Abstract
This study describes the occurrence of unique dissimilatory sulfite reductase (DSR) genes at a depth of 1,380 m from the deep-sea hydrothermal vent field at the Suiyo Seamount, Izu-Bonin Arc, Western Pacific, Japan. The DSR genes were obtained from microbes that grew in a catheter-type in situ growth chamber deployed for 3 days on a vent and from the effluent water of drilled holes at 5°C and natural vent fluids at 7°C. DSR clones SUIYOdsr-A and SUIYOdsr-B were not closely related to cultivated species or environmental clones. Moreover, samples of microbial communities were examined by PCR-denaturing gradient gel electrophoresis (DGGE) analysis of the 16S rRNA gene. The sequence analysis of 16S rRNA gene fragments obtained from the vent catheter after a 3-day incubation revealed the occurrence of bacterial DGGE bands affiliated with the Aquificae and γ- and ɛ-Proteobacteria as well as the occurrence of archaeal phylotypes affiliated with the Thermococcales and of a unique archaeon sequence that clustered with “Nanoarchaeota.” The DGGE bands obtained from drilled holes and natural vent fluids from 7 to 300°C were affiliated with the δ-Proteobacteria, genus Thiomicrospira, and Pelodictyon. The dominant DGGE bands retrieved from the effluent water of casing pipes at 3 and 4°C were closely related to phylotypes obtained from the Arctic Ocean. Our results suggest the presence of microorganisms corresponding to a unique DSR lineage not detected previously from other geothermal environments.
Deep-sea hydrothermal vent sites exhibit much microbial diversity: due to their sharp physical and chemical gradients, they harbor hyperthermophiles, mesophiles, psychrophiles, chemolithotrophs, heterotrophs, anaerobes, microaerobes, and aerobes (29). The use of molecular techniques of the 16S rRNA gene, combined with the physiological characterization of isolated strains, has allowed researchers to study the phylogenetic diversity and ecosystems at deep-sea hydrothermal vents (17, 18, 19, 35, 45, 48, 49). Furthermore, molecular ecological approaches to studying the 16S rRNA gene have revealed the presence of phylogenetically novel archaeal and bacterial sequences at these sites (44, 50).
Sulfate-reducing prokaryotes (SRP) that obtain energy from dissimilatory sulfate reduction play an important role in the mineralization of organic matter in organic sulfate-rich marine anaerobic sediments, in addition to being key organisms in the sulfur cycle (25, 59). Biological sulfate reduction at temperatures up to 110°C was observed in deep-sea hydrothermal vent sediments of the Guaymas Basin in the Gulf of California, Mexico (26). Recently, a strain belonging to the genus Thermodesulfobacterium was isolated from a vent at Guaymas Basin (24). Sequence analysis of the dissimilatory sulfite reductase (DSR) gene, coding for an enzyme that catalyzes the reduction of sulfite to sulfide during anaerobic sulfate respiration, was found to be useful in the detection of SRP within complex microbial populations. Indeed, molecular ecological approach using the DSR gene has even been successfully used to detect SRP in complicated habitats, such as estuarine and marine sediments (12, 27, 53), hypersaline microbial mats (33), deep-sea hydrothermal vent polychaete annelids (11, 13), terrestrial hot springs (14, 37), a uranium tailing site (8), and a hydrothermal subsurface mine (3, 38). However, relatively little is known about the phylogenetic relationships between the DSR genes from deep-sea hydrothermal vent environments and those from other terrestrial geothermal environments, such as high-temperature hot springs and subsurface habitats.
The Suiyo Seamount is an active submarine volcano located on the volcanic front of the Izu-Bonin Arc, Western Pacific, Japan (61). Vigorous hydrothermal activity has occurred on the caldera floor atop the west peak of the Suiyo Seamount (58). The caldera floor is predominantly covered with sandy sediment and hydrothermal precipitations and lacks any evidence of muddy pelagic sediment. Organic geochemical studies on its surface sediments have revealed only trace amounts of long-chain fatty acids originating from a higher plant wax of terrestrial origin (T. Yamanaka, H. Naraoka, F. Kitajima, T. Naito, K. Marumo, and T. Urabe, 2002 Ocean Sci. Meet., abstr. OS32O-04, 2002). This is to be expected, since the Suiyo Seamount is located about 1,000 km from land. It also lacks pelagic sediment, since at less than one million years old, it formed too recently for sediment accumulation on a geological time scale.
In this study, we first report unique α-subunit DSR-deduced amino acid sequences from the deep-sea hydrothermal sites of the Suiyo Seamount. In order to compare phylogenetic relationships between the DSR and 16S rRNA gene sequences and to investigate the temperature-related distribution of microorganisms, we describe the microbial community structure in several samples, including effluent water from casing pipes at temperatures of 3 or 4°C and from a hole drilled with a tethered marine rock drill at 33 to 55°C, natural vent fluids at various temperatures (7 to 300°C), and the substratum of a catheter-type in situ growth chamber (vent catheter) placed on the hydrothermal vent.
MATERIALS AND METHODS
Sample collection.
