression in fishes upon hypoxia exposure has been made using cDNA microarray technology (reviewed in Ju et al., 2007a). Microarray studies have identified hundreds of genes that respond to hypoxia by modulating expression levels in several different fish species and from different embryonic stages or tissues. For example, studies have identified fish genes responding to hypoxia in liver and muscle (Gracey et al., 2001), embryo (Ton et al., 2003), gill (van der Meer et al., 2005), non-muscle tissues (Fraser et al., 2006), liver, gill and brain (Ju et al., 2007a,b; Boswell et al., 2009), and the heart (Marques et al., 2008). In previous studies, we have detailed the fabrication and evaluation of a Japanese ricefish (medaka, Oryzias latipes) 8K oligonucleotide microarray and have shown this array reproducible and useful (Zhang et al., 2009; Boswell et al., 2009; Ju et al., 2007a.b) to screen hypoxia responses of medeka brain, liver and gills. In addition, sets of hypoxia responsive genes have been cloned and some of these genes include hypoxia-responsive glucose transporters (Hall et al., 2004; Ng et al., 2003; Zhang et al., 2003), hypoxia inducible factors (Law et al., 2006), telomerase reverse transcriptase (Yu et al., 2006), hypoxia-responsive leptin receptor (Wong et al., 2007), and heme oxygenase 1 (Wang et al., 2008). These studies have greatly facilitated the identification of potential molecular biomarkers for hypoxia.
An ideal biomarker should be easy to assay and secondary stimuli induced in the sampling procedure should be minimal. The fin is in direct contact with the external environment and fin sampling does not require sacrifice of the organism as would sampling of internal organs. A “fin clip” is a rapid and standard non-lethal sampling procedure used in fisheries management (Thompson et al., 2005), and research has documented that fin clipping is a simple, non-lethal technique for predicting muscle tissue mercury concentrations in largemouth bass (Ryba et al., 2008). Thus, reliable fin hypoxia biomarkers may be useful to develop a facile method for hypoxia testing in fisheries management.
With the exception of one report showing the basic response of larval trout exposed to hypoxia is to increase in fin movement (Holeton, 1971), there is little information available regarding how the fish fin responds to hypoxia at the molecular genetic level. Although a main component of fish fins is bone, the musculature of the fins is formed from a dorsal lateral muscular mass (Nakatani et al., 2007). The fin is a complex organ composed of epidermal cells, pigment cells, mesenchymal cells, fibroblasts, scleroblasts, nerve fibers and blood vessels, and bone secreting cells. The fin is used for movement, stability, nest-building, spawning, and as tactile organs (Luther et al., 1995). The fish fin has the capacity for rapid cell proliferation when partial amputation is carried out. Based on these facets of fin biology, we may expect fish fin to produce a dynamic response to hypoxia exposure resulting from modulation of gene expression patterns. Herein we provide results from studies undertaken to compare the fin response to other tissues and to determine if the fin produces a consistent molecular genetic response to hypoxia that may be used to identify potential fin hypoxia biomarker candidates.
2. Materials and Methods
2.1. Hypoxia treatment and RNA extraction, Microarray Probe Preparation and Hybridization, Data Acquisition and Statistical Analysis
Adult male Japanese ricefish (medaka; Oryzias latipes) were provided by the Aquatic Biotechnology and Environmental Laboratory at the University of Georgia (Athens, GA, USA). Stocks were raised to adulthood on a diet of brine shrimp and liver-based paste. Fish utilized in treatment were 9–12 months old.
The system used in this study to induce and maintain hypoxic conditions has been described elsewhere (Boswell et al., 2009; Ju et al., 2007a,b). Control aquaria were continuously bubbled with compressed air and were thus oxygen saturated at 8.0 mg O2/L. The oxygen levels in the hypoxic exposure aquaria were stepped-down from normoxic levels of 8.0 mg O2/L over the course of several days (1 day at 2.4 mg O2/L, 1 day at 2.0 mg O2/L, 2 days at 1.6 mg O2/L) to 0.8 mg O2/L for 5 additional days. At the end of the extensive period, the number of surviving medaka was assessed and fish were sacrificed for dissection of the brain, gills, liver, and fin (fin samples included dorsal, caudal and ventral fins). All organs were immediately flash frozen upon dissection in dry ice/ethanol and like organs from three fish were pooled into each sample tube for freezing.
Total RNA was extracted from frozen tissues using TRI Reagent (Sigma-Aldrich, St. Louis, MO, USA) and purified to remove residual phenol and minimize DNA contamination with the RNeasy Kit (Qiagen, Valencia, CA, USA), both according to the manufacturer’s instructions.
Microarray probe preparation and hybridization, data acquisition and statistical analysis were performed as previously reported (Boswell et al., 2009; Ju et al., 2007a,b) with three biological repeats and six technical repeats. Hypoxic and control (normoxic) RNAs differentially labeled with fluorescent dyes were hybridized to a medaka 8,046-feature oligonucleotide microarray slide and amplified as previously described (Boswell et al., 2009; Ju et al., 2007a,b). After hybridization, all slides were washed by a series of four SDS and SSC wash buffers with increasing stringency. First, array slides were dipped 10x in low stringency wash buffer (1x SSC and 0.5% SDS) at a 42 °C, then the slides were rocked in 1x SSC and 0.2% SDS at 42 °C for 5 min, followed by high-stringency buffer (0.1x SSC and 0.2% SDS) at room temperature for 5 min, and finally rocked in 0.05x SSC for 5 min. Centrifugation at 1,500 rpm for 3 min was employed to dry the array slides prior to data acquisition.
