Abstract
Identification of the sources and methods of transmission of Escherichia coli O157:H7 in feedlot cattle may facilitate the development of on-farm control measures for this important food-borne pathogen. The prevalence of E. coli O157:H7 in fecal samples of commercial feedlot cattle in 20 feedlot pens between April and September 2000 was determined throughout the finishing feeding period prior to slaughter. Using immunomagnetic separation, E. coli O157:H7 was isolated from 636 of 4,790 (13%) fecal samples in this study, with highest prevalence earliest in the feeding period. No differences were observed in the fecal or water trough sediment prevalence values of E. coli O157:H7 in 10 pens supplied with chlorinated drinking water supplies compared with nonchlorinated water pens. Pulsed-field gel electrophoresis of XbaI-digested bacterial DNA of the 230 isolates obtained from eight of the pens revealed 56 unique restriction endonuclease digestion patterns (REDPs), although nearly 60% of the isolates belonged to a group of four closely related genetic subtypes that were present in each of the pens and throughout the sampling period. The other REDPs were typically transiently detected, often in single pens and on single sample dates, and in many cases were also closely related to the four predominant REDPs. The persistence and predominance of a few REDPs observed over the entire feeding period on this livestock operation highlight the importance of the farm environment, and not necessarily the incoming cattle, as a potential source or reservoir of E. coli O157:H7 on farms.
Escherichia coli O157:H7 is an important human pathogen worldwide (17, 36). The primary route of human infection is via foods of bovine origin and water contaminated with bovine fecal material (3). Not surprisingly, the prevalence of E. coli O157:H7 contamination on bovine carcass surfaces at slaughter is correlated with the fecal prevalence in the live cattle before slaughter (13). The peak incidence of human disease associated with this pathogen occurs during the summer months, the same season that the prevalence of bovine fecal carriage of this organism is also at its highest (6, 20, 45). It has been proposed that reducing both the magnitude of fecal excretion and the fecal prevalence of this organism in cattle immediately prior to slaughter could significantly reduce the incidence of human disease associated with this pathogen (24, 31). Unfortunately, methods to reduce the fecal prevalence of E. coli O157:H7 in cattle are not currently available. Given the typically transient fecal shedding of E. coli O157:H7 by cattle, a better understanding of the epidemiology and ecology of E. coli O157, particularly the source of exposure of cattle, may provide valuable information that could be used in the design of preharvest control measures for E. coli O157:H7.
The purpose of this study was to determine the pattern of fecal carriage of E. coli O157:H7 in cattle fattened under modern intensive feedlot management conditions. A temporal association between fecal shedding of E. coli O157:H7 among cattle drinking from E. coli O157-contaminated water supplies has been described (14, 38). Furthermore, experimental studies have demonstrated the potential role of waterborne transmission of E. coli O157:H7 to cattle (28, 39). Thus, it was of interest to determine if improved livestock drinking water quality through chlorination would influence fecal prevalence of this organism among feedlot cattle. Pulsed-field gel electrophoresis (PFGE) was used to determine the dynamics of E. coli O157:H7 strains present in pens of cattle drinking chlorinated or unchlorinated water.
MATERIALS AND METHODS
Feedlot description.
Cattle on a commercial beef feedlot were studied throughout the entire feeding period. Twenty lots of animals were enrolled in the study between April and May 2000. Cattle were maintained in pens of approximately 500 animals each. The total cattle population of the feedlot was approximately 75,000 animals throughout the study. Incoming animals in each lot were obtained from multiple sources. Incoming lots of animals (shipment of animals from a common source arriving at the same time) were alternately assigned to pens that were supplied with either chlorinated drinking water or unchlorinated drinking water. Chlorine was added to the water supply to achieve 1 ppm residual free chlorine in the chlorinated section of the feedlot. The remaining pens received water from the same surface reservoir source, without chlorination. Water access was ad libitum from single float-regulated 450-liter water troughs (model 90 Super; Headstrom, Woodbine, Iowa) centrally located in each pen. Upon arrival animals were fed a 20% grain-based diet that was gradually increased to 92 to 94% grain on a dry matter basis by 15 days on feed and maintained at that level throughout the finishing feeding period.
Fecal E. coli O157:H7 detection.
Within 1 week of filling each pen and at 14-day intervals thereafter and the week of shipment to slaughter (up to eight additional sampling dates), 10-g samples of feces were collected from 30 fresh fecal pats in each pen. Samples were immediately shipped overnight on ice to the laboratory for analysis. Due to laboratory capacity, half of the samples from each pen were cultured immediately upon arrival at the laboratory as described below. The remaining samples were refrigerated for 1 day before culturing for E. coli O157:H7 in the same manner. Half of one shipment of samples (collected on 24 July) was extensively delayed in transit, and therefore the culture results from this sample date were not included in the analysis. Instead, these same pens of animals were resampled the following week (1 August), and the biweekly sampling scheduled resumed from that date.