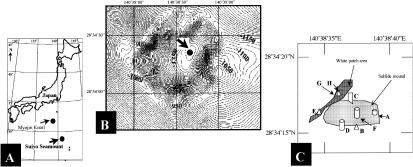
Water samples at various temperatures at a depth of approximately 1,380 m were collected from the natural vents and drilled holes in the hydrothermal sites of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific, Japan (28°34′N, 140°38′E) (Fig. 1 and Table 1). A remotely operated vehicle (ROV), the Hakuyo 2000, was used during an SR02 cruise of the mother ship Shinryumaru (3 to 15 August 2002). A manned research submersible, Shinkai 2000, was used during an NT02-09 cruise of the R/V Natsushima (22 August to 18 September 2002). To collect microorganisms, a peristaltic pump was used in situ to pump water through a 0.2-μm-pore-size, 142-mm-diameter filter (Supor-200; Pall Corporation, Inc., Ann Arbor, Mich.) attached to the vehicles (drawing speed about 0.15 liter/min). At each site, water samples for chemical analyses were pumped into the collection syringe and filter assembly. A portion of the filter was aseptically placed into a sterilized 2-ml screw-cap microcentrifuge tube and then frozen at −20°C on the ship until processed.
FIG. 1.
(A) Map of the Western Pacific, showing the locations of the Suiyo Seamount and the Myojin Knoll. (B) Map of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific, Japan (28°34′N, 140°38′E). Samples were taken from a depth of 1,380 m. (C) Map showing sampling sites A to H.
TABLE 1.
Environmental samples used in this study
| Sitea | Dive no. | Filter no. | Location | Depth (m) | Filter tempb (°C) | Filter vol (liter) | Chemical analysisc
|
Sample | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Mg concn (mM) | Si concn (μM) | H2S concn (mM) | ||||||||
| A | HY30 | F1 | 28°34′16.9"N 140°38′38.0"E | 1,385 | 33-55 | 0.8 | 5.2 | 38.0 | 3.8 | 0.51 | Effluent water from drilled hole (APSK07) |
| B | HY32 | F2 | 28°34′15.2"N 140°38′37.5"E | 1,384 | 299-300 | 4.0 | 3.7 | 2.1 | 12.2 | 1.50 | Effluent water from hydrothermal vent |
| C | HY25 | 28°34′17.0"N 140°38′37.0"E | 1,384 | NDd | ND | ND | ND | ND | ND | Catheter-type in situ growth chamber placed on hydrothermal vent | |
| D | 1383 | F-1 | 28°34′15.2"N 140°38′36.6"E | 1,380 | 3 (maximum, 4)e | 6.5 | 6.6 | 51.8 | 0.43 | 0.74 | Effluent water from casing pipe (APSK09) |
| E | 1385 | F-1 | 28°34′16.9"N 140°38′38.8"E | 1,366 | 25-36 | 14.0 | 7.2 | 53.1 | 0.2 | ND | Diffuse flow from white patch area |
| F | 1386 | F-1 | 28°34′16.1"N 140°38′37.5"E | 1,380 | 4 | 15.5 | 7.1 | 52.7 | 0.17 | ND | Effluent water from casing pipe (APSK10) |
| G | 1388 | F-2 | 28°34′17.4"N 140°38′36.3"E | 1,376 | 310 | 1.8 | 4.0 | 7.1 | 11.1 | 1.40 | Effluent water from hydrothermal vent |
| H | 1389 | F-1 | 28°34′17.4"N 140°38′36.0"E | 1,383 | 3-50 | ca. 20 | 6.2 | 49.7 | 0.71 | 0.03 | Diffuse flow from white patch area |
Site locations are shown in Fig. 1C.
The temperatures were measured in situ while the water was filtered to collect microorganisms. Fluid temperature occasionally fluctuated due to mixing with ambient seawater.
Samples for chemical analysis were collected by the same sampling procedure at each site [J. Ishibashi, Y. Morimoto, Y. Umeki, F. Kouzuma, T. Toki, U. Tsunogai, K. Namba, M. Utsumi, T. Yamanaka, H. Chiba, and K. Okamura, EOS Trans. AGU, 83(41), abstr. v11c-05, 2002].
ND, not determined.
The temperature was measured when the water was filtered for chemical analyses.
Steel casing pipes, APSK07 (length, approximately 1,600 mm; outside diameter, 70 mm) at site A, titanium casing pipes, APSK09 (600 mm above the sea bottom and approximately 2,600 mm below the surface; outside diameter, 70 mm) at site D, and APSK10 (450 mm above the sea bottom and approximately 1,200 mm below the surface; outside diameter, 70 mm) at site F, were inserted into the holes (at depths of 2,690 mm at site A, 8,992 mm at site D, and 7,035 mm at site F) bored with a tethered marine rock drill (the so-called benthic multicoring system [BMS]) to approach the subvent biosphere directly beneath the seafloor during the 2001 and 2002 BMS cruises (site A, 18 to 27 June 2001; sites D and F, 15 to 23 July 2002). The temperatures of effluent water from casing pipes APSK07, APSK09, and APSK10 after insertion were 156, 6, and 65 to 89°C, respectively. Steel casing pipe APSK07 had been destroyed by corrosion at sampling time.
A vent catheter of stainless steel pipe (length, 500 mm; inner diameter, 20 mm) with tipping porous glasses wrapped with titan mesh (length, 100 mm) was designed for in situ collection and incubation of microbes in venting fluid below the seafloor using porous inorganic grains as a substratum (18a). The apparatus was sterilized by autoclaving and transferred to the seafloor in a plastic cylinder containing filtered salt water to minimize contamination. It was inserted into a vent orifice at site C using a manipulator of the Hakuyo 2000 and left there for 3 days. After the apparatus was recovered (measures were taken to prevent contamination from the surrounding seawater during its ascent), a portion of porous grain samples that had changed from white to gray was aseptically placed into a sterilized 2-ml screw-cap microcentrifuge tube and frozen at −20°C aboard the ship until processed.