Data acquisition was also performed as previously reported (Boswell et al., 2009; Ju et al., 2007a,b) using a 400B GenePix array scanner (Axon Instruments). After the Pearson Correlation analysis (r ≥ 0.85), a one-sample t-test was applied to identify statistically significant results (p ≤ 0.05).
All expression data were submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Differentially expressed genes were identified via BLAST searching against GenBank (Altschul et al., 1997) and mined for annotation and ontology using Swiss-Prot (Bairoch and Boeckmann, 1994; Boeckmann et al., 2003), Entrez Gene (Maglott et al., 2007). The GO term and pathway associations of differentially expressed genes were investigated using blast2go (www.Blast2GO.de) (Gotz et al., 2008).
2.2. Quantitative real-time PCR
Medaka EST sequences were initially used to fabricate 60-mer oligonucleotides for each array feature (Ju et al., 2007a,b). To validate microarray results, a subset of six features that appeared from microarray data to yield robust (≥ 4 fold change) response to hypoxia in fin were chosen for quantitative real-time PCR (qRT-PCR) analysis. The 60-mer sequences for each of these hypoxia responsive features were used to generate qRT-PCR primers. The sequences of the qRT-primers are shown in Table 1 and were designed using an online qRT-PCR primer design tool (http://www.genscript.com/cgi-bin/tools/primer_genscript.cgi).
Table 1.
Sequences of qRT-PCR Primers employed in this study
| Gene ID | Forward Primer | Reverse Primer |
|---|---|---|
| 18SRNA | TCTCGATTCTGTGGGTGGT | TAACCAGACAAATCGCTCCA |
| AB111381 | AGAAGGAGAATGGAAAGGCA | AGCATGGTTAGCCACAAACA |
| AB111384 | GAGTTACAGGAGGCCTGGAG | TCCTCATAAGGTTGTCTGATTCAT |
| AB111386 | CCTCAACAACATGGAGATCG | AATCTTCACGCCGTTCTTCT |
| AU169680 | GTTCATGATCCTGCCTGTTG | ACATTAGTGGCGTCCTTTCC |
| BJ514046 | TGCAGAATGGTGCTTAAGGT | TTTCCAAACGACTGCTATGG |
| BJ735388 | AGGACACCTCCGGTAATCTG | TCCCAAGACTGCAAGAAAGA |
| BJ742144 | TCTTCAAAGCGTCCATTCAC | TGCAAGCTCTGCTTGTCAC |
To perform qRT-PCR, 2 μg of medaka RNA was reverse transcribed (MMLV reverse transcriptase, Invitrogen) and qRT-PCR performed using an Applied Biosystems Prism 7500 fast real-time PCR system employing Syber green master mix (see Boswell et al., 2009; Ju et al., 2007a,b). The comparative threshold cycle (CT) method was used to calculate the relative concentrations. This method involves obtaining CT values for the target gene and normalizing to 18S rRNA housekeeping gene from the same organ; followed by comparison of the relative expression levels between hypoxia and control samples. The CT values of 18S from controls and hypoxia exposed samples of the same organ with same quantity of RNA are consistent. Quantification of gene expression is reported as a relative quantity to the control value (normoxic) in the same tissue. We used the 18S rRNA as an endogenous control as extensively reported in other hypoxia (Fuchshofer et al., 2009; Zhang et al., 2003) and gene expression studies (Shen et al., 2009; Zhang et al., 2007). All experiments were repeated with three biological replicates and each biological replicate was repeated with four technical replications. The mean values of the technical replicates in hypoxia and control samples were used to calculate the fold-change in gene expression for qRT-PCR. The averages of the relative quantities for the biological replications were used in a 2-tailed students t-test at a 90% confidence level (p<0.05) to determine the significance of the difference between gene expression in hypoxic verses normoxic (i.e. control) organs.
3. Results
3.1. Hypoxia responsive genes from various tissues
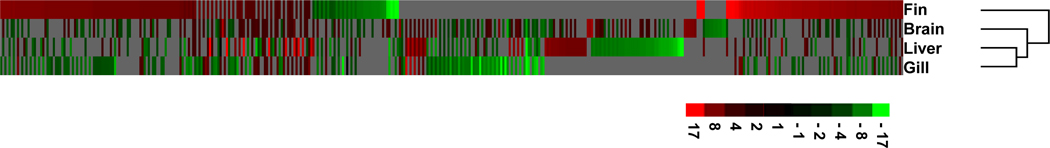
An 8k medaka microarray was used to compare gene expression in four individual tissues including fin, gill, liver, and brain of medaka that were either maintained under normoxic (8.0 mg O2/L) conditions or acclimated to hypoxic (0.8 mg O2/L) conditions. Features that hybridized when probed with RNAs derived from each tissue were streamlined using the Pearson correlation and a one-sample t-test for each hybridized feature. This analysis resulted in a significant number of features with differential expression in each tissue (i.e., 2,011 in brain, 1,163 in liver, 740 in fin, and 506 in gill). After removing features duplicated in more than one tissue, we were able to identify 3,206 unique and statistically significant hypoxia responsive features. The relative patterns of the 3,206 hypoxia responsive features are presented in the heat map shown in Figure 1.
Figure 1. Hierarchical clustering analysis of 3,206 hypoxia responsive features in different organs of medaka.