For detection of E. coli O157, each sample was enriched overnight at 37°C in 90 ml of Trypticase soy broth (Difco Laboratories, Detroit, Mich.) containing cefixime (50 ng/ml; Wyeth-Ayerst Laboratories, Pearl River, N.Y.) and vancomycin (40 μg/ml; Sigma Chemical Company, St. Louis, Mo.). E. coli O157:H7 was concentrated from enriched cultures using immunomagnetic separation as per the manufacturer's (Dynal, Oslo, Norway) recommendations. Immunomagnetic separation beads were plated on sorbitol MacConkey agar plates (Difco Laboratories) containing cefixime (50 ng/ml) and potassium tellurite (2.5 μg/ml; Sigma Chemical Company) (SMACCT). Up to 10 sorbitol-negative (white) colonies were picked from each plate. Suspect colonies picked from the SMACCT plates were further identified as E. coli O157:H7 based upon lactose fermentation and the inability to cleave 4-methylumbeliferyl-beta-d-glucuronide to a fluorescent product (2, 44). The presence of the O157:H7 antigen was determined using a particle agglutination test (Oxoid, Basingstoke, Hampshire, United Kingdom).
E. coli O157:H7 detection in water.
Commencing at the first fecal sampling date and at 2-week intervals thereafter, water from the bottom of each trough was collected in 100-ml polystyrene vials containing 15 to 30 mg of sodium thiosulfate. A 30-ml aliquot of water from each trough was mixed with 30 ml of double-strength Trypticase soy broth and incubated overnight at 44.5°C. One-milliliter aliquots of enriched broth were concentrated using immunomagnetic beads specific for the O157:H7 antigen following the manufacturer's (Dynal) recommendations and plated on SMACCT as described above. After overnight incubation at 37.0°C, E. coli O157:H7 was identified on the SMACCT plates using the biochemical and immunological tests described in the previous section. In a similar fashion, E. coli O157:H7 was cultured from drainage areas of several continuous-flow water troughs and from the source water reservoir approximately 1 km away from the feedlot.
Characterization of E. coli O157:H7 isolates.
One E. coli O157:H7 colony obtained from each positive sample was further characterized using a multiplex PCR with primers specific for the fliCh7, eaeA, and stx2 and stx1 genes (16, 46). Briefly, isolated colonies were boiled in 200 μl of sterile deionized distilled water for 20 min. Two microliters of this solution was used as a template for amplification in a 50-μl PCR-buffered (Gibco-BRL) reaction volume containing primers (0.02-nmol/μl solutions; 0.8 μl of fliCh7, 1.0 μl of stx1, 1.2 μl of stx2, and 1.6 μl of eaeA) and 0.02 mM deoxynucleoside triphosphates, 2 mM MgCl2, and 2.5 U of Taq polymerase (Gibco-BRL). Following denaturation for 3 min at 94°C, DNA was amplified using 35 cycles of denaturation, annealing, and elongation at 94, 58, and 72°C for 1, 1.5, and 2.5 min, respectively (Perkin-Elmer 9600). A final elongation step was included at the end of the cycle (72°C for 10 min). PCR products were separated by electrophoresis in 1% agarose gel and visualized with UV illumination following staining with ethidium bromide. Only isolates that encoded fliCh7 and eaeA were considered E. coli O157:H7.
E. coli O157:H7 isolates obtained from cattle and water troughs in eight pens (a subset comprised of four adjacent pens each in chlorinated and unchlorinated drinking water sections of the study) were analyzed by PFGE using XbaI restriction and the Pulse-Net separation protocol (7, 42). Banding patterns in digital images of gels were clustered into restriction endonuclease digestion patterns (REDPs), using Dice similarity coefficients calculated with the unweighted pair group methods arithmetic average algorithm, employing bands between 50 and 600 kb, 0.7% optimization, and 1.25% tolerance (BioNumerics 2.0; Applied Maths, Kortrijk, Belgium), similar to that done by Davis et al. (11).
Statistical analyses.
Differences in the fecal prevalence in cattle drinking chlorinated water and those drinking unchlorinated water were determined by a repeated-measures analysis of variance using the SAS PROC MIXED function (Statistical Analysis Systems, Cary, N.C.). Significance of differences in prevalence among sample dates was assessed using the Wilcoxon sum rank test (α = 0.05). Significance of differences in the frequency of E. coli O157:H7 recovery from chlorinated and unchlorinated water troughs was assessed using Fisher's exact test (α = 0.05).