Chemical analysis.
Concentrations of major chemical species were analyzed using the separate aliquots obtained by the same sampling procedure in order to characterize the signatures of the samples obtained. Immediately after sample recovery, the pH and hydrogen sulfide concentration were determined onboard using an electrode and the colorimetric technique (9), respectively. Samples were filtered with a 0.45-μm-pore-size filter and stored in a refrigerator. The silica concentration was also analyzed onboard by the colorimetric method (16) within 24 h. Magnesium concentration was analyzed after a 200-fold dilution using inductively coupled plasma atomic emission spectroscopy in an onshore laboratory. Both magnesium and silica concentrations were conventionally employed as indicators for mixing between pure hydrothermal fluid and ambient seawater (54). Magnesium is thought to be absent in high-temperature hydrothermal fluid. Silica concentrations varied from 0.12 μmol/kg in ambient seawater to 13.0 mmol kg−1 in pure hydrothermal fluid (hydrothermal end member) from the Suiyo hydrothermal field. Hydrogen sulfide concentration of the hydrothermal end member was estimated as 2.0 mmol kg−1 [J. Ishibashi, Y. Morimoto, Y. Umeki, F. Kouzuma, T. Toki, U. Tsunogai, K. Nanba, M. Utsumi, T. Yamanaka, H. Chiba, and K. Okamura, EOS Trans. AGU 83(47), abstr. V11C-05, 2002].
DNA extraction.
Nucleic acids of microorganisms were extracted by a bead-beating, phenol-chloroform extraction procedure as follows. The filter and crumbled porous glasses were mixed with 850 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), 850 μl of chloroform-isoamyl alcohol (24:1 [vol/vol]), 30 μl of 20% sodium dodecyl sulfate, and 0.5 g of glass beads (with diameters of 0.1 and 0.05 mm). After the bead-beating step and centrifugation, the aqueous phase was transferred to a new tube and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]). A 0.6 volume of isopropanol was added to the aqueous phase that had been transferred to a new tube. After centrifugation, the precipitated DNA was rinsed with 1 ml of 70% (vol/vol) ethanol (−20°C), dried, resuspended in double-distilled water (RNase- and DNase free), and then stored at −20°C.
Amplification and cloning of DSR gene fragments.
DSR gene fragments were amplified with a primer pair, DSR1F and DSR4R, as described previously (56). Amplification was performed with a DNA thermal cycler (PCR EXPRESS; HYBAID, Franklin, Mass.) as follows: 30 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 54°C, and 3 min at 72°C. The reaction was completed by a final extension step of 10 min at 72°C. When there were several nonspecific bands, a touchdown PCR was performed (annealing temperature from 65 to 55°C for 20 cycles). When there was no specific band, the PCR products were reamplified at 10, 20, and 30 cycles with 1 μl of the amplicons as template DNA to obtain DSR gene fragments. For the DSR PCR amplification, 1× EX Taq buffer (Takara Shuzo, Kyoto, Japan), 1 to 100 ng of template DNA, 250 μM (each) of the four deoxynucleoside triphosphates, 25 pmol (each) primer, and 2.5 U of EX Taq DNA polymerase (Takara) were combined in a final volume of 50 μl. The DSR gene fragments were analyzed by electrophoresis in 1.1% (wt/vol) agarose S gels (Nippon Gene) containing ethidium bromide (1 μg ml−1). The DSR gene products were cloned as described previously (38), and insert-containing clones reamplified with the vector primers M13 reverse and M13 forward were screened by restriction fragment length polymorphism analysis using restriction enzyme AluI as described previously (38). By using a CONCERT Rapid PCR purification kit (Invitrogen, Carlsbad, Calif.), the PCR insert-containing amplicons were purified for use as the template DNA for sequencing.
Amplification and DGGE of 16S rRNA gene fragments.
The bacterial and archaeal 16S rRNA gene fragments were amplified as described previously (7). A touchdown PCR was performed for primer pair Eub341F (with GC clamp) and 907R (annealing temperature decreased from 65 to 55°C in 20 cycles) and for primer pair Arch344F (with GC clamp) and Arch915R (annealing temperature decreased from 71 to 61°C in 20 cycles). PCR products were subjected to DGGE analysis, which was performed with D-code systems (Bio-Rad Laboratories, Hercules, Calif.) with 1.5-mm-wide gels as described previously (35). PCR products were applied directly to 6% (wt/vol) polyacrylamide gels with denaturing gradients from 20 to 60% (100% denaturant is 7 M urea and 40% [vol/vol] deionized formamide). After PCR products of the second amplification were electrophoresed again in a DGGE to check the purity of the bands, the PCR amplicons were purified with a CONCERT Rapid PCR purification kit (Invitrogen) to use as the template DNA for sequencing.
16S rRNA and DSR gene fragment sequencing and phylogenetic analysis.