The gene cluster tool developed by Eisen et al. (1998) was employed for analyses. The gene expression clustering results were visualized by treeview (http://rana.lbl.gov/EisenSoftware.htm). Each row represents an organ and each column a single gene. Green indicates down-regulation response, red indicates up-regulation response, and grey indicates missing spots. The dendrogram on the right indicates the relatedness of the gene expression profiles of individual organs. The longer the dendrogram arm shows the greater the differences.
Since in-depth analyses of each of these 3,206 features to verify their utility as hypoxia biomarkers is both time and cost prohibitive, we refined our list of most interesting features to contain only the strongest biomarker candidates. Two criteria were applied to select features; (1) any feature that exhibited a 2-fold or greater induced or reduced hypoxia-response in at least one of the four tissues studied. This resulted in 382 features. Also, (2) features that exhibited a consistent directional response in at least three of the four tissues analyzed (i.e., always up regulation regardless of the magnitude of change). This second criterion yielded 46 features. Thus, 428 hypoxic responsive features were chosen for further analysis.
3.2. Strongest modulated response to hypoxia in fin
Of the 428 hypoxic responsive features, 255 features exhibited a hypoxia modulated response in fin (214 up, 41 down), 191 in gill (39 up, 152 down), 207 in liver (89 up, 118 down), and 214 in brain (112 up, 102 down). A complete list of all 428 hypoxia responsive features is provided in the supplementary materials (Table S1, available at www.xiphophorus.org). Very few (<5) of these responsive features demonstrate consistent patterns across all four species with the most significant of these hypoxic induced features (≥ 4-fold) being present in fin 17 (11 up, 6 down) out of 29 significant features (Table 2). In addition, significant features were found in liver (7 total, 4 up, 3 down) and gill (4 total, 4 down) tissues.
Table 2.
Genes identified by microarray as exhibiting at least a four fold or greater response to hypoxic exposure. Features in Italics were confirm
| Sequence Ontology Classes | |||||
|---|---|---|---|---|---|
| Feature ID |
Fold Change |
Description | Molecular Function | Biological Process | Cellular Component |
| INDUCED | |||||
| Fin | |||||
| BJ728005 | 16.9 | Hypothetical protein LOC678528 | Structural molecule Activity | Cell migration; Gastrulation | Intermediate filament |
| AU167844 | 8 | Hemoglobin alpha embryonic-1 | Heme & oxygen binding; oxygen transport activity | Oxygen transport | Hemoglobin complex |
| BJ735388 | 7.7 | No hit | - | - | - |
| AU169680 | 6.1 | Enolase 1, (alpha) | Mg+2 binding; serine-type endo-peptidase activity; heterodimer activity; DNA-dependent | Glycolysis; in utero; Embryonic development | Cell surface; synaptosome; plasma Membrane |
| AB111386 | 5 | Phosphoglycerate kinase 1 | ADP/ATP binding, Phosphoglycerate kinase activity | Hypoxia response; Glycolysis; gluconeogensis | Cytosol; Soluble fraction |
| BJ508273 | 4.9 | Unnamed proein [Tetraodon nigroviridis] | - | - | - |
| BJ720277 | 4.6 | Vimetin | Structural molecule activity; Protein binding | Cytolasm; Intermediate filament; Cytoskeleton | |
| BJ742144 | 4.6 | No hit | - | - | - |
| AB111384 | 4.4 | Glucose-6-phosphatedehydrogenase | G6PD activity | Glucose Metabolism; Oxidation/ reduction | |
| BJ715269 | 4.4 | Pyruvate dehydrogenase kinase 2 | Protein binding; ATP binding; Kinase activity | Signal transductin; Peptidyl-histidine posphorylation | Nucleolus; Mitochondrial matrix |
| AV669453 | 4 | Cytochrome C oxidase subunit II | Cu ion binding; Electron carrier; Heme Binding | Transport; Respiratory election transport | Mitochondrial inner membrane chain |
| Liver | |||||
| AB064320 | 6.9 | Vitellogenin A | Transporter activity; Nutrient reservoir activity | Transport | |
| BJ728736 | 5.8 | Apolipoprotein B (ApoB) | Protein binding; Transporter activity; Endoplasmic reticulum | Transport; Lipid metabolic process; Cellular component organization | Extracellular space; Organelle; Intracellular; Endosome |
| AB163431 | 4.3 | Heme oxygenase 1 | Heme degrading activity | Response to stress; Breaks down heme | Cell; Endoplasmic Reticulum |
| BJ736365 | 4.3 | Unnamed protein [T. nigroviridis] | - | - | - |
| SUPRESSED | |||||
| Fin | |||||
| D63796 | −4 | Protamine (LOC100049420) | - | - | - |
| AU311117 | -4.2 | Suppressor of cytokine signaling 3 | Protein binding | Placenta development; Negative regulator of cytokine signaling; JAK-STAT signaling pathway | - |
| BJ715122 | −4.3 | Jun dimerization protein 2 | dsDNA binding; Transcription factor and repressor activity | (−) regulation of transcription from RNA pol II promoter | Nucleus |
| BJ021653 | −5 | No hit | - | - | - |
| BJ514046 | −9.2 | No hit | - | - | - |
| AU311099 | −10.7 | Sperm plasma glycoprotein | - | - | - |
| Gill | |||||
| AB111381 | −4.2 | Fructose-bisphosphate aldolase A | Catalytic activity | Glycolysis | |
| BJ735325 | −4.3 | PREDICTED: similar to protocadherin 1 | - | - | - |
| AU168239 | -4.5 | Myelin basic protein | Structural constituent of myelin sheath | Negative regulation of axonogenesis; Myelination | Myelin sheath; Nucleusinternode region of axon; Neuronal cell body |
| BJ706653 | −4.9 | Carbon catabolite repression 4-like | Transcription factor activity | Transcription | Nucleus |
| Liver | |||||
| AJ457750 | −4.1 | Solute carrier family member 43 | Amino acid Transport activity | Transport | Mitochondrion |
| AF359589 | −4.5 | Insulin-like growth factor binding protein 2 | Insulin-like growth factor carrier protein | Multicellular organismal development; Signal transduction; Cell growth; Response to stress; Reproduction | Extracellular space; Organelle; Cytoplasm; Plasma membrane |
| BJ736564 | −5.7 | Nuclear receptor subfamily 1 group D member 2 | Receptor activity; DNAC binding | Transcription | |
| AU311111 | −11.3 | - | - | - | |
3.3. Function groups and pathways of hypoxia responsive genes
The genes corresponding to all 428 features were analyzed via Blast searching and 173 features were identified based on homology. These features were mined for gene ontology. The GO terms and pathway associations of differentially expressed genes were investigated (Table S1 and Table 2). Most features have more than one functional group or pathway assigned to them.