RESULTS
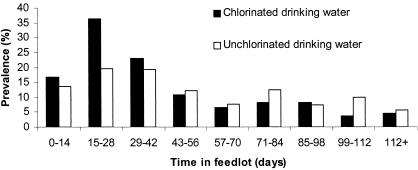
Some pens were shipped sooner than others and therefore could not be sampled nine times each. A total of 4,800 samples were collected, but 10 samples were damaged in transport and could not be analyzed. E. coli O157:H7 was isolated from 636 of 4,790 (13.3%) fecal samples tested. All pens had one or more positive samples. The prevalence of E. coli O157-positive fecal samples at the first sampling, obtained within 7 days of arrival at the feedlot, was 15%. Significant (P < 0.001) differences among prevalence rates on different sample dates were identified. The overall prevalence of E. coli O157-positive fecal samples peaked 2 weeks after entry to the feedlot and then declined below entry-level prevalence by 42 days on feed. The prevalence remained relatively stable at approximately half the entry prevalence for the remaining 70 days of the feeding period prior to slaughter. The pen-wise fecal E. coli O157:H7 prevalence values detected in pens supplied with chlorinated and unchlorinated water did not differ significantly: 342 of 2,483 (13.8%) versus 294 of 2,307 (12.7%) samples, respectively (Fig. 1).
FIG. 1.
Overall prevalence of E. coli O157:H7 in feedlot cattle consuming chlorinated or unchlorinated drinking water.
E. coli O157:H7 was isolated from 37 of 172 (21.5%) water trough specimens during the study. E. coli O157:H7 was isolated somewhat more frequently from troughs supplied with unchlorinated water than from those supplied with chlorinated water (22 of 86 [26%] versus 15 of 86 [17%]); however, this difference was not statistically significant. One of 37 water and sediment samples collected from three water reservoirs within 8 km of the feedlot tested positive for E. coli O157; however, the reservoir from which the isolate was obtained was downstream of the reservoir from which the feedlot water supply was obtained, and no isolates of E. coli O157:H7 were obtained from the source reservoir or from the reservoir upstream from the source reservoir. Three of 10 samples collected from drain areas for collection of the overflow of continuous-flow water troughs (located in pens not included in this study) tested positive for E. coli O157.
E. coli O157:H7 isolates from a subset of eight pens were examined by molecular methods. Isolates from four linearly arranged groups of pens, one group of four pens supplied with chlorinated and the other group of pens supplied with unchlorinated water, were evaluated in this manner. Thus, pens with and without shared fence lines were included in each group. Among the 230 isolates obtained from water and fecal samples in the pens selected for molecular analysis with PFGE, 56 unique XbaI REDPs were identified (Fig. 2).
FIG. 2.