Sequencing reactions were performed using the ABI PRISM Big Dye Terminator Cycle sequencing kit and an ABI model 310 automated sequencer (Applied Biosystems) according to the manufacturer's instructions. 16S ribosomal DNA (rDNA) fragments from DGGE bands were sequenced using primers 341F and 907R. Partial sequences of DSR amplification products were sequenced by using primer DSR1F and vector primers M13 reverse and M13 forward for clones. The sequences from DGGE bands and the deduced amino acid sequences of α-subunits of DSR genes were entered into the BLAST (2) and FASTA programs (32) of the National Center for Biotechnology Information and the DNA Data Bank of Japan (DDBJ) in order to identify phylogenetic relationships. Sequence alignments with portions of the 16S rRNA and deduced DSR amino acid sequences of reference prokaryotes from DDBJ/EMBL/GenBank were performed by using the CLUSTAL W program (52), and matrices of evolutionary distance were constructed by the neighbor-joining method (47). Phylogenetic trees were visualized with TreeView software (41). Bootstrap resampling analysis of 1,000 replicates was performed to estimate the confidence of tree topologies. Maximum-parsimony analysis was accomplished using MEGA2 program (31). The RNA secondary structure was predicted by using the free-energy minimization algorithm with Genetyx-Mac software (Software Development Co., Ltd., Tokyo, Japan).
Nucleotide sequence accession numbers.
The 16S rRNA and DSR gene sequences were submitted to DDBJ/EMBL/GenBank and have been assigned the following accession numbers: AB096615 to AB096630 (16S rRNA genes) and AB095362 and AB095363 (DSR genes).
RESULTS AND DISCUSSION
Environmental characteristics.
During the 3 days the vent catheter was set up, the color of its porous glass turned black due to the buildup of FeS and other sulfide compounds on the inside surface of the stainless steel pipe at site C where the maximum fluid temperature was 279°C. The temperature of effluent water from a drilled hole (APSK07) at site A and that of a diffusive flow from a white patch area at sites E and H were lower than those of hydrothermal fluids at sites B and G (Table 1). The water at sites A, E, and H seemed to be brackish due to mixing with ambient cold seawater. The temperature of effluent water from the casing pipes (APSK09 at site D, APSK10 at site F) was the same as the ambient cold seawater (4°C). Judging from the chemical data of magnesium, silica, and sulfide concentrations, it is likely that effluent waters at sites A, B, and G were mixed with hydrothermal water of the same origin and ambient seawater. The sulfide concentration at site H was markedly low, despite the mixed hydrothermal water.
Clone library characterization.
Gene fragments encoding DSR (ca. 1.9 kb) were detected in the vent catheter after a 3-day incubation at site C and in the water samples at sites F and H. The DSR-PCR products from sites C and F together with those from site H were obtained with touchdown PCR amplification and another PCR for 10 cycles after the first PCR, respectively. No DSR-PCR product was amplified from the negative-control samples using DSR primers. A clone library of the PCR products of DSR gene fragments was used to investigate the diversity of the DSR genes of SRP in the samples at sites C, F, and H. A total of 83 clones were put into two clone families designated SUIYOdsr-A and SUIYOdsr-B on the basis of AluI restriction fragment banding patterns (Fig. 2). The frequency of the DSR clone family SUIYOdsr-A was higher at site C than at other sites (Table 2). However, PCR-dependent techniques have several pitfalls and potential biases (55). PCR biases may have occurred in the present study due to our use of touchdown PCR amplification and reamplification of the first PCR products from hydrothermal water. To clarify the temperature-related SRP in deep-sea hydrothermal vents, it is essential to develop a novel quantitative technique for DSR genes based on sequence information and the accumulated DSR gene sequence data for several deep-sea hydrothermal systems.
FIG. 2.
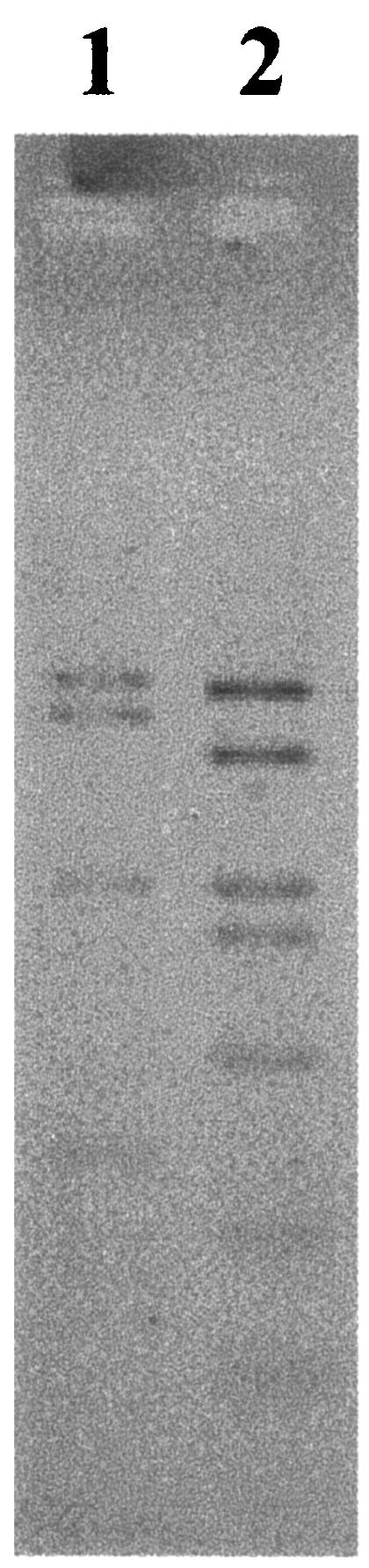
AluI restriction patterns of two clone families identified in the clone library of DSR gene fragments retrieved from Suiyo Seamount. Lane 1, clone family A (SUIYOdsr-A); lane 2, clone family B (SUIYOdsr-B).