In the glycolysis functional group several features displayed alterations in gene expression including AU169680 (enolase, 6.1-fold in fin and 1.7 fold in liver), AB111386 (PGK1, 5.0-fold in fin and 1.8 fold in liver), BJ729352 (phosphoglycerate mutase, 1.3 fold in liver) and BJ729242 (phosphoglucoisomerase, 2.2 fold in liver). In contrast, glycolysis features analyzed from gill (4 total) and brain (5 total) were down regulated in response to hypoxia (Supplemental Figure 1).
Further examination of significantly responsive features in fin (Table 2) revealed features involved in transcriptional signaling or signal transduction pathways, including; BJ715269 (pyruvate dehydorgenase kinase 2, 4.4 fold), AU311117 (suppressor of cytokine signaling, -4.2-fold) and BJ715122 (Jun dimerization protein 2, -4.3 fold). The significantly regulated features that participate in the pentose phosphate, oxidative phosphorylation, oxygen transport pathways, and cell communications were found in fin tissues and included AB111384 (G6PDH, 4.4-fold), AV669453 (cytochrome C oxidase, 4.0-fold), AU167844 (hemoglobin embryonic alpha, 8.0- fold), and BJ720277 (vimetin, 4.6-fold) features, respectively.
3.4 Real time PCR validation of the microarray response
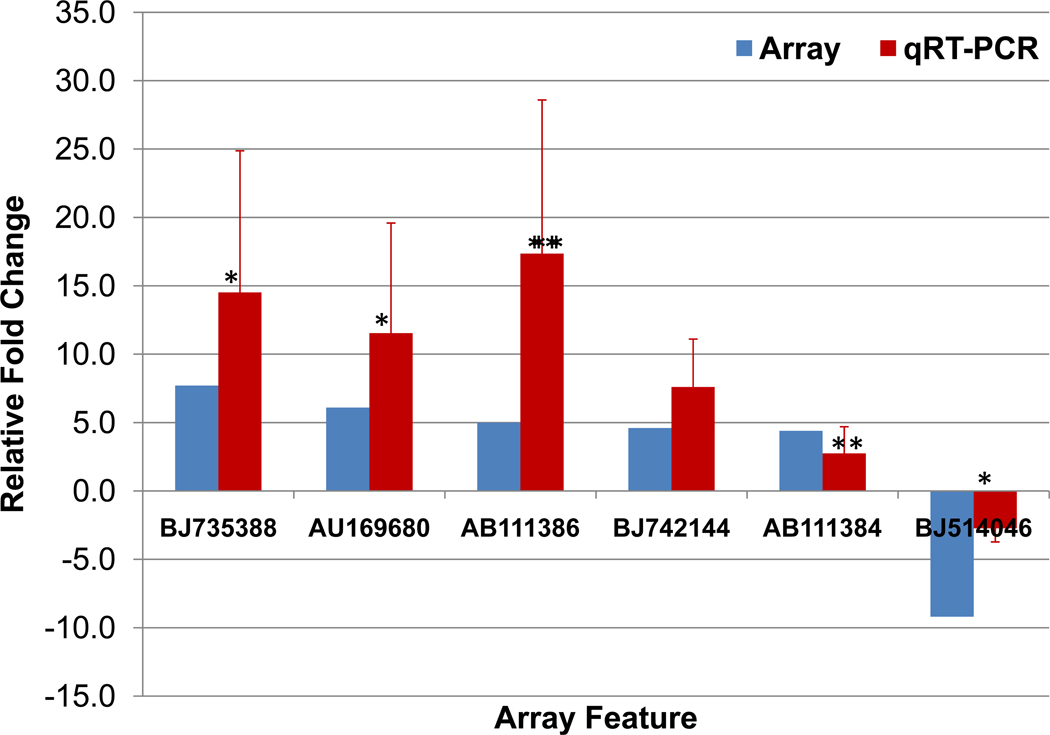
In order to assess the validity of our microarray results, we selected six features that had produced greater than 4.0-fold modulation in fin tissue upon exposure to hypoxia for further analysis with qRT-PCR. The six features subjected to qRT-PCR validation were BJ735388 (array 7.7-fold and qRT-PCR 14.5-fold), AU169680 (array 6.1-fold and qRT-PCR 11.5-fold), AB111386 (array 5.0-fold and qRT-PCR 17.3-fold), BJ742144 (array 4.6-fold and qRT-PCR 7.6-fold), AB111384 (array 4.4-fold and qRT-PCR 2.8-fold), and BJ514046 (array -9.2-fold and qRT-PCR -2.7-fold). The trends in qRT-PCR fold changes in hypoxic samples relative to normoxic samples mirrored that of the array fold changes with the exception of AB111386, which is up-regulated in both, but has the greatest fold change in qRT-PCR (Figure 2).
