Abstract
Nucleotide excision repair (NER) is an important prokaryotic and eukaryotic defense mechanism that removes a large variety of structurally distinct lesions in cellular DNA. While the proteins involved are completely different, the mode of action of these two repair systems is similar, involving a cut-and-patch mechanism in which an oligonucleotide sequence containing the lesion is excised. The prokaryotic and eukaryotic NER damage-recognition factors have common structural features of β-hairpin intrusion between the two DNA strands at the site of the lesion. In the present study, we explored the hypothesis that this common β-hairpin intrusion motif is mirrored in parallel NER incision efficiencies in the two systems. We have utilized human HeLa cell extracts and the prokaryotic UvrABC proteins to determine their relative NER incision efficiencies. We report here comparisons of relative NER efficiencies with a set of stereoisomeric DNA lesions derived from metabolites of benzo[a]pyrene and equine estrogens in different sequence contexts, utilizing 21 samples. We found a general qualitative trend towards similar relative NER incision efficiencies for ~ 65% of these substrates; the other cases deviate mostly by ~ 30% or less from a perfect correlation, although several more distant outliers are also evident. This resemblance is consistent with the hypothesis that lesion recognition through β-hairpin insertion, a common feature of the two systems, is facilitated by local thermodynamic destabilization induced by the lesions in both cases. In the case of the UvrABC system, varying the nature of the UvrC endonuclease, while maintaining the same UvrA/B proteins, can markedly affect the relative incision efficiencies. These observations suggest that, in addition to recognition involving the initial modified duplexes, downstream events involving UvrC can also play a role in distinguishing and processing different lesions in prokaryotic NER.
Keywords: NER, prokaryotic, human, benzo[a]pyrene, equilenin
1. Introduction
1.1 Overview
Nucleotide excision repair (NER) is important for recognition and removal of a large variety of structurally and chemically distinct DNA lesions in prokaryotic [1] and eukaryotic [2] organisms. While the proteins involved share little sequence homology, the mode of action of this repair system in all branches of life is similar in many respects. Since a large range of substrates are removed by both NER systems, it is widely accepted that the recognition process involves the sensing of the structural distortions/destabilizations in the DNA duplex caused by bulky lesions, rather than the lesions themselves. While NER can remove a large variety of DNA lesions, the DNA repair capacity, or efficiency, can vary by two orders of magnitude or more, depending on the lesion [2–5]. The chemical structures of the DNA lesions, the base sequence contexts in which the lesions are embedded, as well as their stereochemical properties can affect the DNA repair efficiencies catalyzed by prokaryotic [6–12] and eukaryotic [4, 5, 13–16] repair systems. While the importance of these factors on NER efficiencies is well documented, the exact molecular origins underlying these differences are still not well understood [17].
Recently, crystal structures of several prokaryotic and eukaryotic NER proteins that recognize and bind to DNA lesions have provided important new insights into the structural features that have an impact on lesion-removal by the two NER systems [16, 18–26] (see also review by Fuss and Tainer in this issue of DNA Repair). Furthermore, we have accumulated a substantial library of NER substrates consisting of structurally defined DNA lesions positioned in similar or different oligonucleotide sequence contexts. These substrates provide unique opportunities for investigating the relationships between the chemical structures of the lesions and the impact of base sequence context effects on their recognition and processing by eukaryotic and prokaryotic NER systems.
These two NER systems (e.g., [1, 2, 27, 28]) share common overall features that include: (1) the initial steps involve the recognition of the local distortions/destabilizations caused by the lesions, rather than the lesions themselves, thus allowing for the processing of a large range of structurally unrelated DNA lesions; (2) the damaged strand is incised by endonucleases on both sides of the lesions, thus removing entire oligonucleotide sequences containing the damage rather than just the lesions; (3) recently published crystal structures suggest that one of the recognition steps in prokaryotic [20, 25, 28] and eukaryotic [23, 25, 28] NER systems involves the insertion of a β-hairpin between the two DNA strands in the immediate vicinity of the lesion. These structural findings are particularly intriguing because they suggest that thermodynamic destabilization of the native B-DNA structure can play an important role [29, 30] in the recognition of DNA lesions in both NER systems, a concept that had been previously proposed [4, 31–33].
In recent years, we have accumulated a body of results on the relationships between the structural properties of DNA lesions and incision efficiencies of prokaryotic and eukaryotic NER systems [9, 17, 34, 35]. Here, we examine the similarities and differences in the processing of bulky DNA lesions by prokaryotic UvrABC NER proteins and the human NER apparatus in vitro. In both cases, the formation of excision products is a result of a complex series of steps that include the critical initial recognition step. Our hypothesis is that the ease of insertion of a β-hairpin into the duplex in the vicinity of a lesion plays an important role in the recognition step in both eukaryotic and prokaryotic NER.
In this article, we first consider some of the key features of these NER systems on which this hypothesis is based. We then compare the relative efficiencies of incision of a series of DNA substrates catalyzed by prokaryotic UvrABC proteins or by the eukaryotic NER system in HeLa cell extracts in vitro. Our studies are focused on two different types of bulky stereoisomeric DNA lesions that are under study in our laboratories. The first type is derived from the reactions of a diol epoxide derivative of benzo[a]pyrene (BP) to DNA [14, 36], and the second is derived from the binding of 4-hydroxyequilenin (4-OHEN), a reactive metabolite of the equine estrogens equilin and equilenin. Both equine estrogens are important components of the hormone replacement therapy preparation Premarin© [37]. The catechol 4-OHEN forms a variety of pre-mutagenic DNA lesions that are suspected to contribute to human cancers by genotoxic pathways [38]. Because of the potential importance of these equine estrogen DNA lesions, we have been interested in their structural properties [39, 40], as well as their response to nucleotide excision repair systems [41, 42].
1.2 Functional and structural characteristics of prokaryotic and eukaryotic NER: similarities and differences
1.2.1. Overview of NER lesion-recognition
It is generally accepted that the recognition and subsequent processing of DNA lesions by NER occur via a two-step process. The first is the recognition of the lesion, and the second is a verification step that ensures that a lesion is actually present (see also review by Naegeli and Sugasawa in this issue of DNA Repair). Remarkable insights into the molecular bases of these recognition phenomena have been obtained from the crystal structures of the key prokaryotic UvrA [20, 43, 44], UvrB [25], UvrC [21, 45], and eukaryotic [23, 24] DNA damage-sensing proteins that initiate NER by binding to the sites of the DNA lesions.
In prokaryotic cells, three proteins, UvrA, UvrB, and UvrC are needed to generate the dual incisions on the two sides of the lesion that generate the 12–13-mer excision products. The distortions/destabilizations caused by the lesion are first recognized by a dimer of UvrA that binds to the damaged site in the form of a UvrA2UvrB2 complex [46]. In an ATP-driven mechanism, the UvrA dimer then dissociates and the UvrB – DNA complex is stabilized by the insertion of a β-hairpin between the two strands in the vicinity of the lesion (Figure 1A). In this damage verification step [28], the local destabilization of the duplex at the site of the damage most likely facilitates the insertion of the UvrB β-hairpin (see also Section 1.2.2). If the latter fails to insert between the two strands, as might be the case in the absence of a lesion, the UvrAUvrB dissociates from the DNA [6, 33]. The stable binding of UvrB to damaged DNA stimulates the activity of its ATPase, which is essential for the recruitment of the endonuclease UvrC by this pre-incision complex [28]. The hydrolysis of one phosphodiester bond on the 3'-side and another on the 5'-side of the lesion results in the 12–13-mer dual incision products.
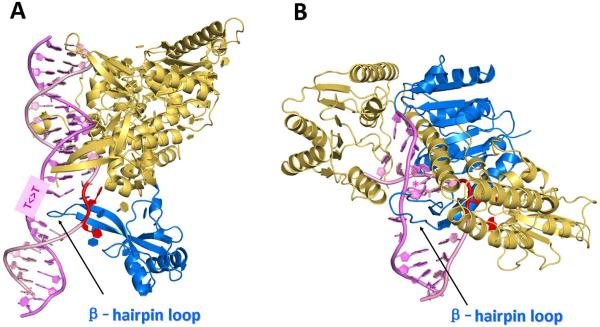
Figure 1.
(A) Crystal structure of Rad4-Rad23, the yeast homologue of human XPC-RAD23B. (PDB ID [74]: 2QSG) [23]; The two flipped-out thymines (red) in the unmodified strand are mismatched with the thymines of the T<>T photodimer, and are within interaction distances of amino acid residues in Rad4. The β-hairpin loop (marine) is inserted between the two strands, thus anchoring Rad4 (or XPC) to the site of the damage. (B) Model of UvrB [35] based on a UvrB-DNA co-crystal structure (PDB ID [74]: 2FDC) [25] with the DNA extended and a (+)-cis-BP-derived adduct (designated by *) modeled into a conserved pocket, concealed behind the β-hairpin. Color code: β-hairpin, marine; flipped-out bases, red. The figures were prepared using the PyMOL Molecular Graphics System [75].