TABLE 2.
Abundance of clone families in the library of DSR gene fragments retrieved from Suiyo Seamount
| Clone family | No. of clones (% of clones) at sample site:
|
||
|---|---|---|---|
| C | F | H | |
| SUIYOdsr-A | 11 (52) | 8 (30) | 0 (0) |
| SUIYOdsr-B | 10 (48) | 19 (70) | 35 (100) |
Phylogenetic analysis of DSR gene fragments.
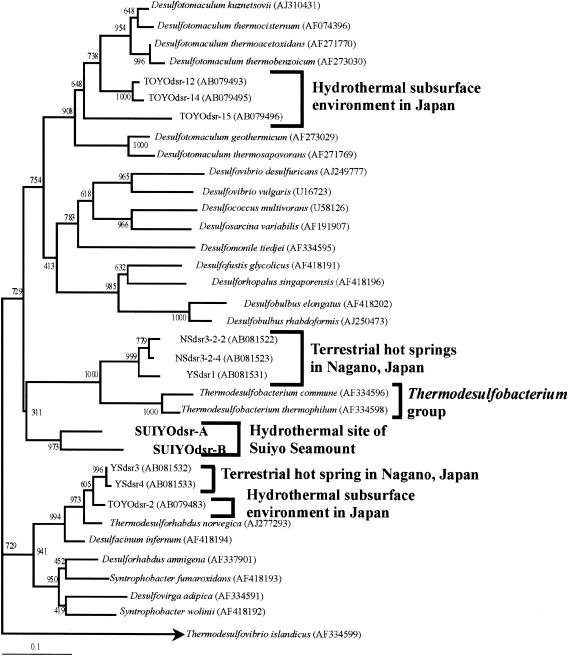
The DSR clones retrieved from the hydrothermal site at the Suiyo Seamount represent a separate but somewhat related lineage with several DSR clones predominantly retrieved from Japanese terrestrial hot springs (37) and with the DSR-deduced amino acid sequences of the Thermodesulfobacterium group, although this position was not strongly supported by a high bootstrap value (Fig. 3). By using the FASTA programs, the designated DSR clones, SUIYOdsr-A and SUIYOdsr-B, were found to be less closely related to cultivated species (their closest relative being Desulfotomaculum thermosapovorans [similarity values, 81 and 75%, respectively]). In addition, these clones were less closely related to environmental clones retrieved from geothermal environments (>75% nucleotide similarity) (3, 12, 14, 37, 38). As shown in Fig. 4, SUIYOdsr-A and SUIYOdsr-B had a major insertion between positions 259 and 273 of α-subunits of the DSR gene (numbered according to the numbering system for Desulfovibrio vulgaris (28), as did δ-Proteobacteria-like SRB as previously described by Klein et al. (30). These results suggest the presence of a unique SRP at the deep-sea hydrothermal sites of the Suiyo Seamount.
FIG. 3.
Phylogenetic relationships of DSR α-subunit fragments (approximately 230 deduced amino acid sequences) obtained from the Suiyo Seamount (SUIYOdsr-A and SUIYOdsr-B) as determined by neighbor-joining analysis. The bar labeled 0.1 represents an estimated 10% sequence divergence. Numbers beside branching points indicate bootstrap values determined from 1,000 iterations.
FIG. 4.
Deduced amino acid alignment of the α-subunits of DSR genes showing that SUIYOdsr-A and SUIYOdsr-B have insertions between positions 259 and 273 of the α-subunits of DSR genes. Amino acid positions correspond to those of the α-subunit of the Desulfovibrio vulgaris DSR gene (28).
No DSR clone related to the α-subunits of DSR genes of the hyperthermophilic marine sulfate reducers Archaeoglobus fulgidus and Archaeoglobus profundus (30) was found in the present study. Similarly, on the basis of 16S rRNA gene sequence analysis, no Archaeoglobales-like phylotype was found for any samples, although such phylotypes have been found in an in situ growth chamber placed at a Mid-Atlantic Ridge hydrothermal vent (44) and in a black smoker chimney at Myojin Knoll, Izu-Ogasawara Arc, Japan (50). However, Takai and Horikoshi (50) reported the small number of microbes and the dearth of archaeal clones that contained an Archaeoglobales-like phylotype compared to that of bacterial clones in a black smoker chimney at the Suiyo Seamount. The Archaeoglobales-like phylotype seems to be a minor population below the detection limit of the methods used during the current investigations.
Phylogenetic analyses of bacterial 16S rDNA fragments.