Figure 2. Comparison of array and qRT-PCR fold changes for select features with gene expression responses (≥ ±4-fold) in medaka fin tissue.
Six features with greater than 4-fold induction or suppression in the array (blue column) were examined by qRT-PCR (red column). The qRT-PCR relative expression for each hypoxic sample was based on normalization to 18 S rRNA. Fold changes were the average hypoxic organ tissue sample (n=3) realitive to normoxic sample from the same organ tissue. * Represents p≤ 0.01 and ** represents p ≤ 0.05
4. Discussion
4.1. Robust gene response to hypoxia in fish fin
Gene expression level studies in fin tissues have not been explored in previous studies of hypoxia exposure (Gracey et al., 2001;Ton et al., 2003; Fraser et al., 2006; Ju et al., 2007a,b; Boswell et al., 2009; Marques et al., 2008). The fish fin contains bone as well as thin layers of tissue that are in direct contact with the exterior environment. Thus, as an assessment target for hypoxic biomarkers or as a response to other external stimuli, the fin may represent an ideal tissue source. We demonstrate that fish fin exhibits a robust and distinct molecular genetic response to hypoxia exposure in regard to the number of genes showing modulated expression, the level of gene expression, and the range and pattern of observed response. Any or all of which may be used to develop a rapid assessment regimen for hypoxic environments.
4.2. Gene expression responses to hypoxia profiled by microarray reveal molecular mechanisms of the hypoxia response
Animal cells require oxygen to generate ATP through oxidative phosphorylation. Under hypoxic conditions, animal cells typically promote glycolysis and suppress oxidative phosphorylation. In fish, up-regulation of anaerobic glycolytic enzymes is a well-documented physiological adaptive strategy to cope with hypoxia in liver and other organs (Randall, 1982; Van den Thillart and van Waarde, 1985), which we see in the up-regulation of four features in liver belonging to the glycolysis functional group. Robust up-regulation of two fin features within glycolysis suggests that this tissue also employs anaerobic glycolytic strategy as one adaptation mechanism to hypoxic stress. Interestingly, down-regulation of these same features and two others in brain and gill tissues may be a damage prevention strategy to avoid accumulation of metabolic by-products (Lutz, 1989).
Activation of glycolysis by hypoxia must be coordinated with other metabolic pathways that also use glucose. AB111384, which corresponds to glucose-6-phosphate dehydrogenase, (G6PDH) was markedly up-regulated (4.4-fold) in fin. To our knowledge this is the first report of the G6PDH response to hypoxia in fish. At the protein level G6PDH activity in erythrocytes of scorpion fish was reported reduced by 62% under hypoxic conditions (15% saturation; Rusinova and Center, 2000) and increased by 26% in goldfish brain tissue (Lushchak and Bagnyukova, 2006) after 8 h of anoxia. In vitro studies using mammalian PC12 cells (a hypoxia sensitive cell line) demonstrated induced G6PDH gene expression, that followed a time that was much slower than that of phosphoglycerate kinase 1 (PGK1; Gao et al., 2004). It was suggested that up-regulation of G6PDH was related to the antioxidant protection due to NADPH synthesis, as an effort to avoid oxygen free radical accumulation. Up-regulation of features related to TCA and oxidative phosphorylation pathway in fin is consistent with this hypoxia adaptive strategy. Thus, significant up-regulation of G6PDH as well as other TCA and oxidative phosphorylation related genes in fish fin may provide an ideal in vivo model to study hypoxia response strategies in short and long term hypoxic exposures at molecular genetic level.
Herein we present novel information regarding the protein synthesis response to hypoxia in fin. In contrast with down-regulation results from liver tissues many features related to protein biosynthesis were up-regulated (between 2 and 4-fold, Table S1) in fin upon hypoxic exposure. As previously reported, the first response to hypoxia of wild fish would be to escape, and since successful escape will ultimately depend on locomotor abilities controlled by use of fin tissues, the observed responses may be reasonable (Domenici, 1997; Howland, 1974; Webb, 1976). For example, many species of fish exposed to hypoxia increase swimming activity, presumably due to an escape response (Lefrancois et al., 2005). Thus, up-regulation of protein biosynthesis genes in fish fin as a response to hypoxia may be a part of the overall escape related strategy.
One desirable characteristic of biomarkers is specificity. Thus, differential gene expression responses among various organs, such as fin compared to other organs, may be used to reflect specific physiological mechanisms employed in adaptation to hypoxia. For example, one adaption strategy is a cellular increase in production of oxygen-transport proteins to increase the oxygen-carrying capacity in the blood (Terwilliger, 1998). Up-regulation of hemoglobin alpha embryonic-1 (AU167844) in fin, which is reported to be down-regulated in blood cells of adult rainbow trout (Iuchi, 1985) and medaka (Mauyama et al., 2002), suggests this physiological adaptation to hypoxia is performed in fish fin. While prior studies examining mRNA expression of globulins in zebrafish (Roesner et al. 2006) and goldfish (Roesner et al. 2008) have reported differential responses to hypoxia (down-regulation and no change, respectively), these studies monitored adult hemoglobin. While it is plausible that the hemoglobin alpha embryonic-1 (AU167844) response is a blood signal, the absence of a response in other blood containing tissues especially liver indicates the potential for other mechanisms within fish fin. Additional studies examining embryonic hemoglobin levels across tissues and within other species are needed to validate its use as a hypoxia biomarker.