In the case of the mammalian NER pathway, the presence of the helix-distorting/destabilizing lesion is sensed by the XPC-RAD23B heteroprotein complex that opens a ~ 6-base pair sequence, as shown with duplexes containing a cisplatin [47] or a bulky BP diol epoxide-N2-dG adduct [36]. This complex initiates the recruitment of other NER factors to the site of the lesion, starting with the multi-protein TFIIH complex which contains the helicases XPB [18] and XPD [19, 22, 26]; the latter induce the unwinding of a 20 – 25 nucleotide-long region around the lesion [48, 49].
1.2.2 Crystal structures: shared feature of β-hairpin insertion
A crystal structure of a complex of the B. caldotenax (Bca) UvrB bound to double-stranded damaged DNA has been solved [25]. In this structure, one DNA strand, containing a 3' overhang, threads behind a β-hairpin motif of UvrB, indicating that this hairpin is inserted between the strands of the double helix, while the nucleotides directly behind the β-hairpin are flipped out and inserted into a small, highly conserved pocket in UvrB. Molecular modeling and molecular dynamics simulations have been utilized to complete the DNA structure and to include a BP-derived lesion (Figure 1A) [35].
Although a crystal structure of an XPC-RAD23B complex containing damaged DNA complex is not yet available, the X-ray crystallographic structure of a truncated form of the yeast S. cerevisiae Rad4/Rad23 homologue of the mammalian XPC-RAD23B in a complex with an oligonucleotide containing a cyclobutane pyrimidine dimer (CPD) lesion has been determined [23]. There are three β-hairpin domains in the Rad4 protein (BHD1, BHD2, and BHD3). While BHD1 binds non-specifically to an unmodified 11-mer sequence on the 3'-side of the CPD lesion, BHD2 and BHD3 are in contact with the DNA in the vicinity of the lesion, although not in direct contact with the CPD. The BHD3 β-hairpin is inserted into the DNA helix, thus separating the lesion from the unmodified strand. The structure shows that this β-hairpin assumes a role similar to the one in the UvrB-damaged DNA complex [25]: it is inserted between the two strands of the DNA duplex and thus assists in the stabilization of the complex. The CPD lesion is positioned in a disordered region of the crystal without any visible contacts with the protein; however, the two mismatched and flipped out thymine bases in the complementary strand opposite the CPD lesion interact with the protein; one thymine interacts with BHD3 and the second with both BHD2 and BHD3 (Figure 1B) [23]. Recently, fluorescence-based imaging techniques have been employed to deduce that the β-hairpin facilitates the mobility of XPC as it scans for DNA damage [50].
1.2.3 A common structural theme of lesion recognition
It is remarkable that the insertion of a β-hairpin between the two strands of DNA at or near the site of the lesion is common to both prokaryotic and eukaryotic NER systems even though the proteins do not share a common fold. The insertion of β-hairpins between the lesion-containing and undamaged complementary strand should be more facile in the case of lesions that cause significant local destabilization of DNA duplexes, and more difficult in the case of lesions that do not (see also review by Fuss and Tainer in this issue of DNA Repair). These considerations suggest that the thermodynamic properties of the lesions are in some way correlated with NER efficiencies in both systems [29, 30]. In order to test this hypothesis we have analyzed a series of defined substrates by the prokaryotic and eukaryotic NER systems.
2. Materials and methods
2.1 Prokaryotic NER proteins
In all experiments with UvrABC proteins, UvrA and UvrB were from Bacillus caldotenax, while UvrC subunits were either from B. caldotenax (UvrCBca), Thermatogamaritima (UvrCTma), or from E. coli (UvrCCho). In a few experiments, all three UvrABC proteins were from E. coli. The UvrABca and UvrBBca, as well as all of the UvrC proteins, were overexpressed in E. coli cells and purified as described by Jiang et al. [51].
2.2 DNA substrates
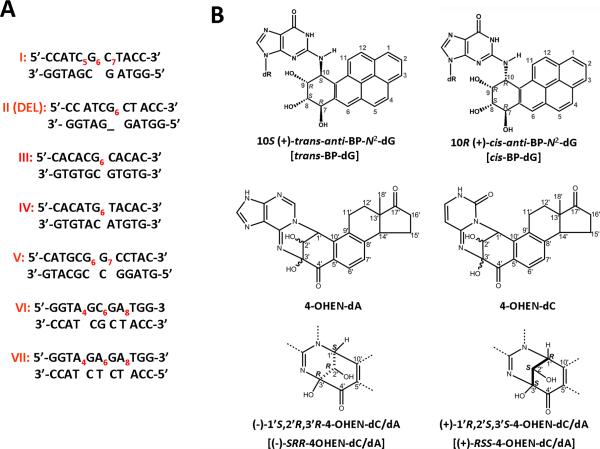
The site-specifically modified single-stranded oligonucleotides (the upper strands shown in Figure 2) were synthesized by reacting (±)-r7,t8-dihydroxy-t9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) with oligonucleotides I, III - V, as described by Pirogov et al. [52] and by Mao et al. [53]. The 4-hydroxyequilenin (4-OHEN)-modified sequences I, VI and VII were synthesized as described [39]. The 135-mer duplexes (See Supporting Information Scheme 1), containing the lesions at or near the center of these duplexes, were synthesized by the annealing and ligation methods described in detail elsewhere [15].
Figure 2.
(A) Sequence contexts and (B) chemical structures of the DNA adducts studied. Subscripted numbers designate the positions of the modified nucleotides (only one lesion per oligonucleotide). In sequences I, VI and VII the labeled adenine and cytosine bases denote 4-OHEN-dA or –dC adducts, respectively. The labeled guanine residues (G6) in sequence I and II (Del) denote either trans- or cis-BP-dG adducts derived from the reactions of (+)-anti-BPDE with the single-stranded sequence I. In sequences III, IV and V, G6 denotes the trans-BP-dG adduct. The abbreviations used for these adducts in the text are shown in square brackets. The full sequences of the oligonucleotide 135-mers containing the indicated duplexes used in HeLa cell NER experiments are defined elsewhere [15], and those of the shorter duplexes used in the UvrABC NER experiments are shown in Supporting Information.
2.3 UvrABC and mammalian cell extract incision assays
Aliquots of the UvrA, UvrB, and UvrC stock solutions were stored at −20 °C. Before use, the enzymes were diluted with x1 UvrABC buffer (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 50 mM KCl, 5 mM dithiothreitol, and 1 mM ATP) to the desired concentrations. The buffer and enzymes were kept at 0 °C during the dilution. The diluted enzymes were then preheated to 55 °C and were kept at that temperature for 5 min in order to inactivate possible E. coli contaminant nucleases. The concentrations employed in the incision assays were 25 nM UvrA, 40 nM UvrB, and 50 nM UvrC. The concentration of oligonucleotide duplex substrates was 2 nM. The incision assays were carried out in 40 μL reaction volumes containing 4 μL of ×10 UvrABC buffer, 6 μL of the diluted UvrA, UvrB, and UvrC solutions and DNA substrates. The reactions were terminated by adding 3 μL of a stop buffer solution (20 mM EDTA, 95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol) to 6 μL aliquots of the reaction mixture taken out at selected time intervals. The solutions were then heated at 90 °C for 3 minutes, then quickly placed on ice, and loaded into a 15% denaturing polyacrylamide gel (19:1). The gel was operated for 2 hours at 45–50 °C in 1 × tris-borate-EDTA (TBE) buffer (89 mM Tris, 89 mM boric acid, 10 mM EDTA, pH 8.4). Upon completion of the gel electrophoresis experiments, the gel was dried and exposed to an imaging screen in a Molecular Dynamics Storm 840 PhosphorImager. The relative radioactivities of the bands were evaluated using Image Quant software.
The eukaryotic NER incision efficiencies were determined in HeLa cell extracts as described earlier [15]. The cell extracts were prepared by standard methods [54], and the fraction of 135-mer duplexes that were incised were determined by measuring the amounts of oligonucleotide incision products 24 – 32 nucleotides in length. The sources of the data presented below are cited within the individual figure captions.