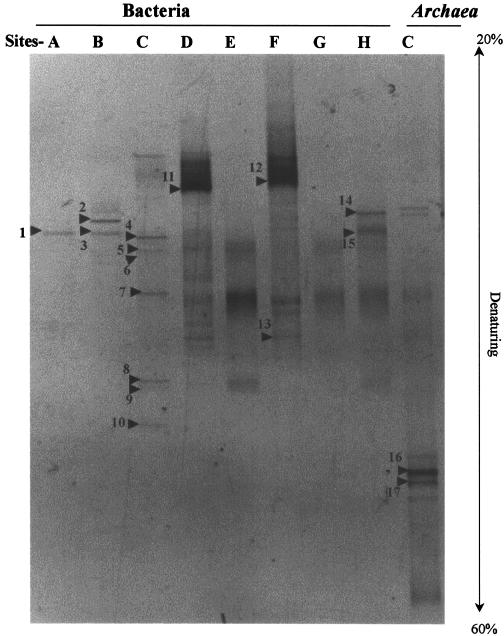
To compare phylogenetic relationships based on the DSR genes to those based on the 16S rRNA gene sequences and to investigate the temperature-related distribution of members of the domain Bacteria, we analyzed the DGGE profiles of PCR-amplified 16S rDNA fragments with a primer set for the domain. There were major differences in the profiles of DGGE bands for samples from the vent catheter (site C), the casing pipes (sites D and F), and the natural vents at other sites (Fig. 5 and Table 3). No PCR product was amplified from the negative-control samples by using bacterial primers.
FIG. 5.
DGGE profiles of 16S rDNA genes derived from filtered hydrothermal water from drilled holes (lanes A, D, and F), natural vents (lanes B, E, G, and H), and the substratum of catheter-type in situ growth chamber (lane C) with primer sets for the domain Bacteria and Archaea. The bands labeled with numbers were sequenced. Bands: 1, SUIYO-E1; 2, SUIYO-E2; 3, SUIYO-E3; 4, SUIYO-E4; 5, SUIYO-E5; 6, SUIYO-E6; 7, SUIYO-E7; 8, SUIYO-E8; 9, SUIYO-E9; 10, SUIYO-E10; 11, SUIYO-E11; 12, SUIYO-E12; 13, SUIYO-E13; 14, SUIYO-E14; 15, SUIYO-E15; 16, SUIYO-A16; 17, SUIYO-A17.
TABLE 3.
16S rDNA sequence similarity between sequences from samples and related taxa
| Phylogenetic group (DDBJ/EMBL/GenBank accession no.) | % Similarity | DGGE band | Presence of the DGGE band at sampling site:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C (279°C) | G (310°C) | B (300°C) | A (55°C) | E (25-36°C) | H (3-50°C) | D (3°C) | F (4°C) | |||
| Bacteria | ||||||||||
| Aquificae | ||||||||||
| Persephonella hydrogenophila (AB086419) | 97 | SUIYO-E8, E9 | + | |||||||
| Thermovibrio ruber (AJ316619) | 95 | SUIYO-E10 | + | |||||||
| “Chlorobia” | ||||||||||
| Pelodictyon luteolum (Y08107) | 98 | SUIYO-E14 | + | |||||||
| α-Proteobacteria | ||||||||||
| Methylobacterium sp. strain SY-2 (AJ278347) | 99 | SUIYO-E13 | + | + | ||||||
| γ-Proteobacteria | ||||||||||
| Thiomicrospira sp. strain MA2-6 (L40811) | 100 | SUIYO-E1, -E3 | + | + | ||||||
| Arctic96AD-3 (AF354607) | 92 | SUIYO-E5, -E6 | + | |||||||
| 98 | SUIYO-E12 | + | ||||||||
| Arctic96AD-9 (AF354608) | 97 | SUIYO-E11 | + | |||||||
| Strain BD1-1 (AB015514) | 89 | SUIYO-E4 | + | |||||||
| δ-Proteobacteria | ||||||||||
| Bdellovibrio sp. strain JS10 (AF084863) | 92 | SUIYO-E2 | + | |||||||
| Ethylbenzene-degrading consortium DGGE band C (AB062689) | 97 | SUIYO-E15 | + | |||||||
| ɛ-Proteobacteria | ||||||||||
| Clone P. palm C/A 26 (AJ441213) | 92 | SUIYO-E7 | + | |||||||
| Archaea | ||||||||||
| Crenarchaeota | ||||||||||
| Thermococcus sp. strain MZ11 (AY017179) | 94 | SUIYO-A16 | + | |||||||
| Not determined | 82 | SUIYO-A17 | + | |||||||
The SUIYO-E8, -E9, and -E10 DGGE bands obtained from the vent catheter were affiliated with the phylum Aquificae. The Aquificae-like phylotypes, SUIYO-E8 and -E9, were closely related to a thermophilic hydrogen-oxidizing bacterium, Persephonella hydrogenophila, isolated from a hydrothermal vent chimney at the Suiyo Seamount (36). Recently, the thermophilic, strictly chemolithoautotrophic, microaerophilic, and hydrogen-oxidizing strains of the genus Persephonella have been obtained from deep-sea hydrothermal vent sites in the Pacific Ocean and Guaymas Basin (17). Their optimum temperature for growth was approximately 70°C (17, 36). The Aquificae-like phylotype SUIYO-E10 was closely related to the extremely thermophilic, chemolithoautotrophic, nitrate-reducing bacterium, Thermovibrio ruber (22). The optimum temperature for its growth was approximately 75°C. The presence of DGGE bands within the Aquificae suggests that there had been a period of optimum temperature for in situ growth of Aquificae-like microbes in the vent catheter, despite a measured temperature of 279°C.