4.3. Robust gene regulation changes in response to hypoxia in fin are also reflected in internal organs
Another desirable characteristic of an informative biomarker is robustness, which implies the response may be easy to measure reproducibly using simple analytical approaches. Many of the gene targets identified here that respond to hypoxia in fin tissues fit these characteristics. For example, the strongest (≥ 4-fold) hypoxia responsive gene expression changes were detected in fin. Feature BJ728005 exhibited the largest up-regulated fold-change among 428 hypoxia responsive features detected in this study.
AB111386 (PGK 1) was also strongly up-regulated in fish fin in our study. This gene is also up-regulated in adult zebrafish (Roesner et al., 2006) as well as in zebrafish embryos (Ton et al., 2003) upon hypoxic exposure. Marques et al., found a suppressor of cytokine signaling 3 (socs3, AU311117) is down-regulated in zebrafish heart and this agrees with a strong downregulation of this gene in medaka fin, gill and liver in these studies (Marques et al., 2008). Enolase I (alpha) (AU169680) was down regulated in the gills of adult zebrafish (van der Meer et al., 2005), up-regulated in zebrafish embryos (Ton et al., 2003), and up-regulated in liver but down-regulated in muscle of the long-jaw mudsucker (Gillichthys mirabilis) (Gracey et al., 2001). Enolase I showed a strong up-regulation in the fin of our adult medaka consistent with our prior results (Boswell et al., 2009; Ju et al., 2007a.b; Oehlers et al., 2007).
Comparing gene expression pattern responses to hypoxia in fin, brain, gill and liver of medaka indicate fish fin exhibited the largest and most varied gene response to hypoxia in all four tested organs (Figure 1). When the features on the array response for each of the four organs are compared and clustered the fin exhibits a response different from the internal organs. The genes with the strongest regulatory change in response to hypoxia were also present in fin and may hallmark potential molecular mechanisms employed by fish fin to adapt to hypoxic stress. Based on these results and combined with comparison analysis, future work will be necessary to develop distinct patterns of gene expression in fish fin for hypoxia biomarker assessment.
Supplementary Material
Acknowledgements
The authors would like to thank Markita Savage, Leona Hazlewood and the other employees of the Xiphophorus Genetic Stock Center, Texas State University, for maintaining the fish used in this study. This work was supported by the National Institutes of Health, National Center for Research Resources grant award R24-RR024790 and a grant award from the National Oceanic and Atmospheric Administration, National Ocean Program # NA06-NOS4260118.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This paper is based on a presentation given at the 5th Aquatic Annual Models of Human Disease conference: hosted by Oregon State University and Texas State University-San Marcos, and convened at Corvallis, OR, USA September 20–22, 2010.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank: current status. Nucleic Acids Res. 1994;22:3578–3578. [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell MG, Wells MC, Kirk LM, Ju Z, Zhang ZP, Booth RE, Walter RB. Comparison of gene expression responses to hypoxia in viviparous (Xiphophorus) and oviparous (Oryzias) fishes using a medaka microarray. Comp. Biochem. and Physiol. C. 2009;149:258–265. doi: 10.1016/j.cbpc.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Diaz RJ. Overview of Hypoxia around the world. J. Environ. Qual. 2001;30:275–281. doi: 10.2134/jeq2001.302275x. [DOI] [PubMed] [Google Scholar]
- Diaz RJ, Rosenberg R. Marine benthic hypoxia: A review of its ecological effects and the behavioral responses of benthic macrofauna. Oceanogr Mar Biol. 1995;33:245–203. [Google Scholar]
- Domenici P. The kinematics and performance of fish fast-start swimming. J. Exp. Biol. 1997;200:1165–1178. doi: 10.1242/jeb.200.8.1165. [DOI] [PubMed] [Google Scholar]
- Fraser J, Vieira de Mello L, Ward D, Rees HH, Williams DR, Fang Y, Brass A, Gracey AY, Cossins AR. Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc Natl Acad Sci USA. 2006;103:2977–2981. doi: 10.1073/pnas.0508270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchshofer R, Yu AL, Teng HH, Strauss R, Kampik A, Welge-Lussen U. Hypoxia/reoxygenation induces CTGF and PAI-1 in cultured human retinal pigment epithelium cells. Exp. Eye Res. 2009;88:889–899. doi: 10.1016/j.exer.2008.11.036. [DOI] [PubMed] [Google Scholar]
- Gao L, Mejías R, Echevarría M, López-Barneo J. Induction of the glucose-6-phosphate dehydrogenase gene expression by chronic hypoxia in PC12 cells. FEBS lett. 2004;569:256–260. doi: 10.1016/j.febslet.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey AY, Troll JV, Somero GN. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc Natl Acad Sci USA. 2001;98:1993. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JR, MacCormack TJ, Barry CA, Driedzic WR. Sequence and expression of a constitutive, facilitated glucose transporter (GLUT1) in Atlantic cod Gadus morhua. J. Exp. Biol. 2004;207:4697–4706. doi: 10.1242/jeb.01346. [DOI] [PubMed] [Google Scholar]