Schematic of all 56 different REDPs isolated from eight pens of feedlot cattle over a 4-month period.
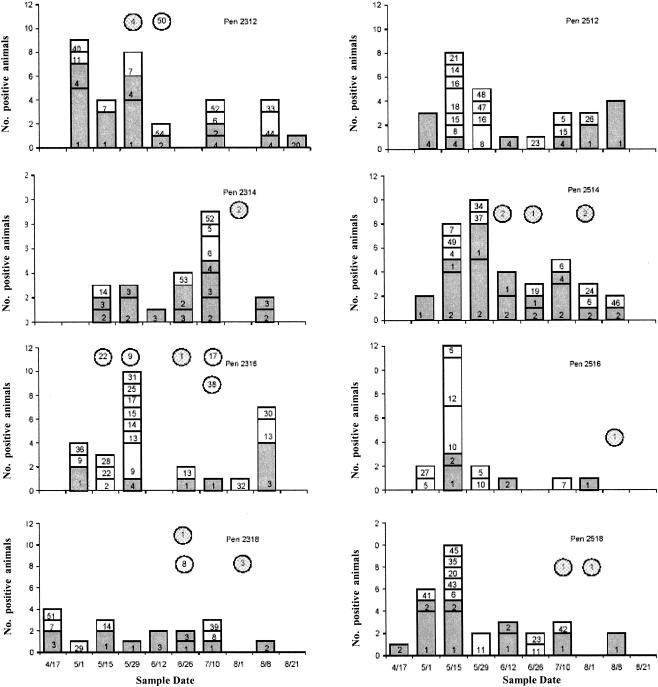
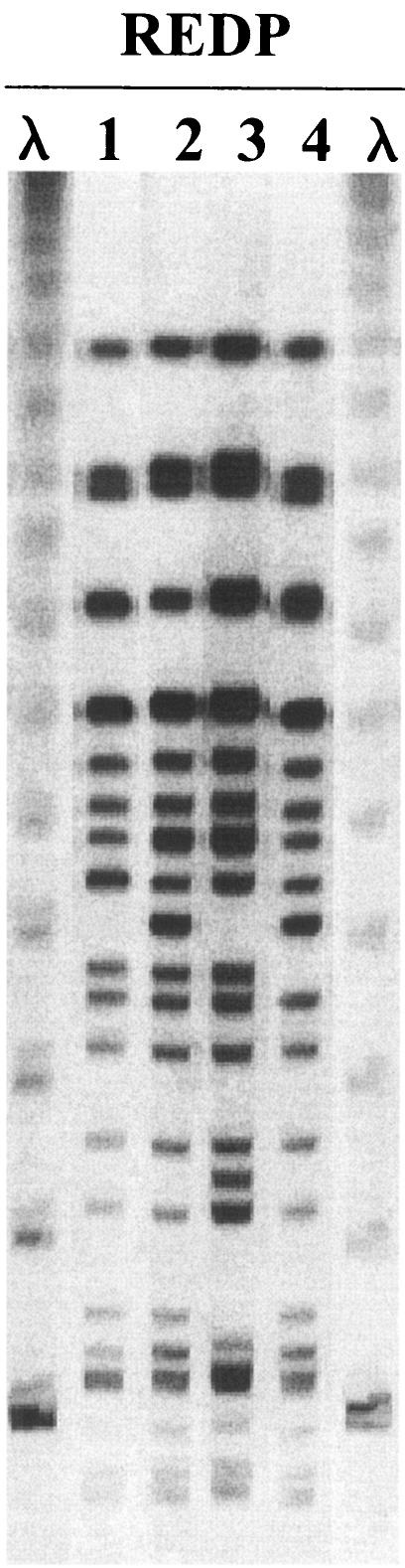
An average of 2.5 REDPs per pen per sample date were identified, with an average of 11.4 different REDPs per pen over the entire feeding period. REDPs were neither equally represented nor evenly distributed spatially or temporally (Fig. 3). Four very closely related REDPs (REDPs 1 to 4; Fig. 4) accounted for 57% of the total bovine isolates obtained from cattle during the study. One of the four predominant REDPs was identified in every pen on at least one sample date and at least one pen on each sample date.
FIG. 3.
Prevalence and REDPs of E. coli O157:H7 isolated from eight pens of feedlot animals. 2300 series, unchlorinated drinking water; 2500 series, chlorinated drinking water. Circles represent water trough cultures positive for E. coli O157. Numbers represent REDP patterns. Shaded areas represent predominant REDPs.
FIG. 4.
PFGE gel of the four most common REDPs identified among eight pens of feedlot cattle over a 4-month period (representative of 60% of all isolates tested). λ, concatemers of lambda cI857 Sam7 (Bio-Rad).
The remaining fifty bovine REDPs were identified far less frequently. No other single subtype accounted for more than 3% of the total. Minority REDPs were usually limited to one or two pens and were only transiently detected in either the cattle or the water. Sixty percent of the subtypes (31 of 50) were identified on only a single sampling date and identified in only a single pen. Most of these REDPs were closely related (i.e., one to three band differences) to one of the predominant four REDPs.
Nine different REDPs were isolated from water troughs. All water trough isolates but two (REDPs 38 and 50) matched REDPs of bovine fecal isolates collected on or around the same sample date; these two exceptions were identified in separate water troughs on different sampling dates and matched no fecal isolates by REDP. E. coli O157:H7 was isolated from surface water collecting water trough overflows on three occasions, 26 June, 10 July, and 8 August. REDPs of isolates obtained on these dates were 5, 1, and 1, respectively. The single water reservoir isolate (REDP 55) isolated on 12 June 2000 was unique and did not match any other REDP identified in this study.
A limited number of isolates were obtained from the study feedlot in the course of prior and subsequent studies: E. coli O157:H7 with REDP pattern 1, 2, 4, or 5 was identified among livestock drinking water and feedstuff isolates obtained in 1999, and REDP 1 was identified among bovine fecal isolates obtained in 2001. Interestingly, REDP 1 was also identified among isolates obtained in 1998 and 1999 from another feedlot approximately 300 km distant.