The SUIYO-E7 DGGE band was related to several uncultured ɛ-Proteobacteria clones retrieved from sediments at the Guaymas Basin (51) and from mucous secretions of a hydrothermal vent polychaete (1). Recent studies of the ɛ-Proteobacteria 16S rRNA gene indicate a high bacterial diversity at deep-sea hydrothermal vents (1, 6, 10, 34, 43, 44, 49, 51). Furthermore, bacteria within the ɛ-Proteobacteria subclass that oxidize hydrogen or sulfur compounds (elemental sulfur or thiosulfate) have been isolated from deep-sea hydrothermal vent samples (49). Although it is impossible to determine the function (ferrous and/or sulfur-utilizing) of the ɛ-Proteobacteria phylotype SUIYO-E7 without more detailed physiological data, such ɛ-Proteobacteria-like microbes may contribute to iron or sulfur metabolism within the vent catheter at deep-sea hydrothermal vents.
DGGE bands SUIYO-E11 and -E12, which were closely related to phylotypes obtained at a depth of 131 m in the Arctic Ocean (4), were observed only for organisms isolated from casing pipes at sites D and F. The maximum temperatures measured at sites D and F were 4 and 5°C, respectively, whereas the other site temperatures were above 18°C. It is likely that these DGGE bands were derived from the psychrophilic microbes growing in the casing pipe at sites D and F.
The DGGE bands SUIYO-E1 and -E2 matched the sequence of the Thiomicrospira sp. strain MA2-6 (35). Sulfur-oxidizing bacteria of the genus Thiomicrospira and related phylotypes have been frequently isolated from deep-sea hydrothermal vent samples (5, 23, 35, 46, 60). Muyzer et al. (35) demonstrated that Thiomicrospira spp. were dominant community members in hydrothermal vent sites at the Mid-Atlantic Ridge. Similarly, it is likely that members of the Thiomicrospira species played a key role in the sulfur oxidation of hydrothermal sites in the Suiyo Seamount.
The DGGE band SUIYO-E14 was closely related to an obligately anaerobic and phototrophic bacterium, Pelodictyon luteolum, that utilizes sulfide and sulfur as an electron donor (15). The SUIYO-E14 band was found only in the sample from site H where sulfide was depleted in hydrothermal water. Nisbet et al. (40) hypothesized that deep-sea hydrothermal vent systems appear to support the growth of phototrophic microorganisms due to the potential excitation of bacteriochlorophyll (BChl) a and b by black-body emission. However, members of the genus Pelodictyon contain BChl c, d, or e as their major photosynthetic pigments (15). Likewise, a thermophilic green sulfur bacterium, Chlorobium tepidum, whose optimum growth temperature is 47 or 48°C, contains BChl c as its major photosynthetic pigment (57). Therefore, it is difficult to determine the in situ function of the microorganism corresponding to the SUIYO-E14 band without more detailed physiological data.
The SUIYO-E15 band was closely related to the mesophilic ethylbenzene-degrading sulfate-reducing bacterium in an ethylbenzene-degrading consortium (39). The microorganisms corresponding to the SUIYO-E15 band appear to be the mesophilic bacteria. Although the occurrence of a DSR gene related to the Desulfobacter-like DSR genes (42) was anticipated, no DSR gene was related to the SRP in the δ-Proteobacteria. PCR biases may have occurred in the present study.
Phylogenetic analyses of archaeal 16S rDNA fragments.
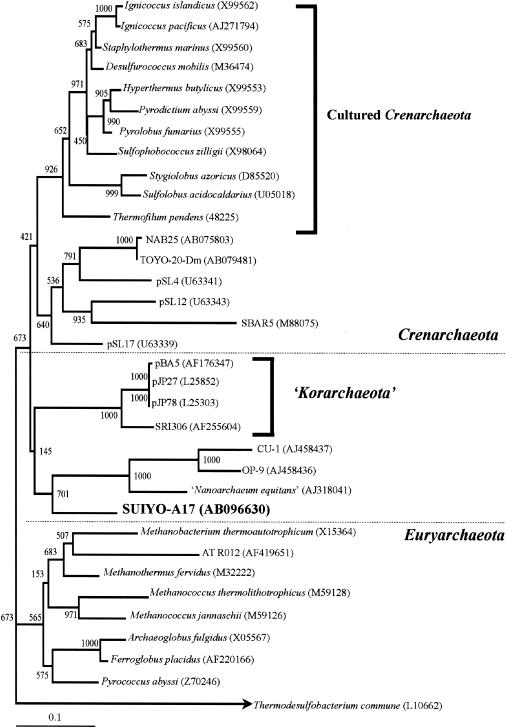
Archaeal PCR-amplified 16S rDNAs were obtained from sites C to H. However, the DGGE bands successfully sequenced were the two DGGE bands, designated SYUIYO-A16 and -A17, that were obtained only from the vent catheter at site C. This gap is most likely due to the use of the chamber. No PCR product was amplified from the negative-control samples using archaeal primers.