3. Results and discussion
3.1 Overview of experimental studies
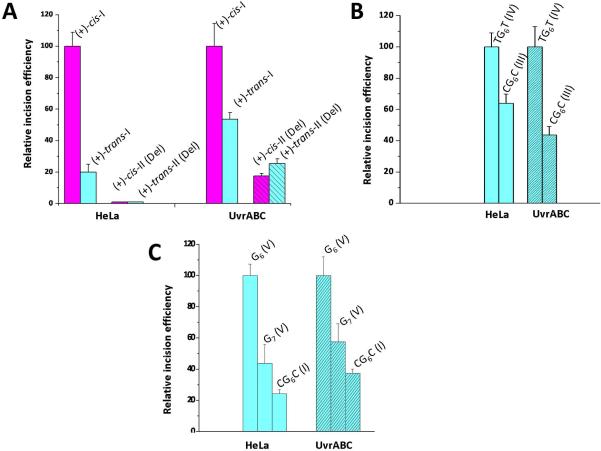
In this study, the incision efficiencies of the same substrates catalyzed by UvrABC proteins and human NER proteins in vitro were evaluated separately. The stereochemical properties of the DNA lesions and the sequence contexts utilized are summarized in Figure 2, while selected NMR solution structures of the adducts investigated are shown in Figure 3. In all of these experiments, the UvrA and UvrB subunits were from B. caldotenax, while the UvrC subunits were either from B. caldotenax, Thermatoga maritima, or E. coli (UvrCBca, UvrCTma, and UvrCCho, respectively). Cho is a UvrC homolog of E. Coli as first reported by Moolenar et al. [55]. As demonstrated by Jiang et al., UvrCBca incises the damaged strands only on the 5'-side of the lesion, UvrCTma incises the damaged strands on both sides [51], and the UvrCCho incisions occur only at the ninth phosphodiester bond on the 3'-side of the lesion, some four nucleotides further away than the normal 3'-incision sites catalyzed by E. coli UvrC. It was suggested that Cho acts as a backup nuclease for NER acting on very bulky substrates that block the 3′ incision site of UvrC [55]. Furthermore since the Tma and Bca proteins are thermostable, they can be used at different temperatures to probe the effects of DNA duplex stability on DNA damage recognition. In order to compare the relative incision rates of different substrates, we assigned the same value of 100 for one of the substrates incised both in HeLa cell extracts and in UvrABC incubation experiments. The incision rates of other substrates are expressed relative to this standard substrate in the series of bar graphs described below.
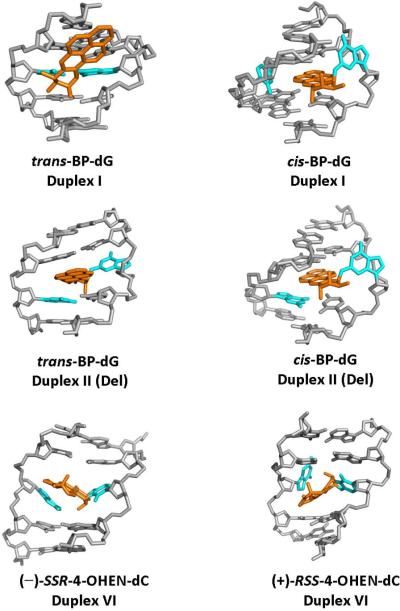
Figure 3.
NMR solution structures of the DNA adducts studied. The trans-BP-dG adduct in the full duplex (Duplex I) has the pyrenyl ring system positioned in the B-DNA minor groove directed 5'-along the modified strand with only minor perturbations of Watson-Crick base pairing [61]. In the cis-BP-dG full duplex I [57] and the deletion duplex [58], the conformation is base-displaced/intercalated, with the pyrenyl ring system inserted into the duplex. In the full duplex I, the modified G and its partner C are displaced into the minor groove and major groove, respectively, and the benzylic ring is directed toward the minor groove. In the (+)-cis-BP-dG duplex II (Del), the structure is very similar except that the partner C to the modified G is missing. However, the conformation of the trans-BP-dG duplex II (Del) differs from that of the full duplex in that it is base-displaced/intercalated, the modified G is displaced into the major groove, and the benzylic ring is directed toward the major groove [76]. In the (−)-SRR-4-OHEN-dC and the (+)-RSS-4-OHEN-dC adducts (Duplexes I and VI), the modified dC is in the anti-glycosidic bond conformation, the equilenin rings are in the minor groove with the distal rings protruding through the helix to the major groove; the rings are directed 5' along the modified strand in the case of (−)-SRR-4-OHEN-dC and 3'-oriented in the case of the (+)-RSS-4-OHEN-dC adduct [70].
3.2 Effects of BP-dG adduct stereochemistry in full duplexes I and deletion duplexes II (Del)
3.2.1. Repair efficiencies in HeLa cell extracts
The efficiency of repair of the BP-dG adducts by the human NER system in duplexes I and II (Del) was first reported by Hess et al. [56]. The (+)-cis and (+)-trans adducts differ from one another only by the absolute configuration of substituents about the C10 carbon atom (Figure 2), but the NER efficiency of the (+)-cis adduct is ~ 5 times greater than that of the (+)-trans adduct [36]. This difference has been attributed to the greater disruption of base pairing in the base-displaced/intercalated (+)-cis adduct, and the smaller extent of structural disruption caused by the minor groove (+)-trans adduct (Figure 3) [36]. Removing the cytidine opposite these stereoisomeric lesions almost completely abolishes the repair of both these BP-dG adducts in II (Del) duplexes in HeLa cell extracts ([56] and present study). Here, we also examined whether similar effects occur in the case of prokaryotic NER proteins in vitro (Figure 4A).
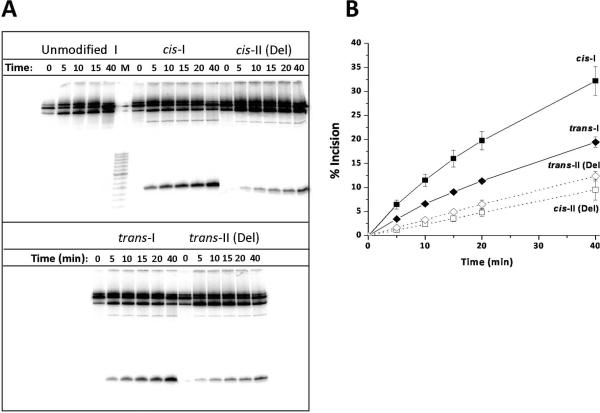
Figure 4.
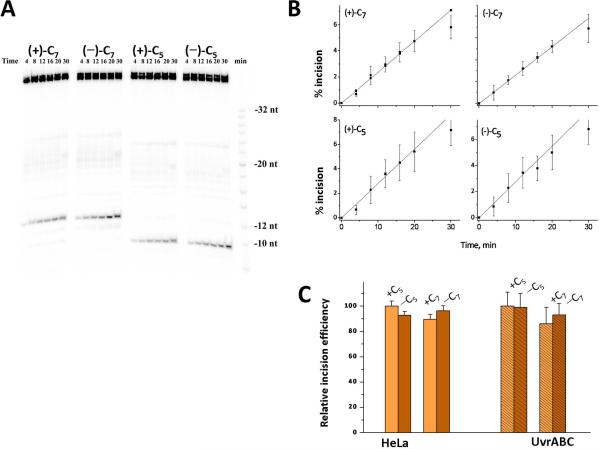
(A) Autoradiogram of typical incisions catalyzed by UvrABcaBBcaCTma at 55 °C using the cis- and trans-BP-dG lesions embedded in the sequence contexts of duplexes I or II (Del) positioned in the middle of otherwise identical 135-mer DNA duplexes; these duplexes were 32P-endlabeled at the 6th phosphodiester bond on the 5'-side of the adduct. Lane M shows the migration distances of oligonucleotide markers 8, 10, 12, 14……28, 32 nucleotides in length (the dark band near the middle represents a 20-mer). The dual incision products migration distance is intermediate between those of the 12 and 14-nucleotide markers. (B) Kinetics of accumulation of incision products as a function of incubation time. Time dependence of formation of dual incision products. The average values and standard deviations are based on seven independent experiments of this type.
3.2.2. Repair efficiencies in the UvrABC system relative to human HeLa cell extracts
The kinetics of dual incisions of the same (+)-cis- and (+)-trans-BP-dG lesions in identical 135-mer DNA sequences (See Supporting Information Scheme 1) catalyzed by prokaryotic UvrABcaBBcaCTma proteins at 55 °C are depicted in Figure 4. A typical gel is shown in Figure 4A, and the densitometric analyses of these gel autoradiograms yields the kinetic incision curves shown in Figure 4B. The initial rates, deduced from the initial linear portions of these kinetic curves, are compared in Figure 5A. In this bar graph, the initial incision rate of the cis-BP-dG adduct in duplex I was assigned a value of 100. The relative incision rate of the stereoisomeric trans adduct is 20 ± 5% on this scale in HeLa cell extracts [36], and 54 ± 4% in the case of UvrABcaBBcaCTma. Thus, the ratios of initial incision rates are larger in the case of the cis-BP-dG than the trans-BP-dG adduct in duplex I in both cases, but the cis-BP-dG/trans-BP-dG ratios are different: ~ 5 in HeLa cell extracts and ~ 1.8 in the case of UvrABC. We note that Zou et al. reported a cis-BP-dG/trans-BP-dG ratio of ~ 2 for the same lesions embedded in 50-mer duplexes with NER assays carried out at 37 °C catalyzed by UvrABC proteins entirely from E. coli [10]. Jiang et al. [51] also studied the repair of the same lesions in 50-mer duplexes, but catalyzed by UvrABca, UvrBBca, and UvrCTma, and reported ratios of 1.4 – 1.6. All of these values are close to the value for the cis-BP-dG/trans-BP-dG ratio reported here (Figure 5A).