- Held M, Gase K, Baldwin IT. Microarrays in ecological research: a case study of a cDNA microarray for plant-herbivore interactions. BMC Ecol. 2004;4:13. doi: 10.1186/1472-6785-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeton GF. Respiratory and Circulatory Responses of Rainbow Trout Larvae to Carbon Monoxide and to Hypoxia. J. Exp. Biol. 1971;55:683–694. doi: 10.1242/jeb.55.3.683. [DOI] [PubMed] [Google Scholar]
- Howland HC. Optimal strategies for predator avoidance: the relative importance of speed and manoeuvrability. J. Theoret Biol. 1974;47:333–350. doi: 10.1016/0022-5193(74)90202-1. [DOI] [PubMed] [Google Scholar]
- Iuchi I. Cellular and molecular basis of the larval-adult shift of hemoglobins in fish. Zool. Sci. 1985;2:1–23. [Google Scholar]
- Ju Z, Wells MC, Walter RB. DNA microarray technology in toxicogenomics of aquatic models: methods and applications. Comp. Biochem. Physiol. C. 2007a;145:5–14. doi: 10.1016/j.cbpc.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Ju Z, Wells MC, Walter RB. Multiple Tissue Gene Expression Analyses in Japanese Medaka (Oryzias latipes) Exposed to Hypoxia. Comp. Biochem. Physiol. C. 2007b;145:134–144. doi: 10.1016/j.cbpc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Law SHW, Wu RSS, Ng PKS, Yu RMK, Kong RYC. Cloning and expression analysis of two distinct HIF-alpha isoforms-gcHIF-1alpha and gcHIF-4alpha-from the hypoxia-tolerant grass carp, Ctenopharyngodon idellus. BMC Mol. Biol. 2006;7:15. doi: 10.1186/1471-2199-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois C, Shingles A, Domenici P. The effect of hypoxia on locomotor performance and behaviour during escape in Liza aurata. J Fish Biol. 2005;67:1711–1729. [Google Scholar]
- Li T, Brouwer M. Hypoxia-inducible factor, gsHIF, of the grass shrimp Palaemonetes pugio: Molecular characterization and response to hypoxia. Comp. Biochem. Physiol. B. 2007;147:11–19. doi: 10.1016/j.cbpb.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Li T, Brouwer M. Bioinformatic analysis of expressed sequence tags from grass shrimp Palaemonetes pugio exposed to environmental stressors. Comp Biochem Physiol. D. 2009a;4:187–195. doi: 10.1016/j.cbd.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Li T, Brouwer M. Gene expression profile of grass shrimp Palaemonetes pugio exposed to chronic hypoxia. Comp. Biochem. Physiol. D. 2009b;4:196–208. doi: 10.1016/j.cbd.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Lushchak VI, Bagnyukova TV. Temperature increase results in oxidative stress in goldfish tissues. 2. Antioxidant and associated enzymes. Comp. Biochem. and Physiol. C. 2006;143:36–41. doi: 10.1016/j.cbpc.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Luther PK, Munro PMG, Squire JM. Muscle ultrastructure in the teleost fish. Micron. 1995;26:431–459. [Google Scholar]
- Lutz PL. Interaction between hypometabolism and acid-base balance. Can J. Zool. 1989;67:3018–3023. [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques IJ, Leito JTD, Spaink HP, Testerink J, Jaspers RT, Witte F, van den Berg S, Bagowski CP. Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. J. Comp. Physiol. B: Biochem. Systemic Environ. Physiol. 2008a;178:77–92. doi: 10.1007/s00360-007-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RP, Hunjan S, Le D, Constantinescu A, Barker BR, Wong PS, Peschke P, Hahn EW, Antich PP. Regional tumor oxygen tension: fluorine echo planar imaging of hexafluorobenzene reveals heterogeneity of dynamics. Int. J. Radiat Oncology Biol Phys. 1998;42:747–750. doi: 10.1016/s0360-3016(98)00306-x. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Yasumasu S, Iuchi I. Characterization and expression of embryonic and adult globins of the teleost Oryzias latipes (medaka) J. Biochem. 132:581–589. doi: 10.1093/oxfordjournals.jbchem.a003260. [DOI] [PubMed] [Google Scholar]
- McLoughlin N, Yin D, Maltby L, Wood RM, Yu H. Evaluation of sensitive and specificity of two crustacean biochemical biomarkers. Environ. Toxicol. Chem. 2000;19:2085–2092. [Google Scholar]
- Nakatani Y, Kawakami A, Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev. Growth Differ. 2007;49:145–154. doi: 10.1111/j.1440-169X.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- Nikinmaa M, Rees BB. Oxygen-dependent gene expression in fishes. Am. J. Physiol., Regul. Integr. Comp. Physiol. 2005;288:R1079–R1090. doi: 10.1152/ajpregu.00626.2004. [DOI] [PubMed] [Google Scholar]
- Ng PKS, Wu RSS, Zhang ZP, Mok HOL, Randall DJ, Kong RYC. Molecular cloning and characterization of a hypoxia-responsive CITED3 cDNA from grass carp. Comp. Biochem. Physiol. B. 2003;136:163–172. doi: 10.1016/s1096-4959(03)00224-0. [DOI] [PubMed] [Google Scholar]