Most isolates (216 of 230) harbored both the stx1 and stx2 genes. A single bovine isolate had only stx1. Thirteen bovine isolates had only stx2. Most isolates within REDPs shared identical Shiga toxin gene profiles; however, 11 isolates within REDPs 1, 2, 3, 5, 9, 13, 14, and 19 had Shiga toxin profiles different from one or more other isolates within the same REDP.
DISCUSSION
This study demonstrates the temporal and spatial dissemination, predominance, and persistence of four E. coli O157:H7 genetic subtypes among cattle on an individual feedlot over a period of 4 months. Furthermore, the similar REDPs of these dominant subtypes suggest that they may be clonally related. Although a diverse population of different E. coli O157:H7 REDPs was identified from study isolates, most were only detected on a few occasions.
The E. coli O157:H7 prevalence in this feedlot was within the range of previously reported prevalence values among feedlot animals (18, 41). Without standardized detection methods, it is impossible to determine whether prevalence differences between studies reflect the effects of different culture and sampling methods or actual differences in E. coli O157:H7 prevalence.
The observed higher prevalence of bovine fecal E. coli O157:H7 early in the feeding period was consistent with previous reports for both E. coli O157:H7 specifically and other Shiga toxin-producing E. coli (10, 18, 33). The reported E. coli O157:H7 prevalence of beef calves at the time of weaning is 7.4% ± 6.2% (mean ± standard deviation) (27), comparable to the prevalence observed during the first sampling period in this study. Two major differences between these cattle and the calves sampled by Laegreid et al. are that some (5 to 10%) of the animals in our study had passed through sales barns and had recently been transported to or from locations where the cattle may have acquired E. coli O157:H7 (27, 40). Furthermore cattle in this study were all yearlings, not calves.
The cause of the higher prevalence early in the feeding period is unknown. Dietary manipulation has been shown to alter the population of gastrointestinal microorganisms (12, 22). The percentage of grain in the ration provided to these cattle was incrementally increased during the first month on feed. It is possible that changes in the ration (not necessarily the ration per se) may contribute to an increased susceptibility of animals to infection or may increase the magnitude of fecal E. coli O157:H7 excretion during the first 4 weeks of feeding. Once the ration composition was stabilized at a high grain concentration, the prevalence stabilized at a lower level than in the early feeding period. Other factors may have also contributed to the observed higher prevalence of E. coli O157:H7 early during the feeding period. For example, it is possible that an age effect was present. It is recognized that the diversity of O-serotypes among E. coli strains in younger calves is higher and less stable than that in adult animals (21). Likewise, the prevalence of E. coli O157:H7 in calves is also typically higher than the prevalence in adult cattle (46). Feedlot animals under 317 kg are more likely to test positive for E. coli O157:H7 than cattle of greater weights (10). Finally, higher fecal prevalence of E. coli O157:H7 has been observed during summer months, and it is possible that at least some of the apparent temporal variation in fecal prevalence in this study represented seasonal variation (8, 9, 19, 35). Consistent with this latter possibility, additional cattle fecal samples were obtained in November following the conclusion of this study. In these additional samples, pens at 2 weeks on feed (the time point where the study pens had the highest fecal prevalence) had a similarly low prevalence as pens after >80 days on feed (data not shown). However, it was not possible to demonstrate any correlation between the fecal prevalence identified in this study and weather data for the weeks preceding each sampling date (data not shown).
As in other studies, periods of highest fecal E. coli O157:H7 prevalence appeared as sporadic events in individual pens (19, 32, 33). The cause of the temporal clusters in prevalence is not known. Since all study pens were located on the same feedlot (within a few 100 m of one another), it is unlikely that climatic changes had a significant impact on the prevalence on a pen-by-pen basis. Furthermore, there was no noted tendency for adjacent pens of cattle to experience simultaneous prevalence peaks, suggesting that cattle-to-cattle transmission across pen fences was not the key factor. Cattle in all pens were managed similarly but, given the number of animal fed in each pen, it is probable that animals in different pens received the same feed components acquired from different loads of feed. Most cattle feeds are contaminated with generic E. coli even before they arrive on the farm, and a small percentage of these feeds are contaminated with E. coli O157:H7 specifically (30). Minority subtypes of E. coli O157:H7 may have been introduced into individual pens via this route. Wildlife, the pen floor, and the initial incoming animals may have also been sources of E. coli O157:H7.
Remarkably, over half (57%) of all bovine isolates analyzed by PFGE shared one of four predominant REDPs. The purpose of the PFGE analysis was to determine if indistinguishable strains of E. coli O157:H7 persisted within individual pens and to determine whether single REDPs appeared in different pens. Indistinguishable REDPs would be expected to occur if the infections were acquired from a common source, if there were sustained transmission of E. coli O157:H7 among animals within a pen, or if there were dissemination of strains from one pen to another. Although four REDP were widespread in the feedlot, there was no apparent temporal or spatial clustering of these subtypes among neighboring pens.