Figure 5.
Comparisons of relative initial incision rates measured in human HeLa cell extracts and catalyzed by UvrABC proteins. In all cases, one of the substrates was assigned a value of 100, and the initial incision rates of the other substrates are expressed relative to the value of 100. The sources of the data are cited below in each case. (A) Effects of adduct stereochemistry and conformation: cis- and trans-BP-dG in the full duplexes I and deletion duplexes II (Del); NER data obtained using HeLa cell extracts and UvrABcaBBcaCTma at 55 °C. The HeLa data is from [36, 56], and the prokaryotic data is from Figure 4. (B) Effects of base sequence context, …TG6T… vs. …CG6C… with G6 denoting the trans-BP-dG adducts in duplexes III and IV (UvrABcaBBcaCBca at 37 °C, 43-mer 5'-endlabeled duplexes). The HeLa data is from [59] and the UvrABC data is from [9]. (C) Effects of base sequence context (G6 or G7) in …CG6G…, …GG7C…, or …CG6C… in sequence contexts V, V, and I, respectively, containing trans-BP-dG adducts. The HeLa data are from [15]. The UvrABC data for UvrABcaBBcaCBca at 55 °C are from Figure S1, Supporting Information (average and standard deviations from four independent experiments using 54-mer duplexes; the sequence is shown in Scheme 2).
In the II (Del) duplexes, the human NER activity is completely abolished, while with UvrABcaBBcaCTma, it is significantly reduced relative to the respective values observed for the cis-and trans-BP-dG adducts in full duplexes I. Overall, in these examples, the relative activities of the human and prokaryotic NER systems in the full (I) and deletion duplexes II (Del) resemble one another qualitatively, but without being directly proportional.
3.2.3. Impact of adduct conformation
The (+)-cis-BP-dG adducts with the base-displaced intercalative conformation are better substrates than the stereoisomeric minor groove (+)-trans-adducts in both NER systems [10, 36, 56], although the difference is larger in HeLa cell extracts. For the II (Del) duplexes, the incision efficiencies relative to the same lesions in the full duplex I are reduced by a factor of ≥ 100 in the (+)-cis-adduct, and by a factor of ~ 10 in the (+)-trans-BP-dG adduct [56]. In contrast, in UvrABcaBBcaCTma experiments, significant levels of incisions of the cis- and trans-BP-dG adducts in II (Del) duplexes are observed although at lower levels than in the full cis- and trans-BP-dG (I) duplexes (Figure 5A). The UvrABcaBBcaCTma experiments were conducted at 55 °C, an optimal temperature for the thermophilic proteins [51]. This incubation temperature is below the melting points of the modified duplexes; spectroscopic experiments indicate that the overall adduct conformations are preserved in oligonucleotide duplexes ~ 43 nucleotide base pairs in length below their melting points of 75 – 80 °C [9].
3.2.4. Comparison of incision efficiencies with thermal stabilities of different DNA duplex substrates
Insights into the repair-resistance of the deletion duplexes have been obtained from thermal melting points (Tm), the temperature that corresponds to the dissociation of 50 % of the DNA duplexes into separate strands (Table 1), and NMR and computational molecular dynamics studies. The Tm of the (+)-trans-BP-dG II (Del) duplex (Figure 2) is + 6 °C higher than the Tm of the modified full duplex I, even though the II (Del) duplex is missing the nucleotide C opposite the lesion. However, the analogous increase is strikingly larger, + 19 °C, in the case of the (+)-cis-BP-dG II (Del) relative to the unmodified II (Del) duplex. Structurally, the (+)-cis-BP-dG-modified full duplex I and the deletion duplex II (Del) adopt base-displaced/intercalated conformations [57, 58], although with opposite orientations of the pyrenyl ring system as it protrudes through the helix (Figure 3). In the II (Del) duplex the stacking interactions between the two guanines surrounding the deletion site and the BP pyrenyl ring system are more favorable than in the full duplex I. According to recent molecular dynamics analyses [16], these enhanced stacking interactions result from the collapse of the two guanines upon each other, due to the absence of the nucleotide between them (Figures 2A and 3) [58]; consequently, the separation of the two strands at the site of modification should be energetically more costly and thus affect the overall incision efficiency if recognition is a key step, as we assume here. Furthermore, two bases opposite the lesion in the complementary strand probably assist in stabilizing the modified DNA –XPC complex [23] and the absence of one of these nucleotides may diminish the stability of this complex and contribute to the observed lack of incisions observed in the cis-BP-dG II (Del) duplex. The absence of the partner base opposite the modified nucleotide also may diminish the binding of the XPC-RAD23B recognition factor to the lesion site. In the case of the (+)-trans-BP-dG adduct in duplex II (Del), there is a major structural rearrangement compared to the full duplex, since the minor groove-alignment of the BP moiety (with Watson-Crick base pairing being intact in the full duplex) changes to a base-displaced/intercalated conformation in the II (Del) duplex (Figure 3). Therefore, similar structural effects may account for the lower incision efficiencies observed in the case of the trans-BP-dG II (Del) duplex as for the cis II (Del) case (Figures 4 and 5).
Table 1.
Summary of thermal melting data for adducts and sequences investigated.
| Sequence a | Adduct | Tm (Tm Unmod) (°C) | ΔTm (°C)h |
|---|---|---|---|
| I | (+)-trans-BP-dG6 | 41 (51)b | −10 |
| I | (+)-cis-BP-dG6 | 40 (51)c | −11 |
| II (Del) | (+)-trans-BP-dG6 | 30 (24)b | +6 |
| II (Del) | (+)-cis-BP-dG6 | 46 (27)c | +19 |
| III | (+)-trans-BP-dG6 | 46 (55) d | −9 |
| IV | (+)-trans-BP-dG6 | 30 (41) d | −11 |
| V | (+)-trans-BP-dG6 | 55 e (60) | −5 |
| V | (+)-trans-BP-dG7 | 57 e (60) | −3 |
| I | (−)-SRR-4-OHEN-dC5 | 27 (51)f | −24 |
| I | (+)-RSS-4-OHEN-dC7 | 27 (51)f | −24 |
| I | (−)-SRR-4-OHEN-dC7 | 26 (51)f | −25 |
| VI | (−)-SRR-4-OHEN-dC4 | 28 (53)g | −25 |
| VI | (+)-RSS-4-OHEN-dC6 | 23 (53)g | −30 |
| VI | (−)-SRR-4-OHEN-dA4 | 43 (53)g | −10 |
| VI | (+)-RSS-4-OHEN-dA8 | 24 (53)g | −29 |
| VII | (+)-RSS-4-OHEN-dA4 | 20 (45)g | −25 |
| VII | (−)-SRR-4-OHEN-dA4 | 30 (45)g | −15 |
| VII | (−)-RSS-4-OHEN-dA6 | 20 (45)g | −25 |
| VII | (+)-RSS-4-OHEN-dA8 | 17 (45)g | −28 |
| VII | (−)-SRR-4-OHEN-dA8 | 19 (45)g | −26 |
3.3. Effects of flanking bases
3.3.1 (+)-trans-BP-dG lesions in full duplexes III (CG6C) and IV (TG6T)
The relative NER efficiencies in HeLa cell extracts of identical trans-BP-dG lesions flanked either by C or by T on both sides, with all other bases in the 135-mer duplexes remaining unchanged, were reported earlier [34]. The initial rates of incisions in HeLa cell extracts were found to be 1.5 ± 0.2 greater in the sequence context of duplex IV than in duplex III. In NER experiments using prokaryotic UvrABcaBBcaCTma proteins, the ratio was reported to be 2.3 ± 0.3 at 37 °C and 1.7 ± 0.4 at 55 °C. This loss of discrimination at the higher temperature is believed be due to the increased dynamic conformational flexibility at the lesion site at 55 °C; this has a greater impact in the more rigid CG6C sequence context of duplex III than on the more flexible TG6T sequence context of duplex IV [59]. In the case of E. coli UvrAE.coliUvrBE.coliUvrCE.coli, however, the ratio at 37 °C was smaller (1.7 ± 0.1) in E. coli UvrABC incision experiments [9]. The relative incision rates catalyzed by human and prokaryotic repair factors are compared in Figure 5B. The TG6T (IV) duplex rates were assigned a value of 100. The relative incision rates of the CG6C-IV duplexes are 65 ± 9% and 44 ± 6 % in HeLa cell extracts and UvrABcaBBcaCBca NER experiments, respectively. Again, there is a similar pattern in the human and prokaryotic relative incision rates even though the relative values for the CG6C (III) duplexes catalyzed by UvrCBca are lower than in human cell extracts.