- Oehlers LP, Perez AN, Walter RB. Detection of hypoxia-related proteins in medaka (Oryzias latipes) brain tissue by difference gel electrophoresis and de novo sequencing of 4-sulfophenyl isothiocyanate-derivatized peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Comp. Biochem. Physiol. C. 2007;145:120–133. doi: 10.1016/j.cbpc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Randall D. The Control of respiration and circulation in fish during exercise and hypoxia. J. Exp. Biol. 1982;100:275–288. [Google Scholar]
- Roesner A, Hankeln T, Burmester T. Hypoxia induces a complex response of globin expression in zebrafish (Danio rerio) J. Exp. Biol. 2006;209:2129–2137. doi: 10.1242/jeb.02243. [DOI] [PubMed] [Google Scholar]
- Roesner A, Mitz SA, Hankeln T, Burmester T. Globins and hypoxia adaptation in the goldfish, Carassius auratus. FEBS J. 2008;275:3633–3643. doi: 10.1111/j.1742-4658.2008.06508.x. [DOI] [PubMed] [Google Scholar]
- Rusinova OS, Center ED. Seasonal changes in glucose-6-phosphate dehydrogenase activity in the tissues of Black Sea mussel. J. GidrobiologicheskII Zh. 2000;36:87–91. [Google Scholar]
- Ryba SA, Lake JL, Serbst JR, Libby AD, Ayvazian S. Assessment of caudal fin clip as a non-lethal technique for predicting muscle tissue mercury concentrations in largemouth bass. Environ. Chem. 2008;5:200–203. [Google Scholar]
- Schlenk D. Necessity of defining biomarkers for use in ecological risk assessments. Mar. Pollut. Bull. 1999;39:48–53. [Google Scholar]
- Shang EH, Wu RS. Auquatic hypoxia is a teratogen and affects fish embryonic development. Environ. Sci. Technol. 2004;38:4763–4767. doi: 10.1021/es0496423. [DOI] [PubMed] [Google Scholar]
- Shen BL, Zhang ZP, Wang YL, Wang GD, Chen Y, Lin P, Wang SH, Zou Z. Differential expression of ubiquitin-conjugating enzyme E2r in the developing ovary and testis of penaeid shrimp Marsupenaeus japonicus. Mol. Biol. Rep. 2009;36:1149–1157. doi: 10.1007/s11033-008-9291-7. [DOI] [PubMed] [Google Scholar]
- Terwilliger NB. Functional adaptations of oxygen-transport proteins. J. Exp. Biol. 1998;201:1085–1098. doi: 10.1242/jeb.201.8.1085. [DOI] [PubMed] [Google Scholar]
- Thompson JM, Hirethota PS, Eggold BT. A comparison of elastomer marks and fin clips as marking techniques for walleye. North Am J. Fisher Manag. 2005;25:308–315. [Google Scholar]
- Ton C, Stamatiou D, Liew CC. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol. Genomics. 2003 doi: 10.1152/physiolgenomics.00128.2002. 1282002. [DOI] [PubMed] [Google Scholar]
- Van den Thillart G, van Waarde A. Teleosts in hypoxia: aspects of anaerobic metabolism. Mol. Physiol. 1985;8:393–409. [Google Scholar]
- van der Meer DLM, van den Thillart GE, Witte F, de Bakker MAG, Besser J, Richardson MK, Spaink HP, Leito JTD, Bagowski CP. Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;289:1512–1519. doi: 10.1152/ajpregu.00089.2005. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhong XP, Qiao ZX, Gui JF. Inductive transcription and protective role of fish heme oxygenase-1 under hypoxic stress. J. Exp. Biol. 2008;211:2700–2706. doi: 10.1242/jeb.019141. [DOI] [PubMed] [Google Scholar]
- Webb DP. Root growth in Acer saccharum Marsh. seedlings: Effects of light intensity and photoperiod on root elongation rates. Bot Gazette. 1976;137:211. [Google Scholar]
- Wong MML, Yu RMK, Ng PKS, Law SHW, Tsang AKC, Kong RYC. Characterization of a hypoxia-responsive leptin receptor (omLepRL) cDNA from the marine medaka (Oryzias melastigma) Mar. Pollut. Bull. 2007;54:797–803. doi: 10.1016/j.marpolbul.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Wu RS. Hypoxia: from molecular responses to ecosystem responses. Mar. Pollut. Bull. 2002;45:35–45. doi: 10.1016/s0025-326x(02)00061-9. [DOI] [PubMed] [Google Scholar]
- Wu RS, Zhou BS, Randall DJ, Woo NY, Lam PK. Aquatic hypoxia is a disrupter and impairs fish reproduction. Environ. Sci. Technol. 2003;37:1137–1141. doi: 10.1021/es0258327. [DOI] [PubMed] [Google Scholar]
- Yu RMK, Chen EXH, Kong RYC, Ng PKS, Mok HOL, Au DWT. Hypoxia induces telomerase reverse transcriptase (TERT) gene expression in non-tumor fish tissues in vivo: the marine medaka (Oryzias melastigma) model. BMC Mol. Biol. 2006;7:27. doi: 10.1186/1471-2199-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Ju Z, Wells MC, Walter RB. Genomic approaches in the identification of hypoxia biomarkers in model fish species. J. Exp. Mar. Biol. Ecol. 2009;381:S180–S187. doi: 10.1016/j.jembe.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Wang YL, Jiang YH, Lin P, Jia XW, Zou ZH. Ribosomal protein L24 is differentially expressed in ovary and testis of the marine shrimp Marsupenaeus japonicus. Comp. Biochem. Physiol. B. 2007;147:466–474. doi: 10.1016/j.cbpb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Zhang ZP, Wu RSS, Mok HOL, Wang YL, Poon WWL, Cheng SH, Kong RYC. Isolation, characterization and expression analysis of a hypoxia-responsive glucose transporter gene from the grass carp, Ctenopharyngodon idellus. Eur. J. Biochem. 2003;270:3010–3017. doi: 10.1046/j.1432-1033.2003.03678.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.