Interpretation of PFGE patterns remains somewhat arbitrary and subjective. Degrees of genetic relatedness have been ascribed to E. coli O157:H7 isolates based upon arbitrary numbers of band differences and by using statistical analyses (23, 29, 38, 43). However, genetically different (based on phage typing and PCR analysis for specific genes) and epidemiologically unrelated isolates may have indistinguishable REDPs, and isolates that are known recent progeny from a single common precursor (i.e., closely related genetically) may have significantly different REDPs (1, 5, 26). Thus, although it is most likely that the epidemiologically related isolates obtained in this study that either matched in all bands or that differed by only one or a few bands may be closely genetically related, this conclusion may not be true in all cases. Despite its shortcomings, PFGE has nonetheless proven to be a valuable tool in discriminating among E. coli O157:H7 isolates (4, 26, 42).
The stringent definition of a distinct REDP used in this analysis resulted in the classification of isolates into a large number of different REDPs. The use of stringent classification criteria is recommended in population-based studies of E. coli O157:H7 (4, 26). Nevertheless, the majority of isolates collected from different pens clustered together, even using these very stringent definitions of genetic subtype. The observed pattern of E. coli O157:H7 REDP distribution is most consistent with the presence of one or two distinct clonal types being endemic at the feedlot and either (i) sporadic introduction and transient carriage by cattle of newly introduced genetic subtypes into the population or (ii) spontaneous mutations in these subtypes to form new genetic subtypes. If one assumes that isolates that share similar (but not identical) REDPs are clonally related, it is possible to attribute some of the observed diversity to clonal turnover (1, 25). On the other hand, it is also possible that some of the isolates with indistinguishable REDPs recovered from different pens on separate sample dates may represent spurious (genetically different) band matches, introduction of the organism from a common source, or a low level of dissemination of particular strains among pens.
Even with the availability of sophisticated gel analysis software, the comparison of banding patterns from isolates run on different gels proved to be challenging. Essentially, it is difficult, if not impossible, to determine if an increase in ethidium bromide staining at a particular distance on a gel represents a single large band or two individual bands with very similar migration patterns. Failure to identify these differing banding patterns, or falsely assigning differing REDPs to isolates actually having the same REDP, could have a significant impact on calculated similarity indices and cluster analyses. To minimize the impact of mislabeling, we standardized the band marking procedure by assigning only one band to areas of ethidium bromide staining, regardless of the width of the band, unless the stain was completely separated by an unstained region on the gel or showed multiple peaks on the densitometric scale. It is probable that many of the denser, thick bands observed on the gels actually represented two comigrating DNA fragments. Picking isolates from different clusters identified on different gels and rerunning them on the same gel resulted in similar clustering patterns (data not shown).
It is interesting that isolates exhibiting indistinguishable REDPs were isolated over several years in the study feedlot and continue to be isolated from this and another feedlot in the geographic area. This is particularly surprising, given the several complete population turnovers per year that occur in these typically managed feedlots. If incoming cattle were the major source of E. coli O157:H7 in the feedlot, one might have expected much more diversity among the E. coli O157:H7 isolates over this time period. It is possible that certain E. coli O157:H7 strain types are better adapted either to survival and persistence in the feedlot environment or to colonization of the bovine gastrointestinal tract in cattle on grain-intensive feedlot-type rations, and either possibility could explain the predominance of these strain types. E. coli O157:H7 isolates of a single REDP or a group of closely related REDPs have been observed to persist on dairy farms from months to years (32, 37, 38), but in the case of dairy farms persistent infection of individual animals is an alternative hypothesis to explain strain type stability, because the rate of dairy animal turnover is vastly lower than is typical of feedlots. The low rate of clonal turnover of E. coli O157:H7 isolates on feedlots as identified in this study tends to exclude persistent individual animal colonization as an explanation for persistent strain types and supports the presence of other, nonanimal stable reservoirs on the farm.
E. coli O157:H7 isolates containing all combinations of stx1 and/or stx2 have been implicated in human disease, but most studies have found some predominance of stx2-only genetic types among isolates from severe human disease (15). The distribution of E. coli O157:H7 toxin genotypes among bovine isolates in this study is similar to the distribution of toxin types reported from 41 cattle farms in another geographic region by our laboratory (37). The loss of Shiga toxin genes has been reported in laboratory-propagated cultures (34). The differing stx profiles observed among isolates having the same REDP may have been the result of such genetic events. If the size of the fragment of DNA that was lost from the genome were the same size as other restricted DNA fragments (comigrating on the PFGE gel), loss of this fragment could occur without altering either the number or distance of migration of the bands on the gel. Alternatively, these may indeed be examples of homoplasy, where isolates of unrelated genetic origin exhibit indistinguishable REDPs.