3.3.2 (+)-trans-BP-dG lesions in full duplexes V (CG6G vs. GG7C) and I (CG6C)
The relative rates of incisions catalyzed by human or UvrABca BBcaCBca NER proteins (55 °C) of the trans-BP-dG adducts in the sequence contexts CG6G (V), GG7C (V), and CG6C (I) in duplexes V and I, respectively, are compared in Fig. 5C. In human cell extracts, the ratio of initial repair rates for trans-BP-dG6 (V) / trans-BP-dG7 (V) is 2.4 ± 0.3 [15], and for the trans-BP-dG6 (V) / trans-BP-dG6 (I) case, it is 4.1 ± 0.3. In Figure 5C, the value of 100 was assigned to the CG6G (V) substrate. The relative incision rates for the three duplexes, CG6G (V) : GG7C (V) : CG6C (I) are 100 : 42 : 24 in HeLa cell extracts and 100 : 58 : 37 in the case of UvrABcaBBcaCBca (55 °C). Again, the trends are similar in both systems, but the incision efficiencies appear to be somewhat more sensitive to sequence context in the case of the human than the prokaryotic repair systems. The differences in repair efficiencies in these three duplexes can be rationalized in terms of their structural features. An in depth NMR analysis of the sequence-dependence of the imino proton linewidths of the 11-mer CG6G (V) duplex as a function of temperature, demonstrates that the Watson-Crick hydrogen bonding of the 5'-base pair adjacent to G6 in the 11-mer CG6G (V) duplex is strongly destabilized; however, in the GG7C (V) duplex, Watson-Crick hydrogen bonding is much less disturbed even though a flexible kink at the site of the lesion of this duplex has been identified [15, 60]. The greater NER efficiency of the CG6G (V) duplex relative to the GG7C (V) duplex has therefore been attributed to its greater local thermodynamic destabilization, stemming from the dynamic, episodic rupturing of the 5'-side base pair adjacent to the lesion at G6 [13, 15]. The CG6C (I) duplex appears to be the least structurally distorted [61]: all Watson-Crick hydrogen bonds are intact in this minor groove adduct (Figure 3) and the bend associated with the lesion is rigid rather than flexible [34].
3.4. Incision efficiencies associated with trans-BP-dG lesions are influenced by UvrC downstream effects
3.4.1. Lesion-recognition versus verification
The binding of a variety of proteins to double-stranded DNA often leads to significant local distortions of the B-DNA double helix (e.g., [62]). These distortions are frequently determined by intrinsic sequence-governed structural parameters of the DNA duplexes [63–65]. The UvrB protein-DNA interaction falls into this pattern since complex formation involves the separation of base pairs near the site of the lesion which is sequence-dependent [66]. Once the β-hairpin has been inserted and the protein-DNA complex has been stabilized as part of the recognition step[28], the DNA is no longer double stranded and the thermodynamic properties of the duplex should be no longer relevant to the further processing of the lesion. Our assumption is therefore that any correlation between the structural features of lesions in double-stranded DNA and NER efficiencies most likely occur at the recognition step, involving XPC-RAD23B in human and UvrA and UvrB in prokaryotic NER. The predominance of the recognition step in determining incision efficiencies is illustrated by the example of the cis-BP-dG adduct in the base-displaced intercalated full and deletion duplexes I and II(Del), respectively. In the full duplex I, the NER efficiency is robust when catalyzed by either the human [56] or by UvrABC [10] NER systems. While the overall adduct conformation is similar in the II (Del) duplex, the incision efficiency in human cell extracts is abolished [56] and considerably diminished in the UvrABC system (Figure 4). Since the lesion is identical in both the full duplex I and the II (Del) duplex, it is unlikely that the post-recognition verification step, perhaps involving helicase activity, could be very different. Instead, the striking difference in NER efficiencies points to the initial recognition step. As discussed, the enhanced BP-DNA Van der Waals base stacking interactions in the cis-BP-dG II (Del) may prevent the β-hairpin from invading the helix at the site of the lesion [16].
3.4.2. The effects of different UvrC proteins on incision efficiencies catalyzed by UvrABcaUvrBBcaUvrC protein system
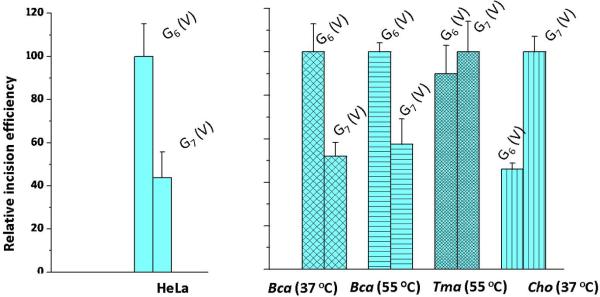
While initial damage recognition is critically important, here we find that the nature of the prokaryotic endonuclease UvrC can also influence the overall efficiencies of incisions. In Figure 6, we compare the effects of temperature and different UvrC proteins (UvrCBca, UvrCTma and UvrCCho) on the incision ratios of the trans-BP-dG adduct in the CG6G (V) and GG7C (V) duplexes. The HeLa cell extract experiments are again shown for reference and, as in Figure 6, the incision rates observed for the CG6G (V) duplex were assigned a value of 100 with the HeLa extract and the UvrCBca experiments. However, the value of 100 was assigned to the GG7C (V) duplex in the UvrCTma and UvrCCho experiments because this duplex is incised with greater efficiency than the CG6G (V) duplex. The relative incision ratios of the CG6G (V) and GG7C (V) duplexes catalyzed by UvrABcaBBcaCBca at 37 °C and 55 °C are similar, within experimental error (Figure 6). However, for UvrABcaBBcaCTma at 37 °C, the trans-BP-dG6 / trans-BP-dG7 ratio is close to unity (experiments at 55 °C were not conducted in this case). Interestingly, when the same substrates are incubated with UvrABcaBBcaCCho at 37 °C, the trans-BP-dG6-V / trans-BP-dG7-V ratio is reversed and is 0.46 ± 0.05. This set of experiments clearly indicates that the bacterial species and therefore the nature of UvrC contributes to the recognition and processing of lesions that are identical, but positioned in different sequence contexts. Thus, the combination of UvrA and UvrB proteins binding to the sites of the lesions, does not alone determine the overall incision efficiencies. The stable binding of UvrB to damaged DNA stimulates the activity of its ATPase, which is essential for the recruitment of the endonuclease UvrC by this pre-incision complex [28]; we speculate that the positioning of UvrC on the UvrB-DNA complex depends on the chemical and/or stereochemical features of the lesion itself and its base sequence context; these factors may govern the dynamic aspects of the UvrB-DNA complex interactions with UvrC. Thus, a downstream effect involving UvrC, when the damaged DNA sequence is in the strand-separated form in the UvrB complex, appears also to be important in processing the lesions and governing incision efficiencies.
Figure 6.
Impact of different UvrC endonucleases on relative initial incision rates catalyzed by UvrABcaUvrBBcaUvrCBca, UvrABcaUvrBBcaUvrCTma, or UvrABcaUvrBBcaUvrCCho proteins of trans-BP-dG adducts in duplex V embedded in 5'-endlabeled 54-mer duplexes. The original data for UvrABcaBBcaCBca and UvrABcaUvrBBcaUvrCCho experiments are presented in Figures S2 and S3, respectively (Supporting Information), while the UvrABcaUvrBBcaUvrCTma data is from [77]. The HeLa cell extract results shown are from Figure 5C.
3.5 NER efficiencies of 4-hydroxyequilenin - dC and dA adducts in duplexes I, VI and VII
3.5.1. Adduct origins and importance
The reactions of 4-OHEN with nucleotides or DNA generates two minor and two major stereoisomeric dC, dA and minor proportions of dG adducts in vitro [38, 39]. Such adducts have been found also in rodent [67], and human tissues [68]. Recently, the existence of 4-OHEN-dC/dA adducts in human cells exposed to 4-OHEN [69] has been reported.