Since the simple chlorination of input water was unable to reduce the prevalence of E. coli O157:H7-contaminated water troughs, the attributable risk of E. coli O157:H7 infection in cattle associated with the consumption of E. coli O157-contaminated water could not be calculated. It was noted that large amounts of sediments composed primarily of feed material rapidly accumulated at the bottom of the water troughs during this study. The accumulation of large amounts of organic matter would be expected to rapidly inactivate the biocidal activity of the chlorine and provide an ideal niche for the survival of E. coli O157:H7 (28). The stirring of water trough sediments either by the drinking activities of cattle or more directly by cattle standing in the water troughs, as was commonly observed during the summer heat, probably exacerbated chlorine neutralization by organic matter. Effective interventions to prevent contamination of bovine water sources with E. coli O157:H7 will seemingly require redesign of troughs to minimize recontamination and to eliminate accumulations of organic sediments.
The heterogeneity among transient minority subtypes is consistent with multiple sources of E. coli O157:H7 that sporadically enter the cattle population from other outside sources during the feeding period. The factors contributing to the sporadic periods of increased E. coli O157:H7 prevalence among individual lots of cattle remain unknown. Due to the complexity of the epidemiology of E. coli O157:H7 in feedlot cattle, preharvest control of this pathogen on feedlots will probably require multiple control measures that address the various sources of exposure of cattle to this important food-borne pathogen.
Acknowledgments
This work was supported by grants from the USDA National Research Initiative program Epidemiologic Approaches to Food Safety and by Lakeside Farm Industries.
REFERENCES
- 1.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1982. Isolation of E. coli O157:H7 from sporadic cases of hemorrhagic colitis—United States. Morb. Mortal. Wkly. Rep. 31:580, 585. [PubMed] [Google Scholar]
- 3.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 4.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 5.Bohm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonardi, S., E. Maggi, G. Pizzin, S. Morabito, and A. Caprioli. 2001. Faecal carriage of verocytotoxin-producing Escherichia coli O157 and carcass contamination in cattle at slaughter in northern Italy. Int. J. Food Microbiol. 66:47-53. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2001. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7 by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, Ga.
- 8.Chapman, P. A., A. T. Cerdan Malo, M. Ellin, R. Ashton, and M. A. Harkin. 2001. Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int. J. Food Microbiol. 64:139-150. [DOI] [PubMed] [Google Scholar]
- 9.Conedera, G., P. A. Chapman, S. Marangon, E. Tisato, P. Dalvit, and A. Zuin. 2001. A field survey of Escherichia coli O157 ecology on a cattle farm in Italy. Int. J. Food Microbiol. 66:85-93. [DOI] [PubMed] [Google Scholar]
- 10.Dargatz, D. L., L. Wells, D. Hancock, and L. Garber. 1997. Factors associated with the presence of Escherichia coli O157 in the feces of feedlot cattle. J. Food Prot. 60:466-470. [DOI] [PubMed] [Google Scholar]
- 11.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez-Gonzalez, F., T. Callaway, M. Kizoulis, and J. Russell. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666-1668. [DOI] [PubMed] [Google Scholar]
- 13.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, A., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 16.Gannon, V. P., S. D'Souza, T. Graham, and R. K. King. 1997. Specific identification of Escherichia coli O157:H7 using a multiplex PCR assay. Adv. Exp. Med. Biol. 412:81-82. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, D., D. Rice, L. A. Thomas, D. A. Dargatz, and T. Besser. 1997. The epidemiology of E. coli O157 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuvelink, A. E., F. L. van den Biggelaar, J. Zwartkruis-Nahuis, R. G. Herbes, R. Huyben, N. Nagelkerke, W. J. Melchers, L. A. Monnens, and E. de Boer. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinton, M., V. Allen, and A. H. Linton. 1982. The biotyping of Escherichia coli isolated from healthy farm animals. J. Hyg. (London) 88:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovde, C. J., P. R. Austin, K. A. Cloud, C. J. Williams, and C. W. Hunt. 1999. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl. Environ. Microbiol. 65:3233-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumiya, H., J. Terajima, A. Wada, Y. Inagaki, K. I. Itoh, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan, D., S. A. McEwen, A. M. Lammerding, W. B. McNab, and J. B. Wilson. 1999. Pre-slaughter control of Escherichia coli O157 in beef cattle: a simulation study. Prev. Vet. Med. 41:55-74. [DOI] [PubMed] [Google Scholar]