3.5.2 Structural and thermodynamic properties
We investigated the set of 4-OHEN-dC and -dA adducts (Figure 2) because they thermodynamically destabilize double-stranded DNA much more extensively than the cis- and trans-BP-dG lesions (Table 1). A common feature of 4-OHEN-dC and -dA adducts is their obstructed Watson-Crick hydrogen bond edge, so base pairing at the lesion site is impossible. The duplexes with 4-OHEN-derived adducts therefore exhibit significantly lower Tm values than the BP-dG adducts (Table 1). Another feature of these equine estrogen-derived adducts is that (−)-SRR and (+)-RSS stereoisomers are oppositely directed along the modified strand [40]. For the dC adducts in duplex VI (Figure 2A), the NMR solution structures [70] (Figure 3) show that the equilenin rings protrude through the helix from the minor to the major groove [40]. Here we synthesized the two major 4-OHEN-dC/dA adducts at the indicated positions in sequences I, VI and VII with either (−)-SRR or (+)-RSS absolute configurations (Figure 2). The (+) and (−)-signs indicate the signs of the CD signals measured at 262 nm [40].
3.5.3. 4-OHEN-dC adducts in duplex I
Typical gel autoradiograms showing the incisions in duplexes I containing stereoisomeric 4-OHEN-dC lesions (5'-end labeled) at either positions 5 or 7 catalyzed by E. coli UvrABC proteins at 37 °C are shown in Figure 7A. The course of the incision reactions are depicted in Figure 7B. The initial rates of incision are determined from the initial linear portions of these plots, and are presented in terms of the bar graphs shown in Figure 7C. These results, after normalization with respect to the NER efficiencies of the (+)-RSS-dC5 sample (assigned a value of 100) in each type of NER experiment, show that the +/-dC5 and +/-dC7 adducts are incised with the same relative efficiencies in HeLa cell extracts and by the E. coli UvrABC NER apparatus. Identical results were obtained when the same samples were incubated with UvrABcaBBcaCTma at 55 °C [41]. The incision efficiencies are thus the same in TC6G (I) and GC6T (I) sequence contexts, and also do not depend on the (−)-SRR or (+)-RSS stereochemistry. Similar results were also obtained when the same samples were incised by the UvrABcaBBcaCTma proteins at either 37 °C or at 55 °C, even though the rate of incisions increased by a factor > 10 as the temperature was raised from the lower to the higher temperature [41].
Figure 7.
Incisions of stereoisomeric 4-OHEN-dC adducts at either position C5 or C6 in sequence I embedded in 5'-end-labeled 43-mer duplexes by E.coli UvrABC proteins at 37 °C (the full sequence is depicted in Scheme 3, Supporting Information). (A) Typical gel autoradiogram. (B) Incision kinetics. The average values are based on three independent experiments (data from Chen [41]). The minus and plus signs correspond, respectively, to the (−)-SRR- or the (+)-RSS-4-OHEN-dC adducts (Figure 2). (C).Comparison of relative initial rates of incisions of stereoisomeric 4-OHEN-dC6 and 4-OHEN-dC7 adducts in the duplex I sequence context embedded in either (1) the middle of 135-mer duplexes incubated in HeLa cell extracts and 32P-internally labeled at the 6th phosphodiester bond on the 5'-side of the adduct, and (2) in the middle of 5'-endlabeled 43-mer duplexes (See Supporting Information Scheme 2) catalyzed by E. coli UvrABC (these data were obtained from the initial linear portions of the plots in Figure 7B).
3.5.4 4-OHEN-dC and –dA adducts in duplexes VI and VII
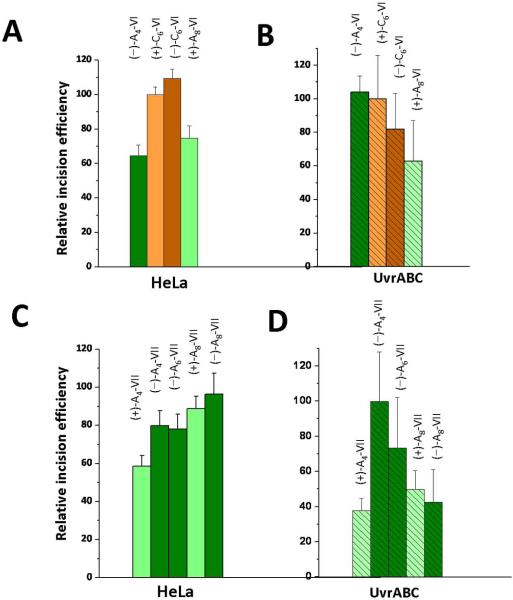
Nine sequences with different adducts derived from the reactions of 4-OHEN with either sequence VI or VII and (−)-SRR or (+)-RSS absolute configurations were synthesized as described [39, 42]. The adducts were positioned either at positions A4, C6, or A8 in duplex VI, or at A4, A6, or A8 of duplex VII. The sequence of these duplexes is identical except for the C or A at the central positions (Figure 2). In designing these experiments, we hypothesized that the duplex VII (top strand) contains a sequence of five consecutive purines which might render this duplex less flexible than duplex VI which has runs of only two purines, and that this difference might affect the NER efficiencies. The initial rates of incisions were measured using UvrABcaBBcaCTma at 55 °C, and typical autoradiograms are shown in Supporting Information. These results are summarized in the bar graphs shown in Figure 8. In this Figure, the (+)-C6-VI adduct served as the reference sample (assigned a value of 100) in each of the two sets of studies using human cell extracts and UvrABC proteins.
Figure 8.
Comparisons of relative NER incision efficiencies of different stereoisomeric 4-OHEN-dC or –dA adducts in sequence contexts VI or VII embedded in the middle of internally labeled 135-mer DNA duplexes (See Supporting Information Scheme 1), in human HeLa cell extracts and prokaryotic UvrABcaBBcaCTma proteins at 55 °C. The value of 100 was assigned to the (+)-4-OHEN-dC6 adducts in sequence context VI (abbreviated as the (+)-dC6-VI adduct in the Figure), and all other incision efficiencies are expressed relative to this value. The HeLa and UvrABC data (typical gels are shown in Figure S4, Supporting Information), are excerpted from Reeves [42]. The minus and plus signs denote the stereochemical properties of the adducts (Figure 2), and the subscripts denote the positions of the modified nucleotides in sequences VI or VII.
In duplex VI, the relative initial rates obtained with all four adducts are similar in value, within experimental error, except for the (−)-dA4-VI sample; the latter exhibits a relative incision rate of ~ 65 % in the human cell extract and ~ 104% in the UvrABC experiments relative to the standard (the (+)-dC6-VI adduct, Figures 8A and 8B). In duplex VII, relative to the same standard, the initial UvrABcaBBcaCTma-catalyzed incision rates are significantly smaller (by up to a factor of ~2) than in human cell extracts in the (+)-dA8-VII and (−)-dA8-VII samples, about ~ 30% smaller in the (+)-dA4-VII sample, and close to one another for the (−)-dA4-VII and (−)-dA6-VII adducts (Figures 8C and 8D). Thus, there are about three 4-OHEN-derived samples out of nine that deviate more markedly in the two repair systems.
In duplex VI and VII in HeLa cell extracts, all of the nine substrates exhibit relative incision values that lie in the range of ~ 60 – 110% (Figures 8A and 8B). In the UvrABC incision experiments, six adducts exhibit incision rates relative to the same (+)-dC6-VI adduct standard in the ~ 60 – 100% range, and the other three in the ~ 40 – 50% range (Figures 8C and 8D). We suggest that the effects of sequence context and stereochemistry may play a lesser role when the lesions themselves are extremely destabilizing as in the 4-OHEN-derived adducts (Table 1).
3.6 Overall comparisons of prokaryotic and eukaryotic NER incision efficiencies
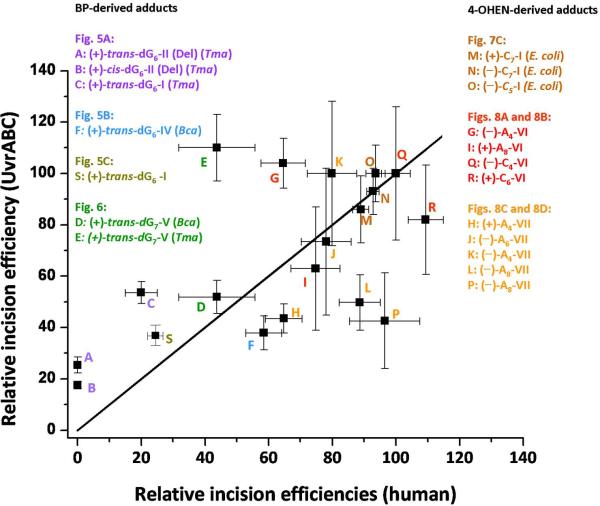
In order to obtain an overview of eukaryotic and prokaryotic NER incision efficiencies of the 21 different substrates collected here, we constructed the plot shown in Figure 9, based on data reported in Figures 5, 6, 7, and 8 (the single UvrCCho result was omitted). In this plot, the relative incision efficiencies observed in HeLa cell extracts are plotted on the horizontal axis, and the relative efficiencies observed in the various UvrABC incision experiments are given on the vertical axis for the same adduct. Of course, the incision efficiency at a particular data point is relative to the same adduct that was assigned a value of 100 in the eukaryotic and prokaryotic NER experiments, as described in each of the Figures. Any data point lying on the straight line shown in Figure 9 would indicate a perfect correlation.
Figure 9.
Comparisons of all relative incision efficiencies in human HeLa cell extracts and UvrABC experiments shown in the individual bar graphs in Figures 5, 6, 7, and 8. The origins of each of the points are also shown. Incision efficiencies for each figure are expressed relative to a designated adduct assigned a value of 100. The straight line corresponds to a hypothetical ideal correlation; any point on this line would signify that the relative incision efficiencies are identical in both NER systems. A correlation coefficient of 0.595 was calculated for all the data points.
The relative incision efficiencies in HeLa cell extracts and in UvrABC experiments obtained with twelve 4-OHEN-derived lesions in duplexes I, VI, and VII, taking into account the error bars, show that only three of the twelve data points (L, P, and G in Figure 9) deviate significantly from the ideal correlation line. In duplex VII, the pattern of incision rates is, overall, much less well correlated with the relative incision rates observed in HeLa cell extracts than in duplex VI.
The BP-dG adducts show reasonable qualitative correlations for all adducts except for point E; this point represents the relative incision efficiency catalyzed by UvrCTma, and is an outlier, showing no difference in relative incision rates of …CG6G… (V) and …GG7C… (V) duplexes (Figure 9). The trans-BP-dG/cis-BP-dG ratios and the relative incision ratios observed with the trans-BP-dG adduct in different sequence contexts do resemble one another in the eukaryotic and prokaryotic NER incision experiments, although there are in some cases better quantitative correlations than in others. For example, with human Hela cell extracts, in contrast to comparatively robust incision efficiencies in full duplexes I [36, 56], the NER activity is practically abolished in the deletion II (Del) duplexes; significantly reduced incision activities with the deletion duplexes are also observed in prokaryotic protein incision experiments. However, the reduction in the II (Del) duplex is not complete, and the resemblance is therefore qualitative (points A and B in Figure 9). In the prokaryotic experiments, the incision rates can also depend remarkably on the nature of the UvrC endonuclease (Figure 6). In general, the prokaryotic repair system seems to be less sensitive than the human HeLa NER system to changes in adduct stereochemistry, adduct conformation, and base sequence context effects. In considering all of the adducts studied, we compute a correlation coefficient of 0.595 for this full data set shown in Figure 9.
A lack of correlation was observed between incision efficiencies catalyzed by human and E.coli NER systems in the case of acetylaminofluorene-dG adducts at different positions in the NarI sequence context [8]. The observations of several outliers in our incision experiments are not surprising. Incision efficiencies are dependent not only on the structural features of the lesions and the base sequence context, but also on downstream damage processing events after recognition, such as the nature of the bacterial species from which UvrC was purified. The structural and functional downstream damage processing steps leading up to efficient incision in eukaryotes are not understood.
4. Conclusions
On the basis of the 21 substrates investigated here (Figure 9), an overall qualitative correlation between NER incision efficiencies in human HeLa cell extracts and UvrABC NER experiments is observable. We find a qualitatively similar pattern in the relative incision efficiencies in human HeLa cell extracts and UvrABC – catalyzed incision experiments in vitro, within ± 30% for two thirds of the different substrates studied. For the BP-dG adducts, the relative incision efficiencies exhibit similar trends for stereoisomeric cis-and trans-BP-dG adducts. The resemblance of NER efficiencies in human and prokaryotic repair systems is consistent with the hypothesis that the recognition step of bulky DNA lesions may involve a common β-hairpin insertion in the eukaryotic XPC-RAD23B and in the prokaryotic UvrB systems. This step should depend on the ease of separation of the two strands at or near the lesion site, determined by local thermodynamic stabilization/destabilization. The thermodynamic properties of the double-stranded region containing the lesion depend on the chemical nature of the lesion, including stereochemistry, topology and physical size, and its sequence context; the impact of these variable factors on the local DNA structure provide the thermodynamic signal. In this concept, the ease of insertion of the β-hairpin between the two strands is facilitated by lesions that destabilize the local region of the duplex, and is hindered by lesions that stabilize the local structure of the duplex by van der Waals stacking and other types of interactions. Hairpin insertion could be facilitated by bending and concomitant unwinding [20, 32], either dynamically due to the lesion or through protein-DNA interactions. As shown in a recent crystal structure [20], UvrA bends and unwinds the DNA, which would facilitate strand opening and recognition by UvrB. The stabilization of the opened DNA helix by the repair protein also needs to be considered. Furthermore, UvrB is believed to interact with the damaged substrate through base stacking with a tyrosine residue at the base of the beta-hairpin and through a base flipping event [25], and the yeast homologue of human XPC-RAD23B [23] also utilizes base-flipping interactions with the protein. Base flipping free energy profiles and repair susceptibility of damaged DNA are currently under investigation [71].
Finally, it is important to realize that, while it is apparent that certain protein motifs such as a β-hairpin are common recognition elements in NER proteins from prokaryotes and eukaryotes, the two systems show remarkable differences. Considering the three orders of magnitude difference between the genomes, the huge degree of genome compaction of the eukaryotic genome into chromatin and the different structural organization of chromatin (transcriptionally active or inactive) within the nucleus of eukaryotes, it is not surprising that eukaryotes have evolved a more complex NER processing system [72] which involves more than 30 proteins as compared to just six in bacterial species. It is likely that eukaryotic chromatin organization of DNA, which is wound around histones in nucleosomes, may provide additional helical stress in repair-susceptible, destabilizing lesions to promote damage recognition, while stabilizing repair-resistant lesions can diminish nucleosome dynamics [78]. Future work on the structure, function, and dynamics of NER proteins, as well as, the role of chromatin reorganization during repair will give clues to both the similarities and differences between these two systems. For example, a co-crystal structure of UvrB bound to a damage site might show structural similarities of damage verification provided by XPD as part of TFIIH-DNA co-crystal [17]. Current work in our laboratories is focused on these exciting and fundamental topics of NER.
Supplementary Material
Acknowledgements
This work was supported by NIH grants CA112412 and CA099194 to N.E.G., CA75449 and CA28038 to S.B., and ES019566 to B.V.H. We also gratefully acknowledge TeraGrid resources provided by the Texas Advanced Computing Center supported by the National Science Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from the Research Facilities Improvement Program (C06RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the Bruker Ultraflex Maldi mass spectrometer at this Facility was supported by the National Science Foundation (CHE-0958457).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: NER, nucleotide excision repair; BP, benzo[a]pyrene; 4-OHEN, 4-hydroxyequilenin; dG, 2'-deoxyguanosine; dA, 2'-deoxyadenosine; CPD, cyclobutane pyrimidine dimer; (+)-anti-BPDE, (+)-7R,8S-dihydroxy-9S,10R-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene; cis-BP-dG, (+)- (7R,8S,9R,10R)-N2-[10- (7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-2'-deoxyguanosine; trans-BP-dG, (+)- (7R,8S,9R,10S)-N2-[10-(7,8,9,10-tetrahydrobenzo[a]pyrenyl)]-2'-deoxyguanosine.
Conflict of interest None.