- 25.Karch, H., H. Russmann, H. Schmidt, A. Schwarzkopf, and J. Heesemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause, U., F. M. Thomson-Carter, and T. H. Pennington. 1996. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J. Clin. Microbiol. 34:959-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laegreid, W. W., R. O. Elder, and J. E. Keen. 1999. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol. Infect. 123:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeJeune, J. T., T. E. Besser, and D. D. Hancock. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl. Environ. Microbiol. 67:3053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louie, M., S. Read, L. Louie, K. Ziebell, K. Rahn, A. Borczyk, and H. Lior. 1999. Molecular typing methods to investigate transmission of Escherichia coli O157:H7 from cattle to humans. Epidemiol. Infect. 123:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynn, T. V., D. D. Hancock, T. E. Besser, J. H. Harrison, D. H. Rice, N. T. Stewart, and L. L. Rowan. 1998. The occurrence and replication of Escherichia coli in cattle feeds. J. Dairy Sci. 81:1102-1108. [DOI] [PubMed] [Google Scholar]
- 31.Marks, H., M. Coleman, C. Lin, and T. Roberts. 1998. Topics in microbial risk assessment: dynamic flow tree process. Risk Anal. 18:309-328. [DOI] [PubMed] [Google Scholar]
- 32.Mechie, S. C., P. A. Chapman, and C. A. Siddons. 1997. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol. Infect. 118:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Midgley, J., N. Fegan, and P. Desmarchelier. 1999. Dynamics of Shiga toxin-producing Escherichia coli (STEC) in feedlot cattle. Lett. Appl. Microbiol. 29:85-89. [DOI] [PubMed] [Google Scholar]
- 34.Murase, T., S. Yamai, and H. Watanabe. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48-50. [DOI] [PubMed] [Google Scholar]
- 35.Paiba, G. A., J. C. Gibbens, S. J. Pascoe, J. W. Wilesmith, S. A. Kidd, C. Byrne, J. B. Ryan, R. P. Smith, M. McLaren, R. J. Futter, A. C. Kay, Y. E. Jones, S. A. Chappell, G. A. Willshaw, and T. Cheasty. 2002. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Vet. Rec. 150:593-598. [DOI] [PubMed] [Google Scholar]
- 36.Reilly, A. 1998. Prevention and control of enterohaemorrhagic Escherichia coli (EHEC) infections: memorandum from a W. H. O. meeting. W. H. O. Consultation on Prevention and Control of Enterohaemorrhagic Escherichia coli (EHEC) Infections. Bull. W. H. O. 76:245-255. [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shere, J. A., C. W. Kaspar, K. J. Bartlett, S. E. Linden, B. Norell, S. Francey, and D. M. Schaefer. 2002. Shedding of Escherichia coli O157:H7 in dairy cattle housed in a confined environment following waterborne inoculation. Appl. Environ. Microbiol. 68:1947-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small, A., C. A. Reid, S. M. Avery, N. Karabasil, C. Crowley, and S. Buncic. 2002. Potential for the spread of Escherichia coli O157, Salmonella, and Campylobacter in the lairage environment at abattoirs. J. Food Prot. 65:931-936. [DOI] [PubMed] [Google Scholar]
- 41.Smith, D., M. Blackford, S. Younts, R. Moxley, J. Gray, L. Hungerford, T. Milton, and T. Klopfenstein. 2001. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64:1899-1903. [DOI] [PubMed] [Google Scholar]
- 42.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. S., D. S. Hodge, and A. A. Borczyk. 1990. Rapid biochemical test to identify verocytotoxin-positive strains of Escherichia coli serotype O157. J. Clin. Microbiol. 28:2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace, D. J., T. Van Gilder, S. Shallow, T. Fiorentino, S. D. Segler, K. E. Smith, B. Shiferaw, R. Etzel, W. E. Garthright, F. J. Angulo, et al. 2000. Incidence of foodborne illnesses reported by the foodborne diseases active surveillance network (FoodNet)—1997. J. Food Prot. 63:807-809. [DOI] [PubMed] [Google Scholar]
- 46.Wells, J. G., L. D. Shipman, K. D. Greene, E. G. Sowers, J. H. Green, D. N. Cameron, F. P. Downes, M. L. Martin, P. M. Griffin, S. M. Ostroff, et al. 1991. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J. Clin. Microbiol. 29:985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]