References
- 1.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–52. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 2.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–76. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 3.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 4.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–98. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 5.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–56. [PubMed] [Google Scholar]
- 6.Snowden A, Kow YW, Van Houten B. Damage repertoire of the Escherichia coli UvrABC nuclease complex includes abasic sites, base-damage analogues, and lesions containing adjacent 5' or 3' nicks. Biochemistry. 1990;29:7251–9. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
- 7.Kow YW, Wallace SS, Van Houten B. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat. Res. 1990;235:147–56. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 8.Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–71. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, Van Houten B, Geacintov NE. Sequence context-and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxideguanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–15. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE. Biochemistry. 1995;34:13582–93. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 11.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–61. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoare S, Zou Y, Purohit V, Krishnasamy R, Skorvaga M, Van Houten B, Geacintov NE, Basu AK. Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures. Biochemistry. 2000;39:12252–61. doi: 10.1021/bi0013187. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J. Mol. Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6664–9. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves DA, Mu H, Kropachev K, Cai Y, Kolbanovskiy A, Kolbanovkiy M, Chen Y, Krzeminski J, Amin S, Broyde S, Geacintov NE. Stabilization of DNA duplexes by stacking interactions between aromatic adducts and DNA bases is correlated with poor NER incision efficiencies. Nucl. Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Y, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Broyde S, Patel DJ, Geacintov NE, Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Wiley-VCH; Weinheim, Germany: 2010. Recognition and removal of bulky DNA lesiosn by the nucleotide excision repair system. [Google Scholar]
- 18.Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Molecular cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat. Struct. Mol. Biol. 2011;18:191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 2007;26:613–22. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–12. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–5. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 24.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–23. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 2006;13:360–4. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 26.Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic. Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 28.Peng Y, Wang H, Santana dos Santos L, Kisker C, Van Houten B. Nucleotide excision repair from bacteria to humans: structure-function studies. In: Penning TM, editor. Chemical Carcinogenesis, Current Cancer Research. Springer Science Business Media, LLC; 2011. [Google Scholar]
- 29.Scharer OD. Achieving broad substrate specificity in damage recognition by binding accessible nondamaged DNA. Mol. Cell. 2007;28:184–6. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Scharer OD. A molecular basis for damage recognition in eukaryotic nucleotide excision repair. Chembiochem. 2008;9:21–3. doi: 10.1002/cbic.200700619. [DOI] [PubMed] [Google Scholar]
- 31.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–10. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 32.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Houten B, Snowden A. Mechanism of action of the Escherichia coli UvrABC nuclease: clues to the damage recognition problem. Bioessays. 1993;15:51–9. doi: 10.1002/bies.950150108. [DOI] [PubMed] [Google Scholar]
- 34.Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J. Mol. Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia L, Kropachev K, Ding S, Van Houten B, Geacintov NE, Broyde S. Exploring damage recognition models in prokaryotic nucleotide excision repair with a benzo[a]pyrene-derived lesion in UvrB. Biochemistry. 2009;48:8948–57. doi: 10.1021/bi9010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–32. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolton JL, Thatcher GR. Potential mechanisms of estrogen quinone carcinogenesis. Chem. Res. Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolton JL, Pisha E, Zhang F, Qiu S. Role of quinoids in estrogen carcinogenesis. Chem. Res. Toxicol. 1998;11:1113–27. doi: 10.1021/tx9801007. [DOI] [PubMed] [Google Scholar]
- 39.Kolbanovskiy A, Kuzmin V, Shastry A, Kolbanovskaya M, Chen D, Chang M, Bolton JL, Geacintov NE. Base selectivity and effects of sequence and DNA secondary structure on the formation of covalent adducts derived from the equine estrogen metabolite 4-hydroxyequilenin. Chem. Res. Toxicol. 2005;18:1737–47. doi: 10.1021/tx050190x. [DOI] [PubMed] [Google Scholar]
- 40.Ding S, Wang Y, Kolbanovskiy A, Durandin A, Bolton JL, van Breemen RB, Broyde S, Geacintov NE. Determination of absolute configurations of 4-hydroxyequilenin-cytosine and -adenine adducts by optical rotatory dispersion, electronic circular dichroism, density functional theory calculations, and mass spectrometry. Chem. Res. Toxicol. 2008;21:1739–48. doi: 10.1021/tx800095f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D. Chemistry Department, Ph.D. Thesis. New York University; New York, NY: 2007. Nucleotide excision repair and translesion synthesis of DNA adducts derived from the equine estrogen metabolite 4-hydroxyequilenin. [Google Scholar]
- 42.Reeves D. Chemistry Department, Ph.D. Thesis. New York University; New York, NY: 2010. Impact of DNA Conformation and Base Sequence Context on Efficiencies of Nucleotide Excision Repair of Bulky DNA Adducts. [Google Scholar]
- 43.Pakotiprapha D, Inuzuka Y, Bowman BR, Moolenaar GF, Goosen N, Jeruzalmi D, Verdine GL. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol. Cell. 2008;29:122–33. doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J. Biol. Chem. 2009;284:12837–44. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–94. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol. Cell. 2010;37:702–13. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–83. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–73. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. The EMBO J. 2009;28:2387–99. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–43. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pirogov N, Shafirovich V, Kolbanovskiy A, Solntsev K, Courtney SA, Amin S, Geacintov NE. Role of hydrophobic effects in the reaction of a polynuclear aromatic diol epoxide with oligodeoxynucleotides in aqueous solutions. Chem. Res. Toxicol. 1998;11:381–8. doi: 10.1021/tx980006q. [DOI] [PubMed] [Google Scholar]
- 53.Mao B, Xu J, Li B, Margulis LA, Smirnov S, Ya NQ, Courtney SH, Geacintov NE. Synthesis and characterization of covalent adducts derived from the binding of benzo[a]pyrene diol expoxide to a -GGG- sequence in a deoxyoligonucleotide. Carcinogenesis. 1995;16:357–65. doi: 10.1093/carcin/16.2.357. [DOI] [PubMed] [Google Scholar]
- 54.Shivji MK, Moggs JG, Kuraoka I, Wood RD. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods in Molecular Biology. 1999;113:373–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- 55.Moolenaar GF, van Rossum-Fikkert S, van Kesteren M, Goosen N. Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1467–72. doi: 10.1073/pnas.032584099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997;17:7069–76. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–55. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 58.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove. Biochemistry. 1994;33:11518–27. doi: 10.1021/bi00204a014. [DOI] [PubMed] [Google Scholar]
- 59.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J. Mol. Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodríguez FA, Cai Y, Lin C, Tang Y, Kolbanovskiy A, Amin S, Patel DJ, Broyde S, Geacintov NE. Exocyclic amino groups of flanking guanines govern sequence-dependent adduct conformations and local structural distortions for minor groove-aligned benzo[a]pyrenyl-guanine lesions in a GG mutation hotspot context. Nucleic Acids Res. 2007;35:1555–68. doi: 10.1093/nar/gkm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1914–8. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Branden C, Tooze J. Introduction to Protein Structure. Garland Publishing, Inc.; New York: 1999. [Google Scholar]
- 63.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences. Biophys. J. 2009;96:2245–60. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–26. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Locasale JW, Napoli AA, Chen S, Berman HM, Lawson CL. Signatures of protein-DNA recognition in free DNA binding sites. J. Mol. Biol. 2009;386:1054–65. doi: 10.1016/j.jmb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen C, Russu IM. Sequence-dependence of the energetics of opening of at basepairs in DNA. Biophys. J. 2004;87:2545–51. doi: 10.1529/biophysj.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F, Swanson SM, van Breemen RB, Liu X, Yang Y, Gu C, Bolton JL. Equine estrogen metabolite 4-hydroxyequilenin induces DNA damage in the rat mammary tissues: formation of single-strand breaks, apurinic sites, stable adducts, and oxidized bases. Chem. Res. Toxicol. 2001;14:1654–9. doi: 10.1021/tx010158c. [DOI] [PubMed] [Google Scholar]
- 68.Embrechts J, Lemiere F, Van Dongen W, Esmans EL, Buytaert P, Van Marck E, Kockx M, Makar A. Detection of estrogen DNA-adducts in human breast tumor tissue and healthy tissue by combined nano LC-nano ES tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2003;14:482–91. doi: 10.1016/S1044-0305(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 69.Okahashi Y, Iwamoto T, Suzuki N, Shibutani S, Sugiura S, Itoh S, Nishiwaki T, Ueno S, Mori T. Quantitative detection of 4-hydroxyequilenin-DNA adducts in mammalian cells using an immunoassay with a novel monoclonal antibody. Nucleic Acids Res. 38:133. doi: 10.1093/nar/gkq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang N, Ding S, Kolbanovskiy A, Shastry A, Kuzmin VA, Bolton JL, Patel DJ, Broyde S, Geacintov NE. NMR and computational studies of stereoisomeric equine estrogen-derived DNA cytidine adducts in oligonucleotide duplexes: opposite orientations of diastereomeric forms. Biochemistry. 2009;48:7098–109. doi: 10.1021/bi9006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng H, Cai Y, Ding S, Tang Y, Kropachev K, Zhou Y, Wang L, Wang S, Geacintov NE, Zhang Y, Broyde S. Base flipping free energy profiles for damaged and undamaged DNA. Chem. Res. Toxicol. 2010:1868–1870. doi: 10.1021/tx1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–46. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 74.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 76.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1994;33:11507–17. doi: 10.1021/bi00204a013. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y. Chemistry, Ph.D Thesis. New York University; New York: 2010. Effects of base sequence on the removal of DNA damage by nucleotide excision repair mechanisms in vitro. [Google Scholar]
- 78.Cai Y, Wang L, Ding S, Schwaid A, Geacintov NE, Broyde S. A bulky DNA lesion derived from a highly potent polycyclic aromatic tumorigen stabilizes nucleosome core particle structure. Biochemistry. 2010;49:9943–9945. doi: 10.1021/bi101560y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.