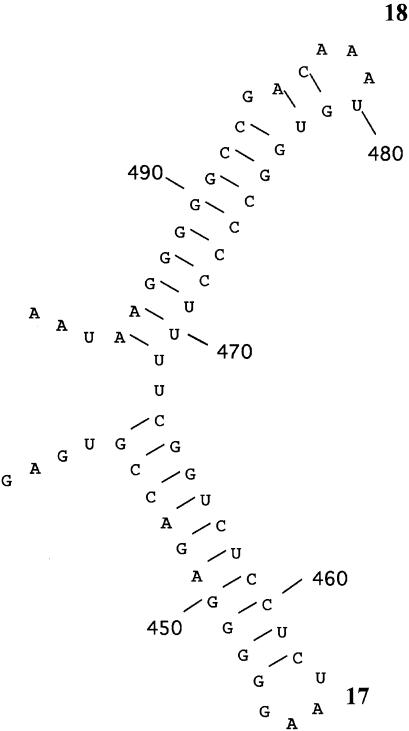
The SUIYO-A17 DGGE band was less closely related to cultivated species and environmental clones belonging to Archaea (Table 3). The SUIYO-A17 DGGE band formed a separate but closely associated lineage with “Nanoarchaeum equitan,” as represented by a nanosized hyperthermophilic symbiont (20), and with the nanoarchaeotal clones CU-1 and OP-9 (21) (Fig. 6). This position was strongly supported by high bootstrap values above 701 (neighbor-joining method) and 82 (maximum-parsimony method), respectively. The SUIYO-A17 DGGE band did not cluster with several “ancient archaeal group” clones representing deeply rooted lineages in the Crenarchaeota that had been found in black smoker chimneys at Myojin Knoll and the Suiyo Seamount (50). Moreover, two secondary structural features of the 16S rRNA of the SUIYO-A17 DGGE band were investigated to confirm its affiliation with Archaea (Fig. 7). This sequence contains the conserved characteristic archaeal secondary structures, structures 17 and 18, which correspond to the helix number (20). These results indicate the potential presence of as-yet-uncultivated Archaea at the deep-sea hydrothermal sites of the Suiyo Seamount. However, the branching position of the cluster consisted of “Nanoarchaeum equitans,” and the SUIYO-A17 position was not strongly supported by high bootstrap values. Therefore, no accurate branching position of the cluster in the domain Archaea could be determined. Further studies, as well as microscopy observations with fluorescence-labeled oligonucleotide probes and culture-based approaches, should be undertaken to clarify whether there is a nanosized hyperthermophilic symbiont corresponding to SUIYO-A17 in the deep-sea hydrothermal vents of the Suiyo Seamount.
FIG. 6.
Phylogenetic relationships of archaeal 16S rDNA fragments (approximately 450 bp) obtained from Suiyo Seamount (in bold type) as determined by neighbor-joining analysis. The bar labeled 0.1 represents an estimated 10% sequence divergence. Numbers beside branching points indicate bootstrap values determined from 1,000 iterations.
FIG. 7.
Conserved characteristic archaeal secondary structures in the region between positions 440 to 498 (Escherichia coli numbering). Boldfaced numbers 17 and 18 correspond to the helix number (20).
Conclusions.
Although there was no evidence of phylogenetic congruence between DSR and 16S rRNA tree topology, the present analysis of DSR gene sequences indicated a notable cluster from the deep-sea hydrothermal sites of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific, Japan. The occurrence of DSR genes may reveal a potential dissimilatory sulfate reduction in the hydrothermal vent sites. Furthermore, 16S rRNA gene analysis provided the temperature-related distribution of the microbial community structure in this hydrothermal site. Further efforts to develop tools for the quantification of DSR gene and its mRNA combined with physiological characterizations (sulfide production and sulfate reduction rate) should provide the temperature-related distribution of the SRP and their role in the sulfur cycle in deep-sea hydrothermal vents.
Acknowledgments
We thank the captains and crews of the Daini-Hakurei, Shinryumaru, and Natsushima as well as the operation group of the vehicles for their technical expertise; Motoo Utsumi for organizing the Shinkai 2000 cruise; Koji Mori for helpful comments on casing pipes; Keizo Shimada and Satoshi Hanada for helpful discussions of this study; Michinari Sunamura, Yousuke Higashi, and Shiho Itahashi for advice on filtered samples; and Akiko Kamono for advice on phylogenetic analyses.
This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science and Technology through the Special Coordination Fund “Archaean Park Project” (International Research Project on Interaction Between Sub-Vent Biosphere and Geo-Environments).
REFERENCES
- 1.Alain, K., M. Olagnon, D. Desbruyeres, A. Page, G. Barbier, S. K. Juniper, J. Quellerou, and M. A. Cambon-Bonavita. 2002. Phylogenetic characterization of the bacterial assemblage associated with mucous secretions of the hydrothermal vent polychaete Paralvinella palmiformis. FEMS Microbiol. Ecol. 42:463-476. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, B. J., D. P. Moser, B. J. MacGregor, S. Fishbain, M. Wagner, N. K. Fry, B. Jackson, N. Speolstra, S. Loos, K. Takai, B. S. Lollar, J. Fredrickson, D. Balkwill, T. C. Onstott, C. F. Wimpee, and D. A. Stahl. 2003. Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ. Microbiol. 5:267-277. [DOI] [PubMed] [Google Scholar]
- 4.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary, S. C., M. T. Cottrell, J. L. Stein, F. Camacho, and D. Desbruyères. 1997. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl. Environ. Microbiol. 63:1124-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamayor, E. O., H. Schäfer, L. Bañeras, C. Pedrós-Alió, and G. Muyzer. 2000. Identification of and spatiotemporal difference between microbial assemblages from two neighboring sulfur lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y.-J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. Anwar Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 10.Corre, E., A.-L. Reysenbach, and D. Prieur. 2001. ɛ-Proteobacterial diversity from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. FEMS Microbiol. Lett. 205:329-335. [DOI] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., and S. C. Cary. 1999. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 65:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubilier, N., C. Mülders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erseus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 14.Fishbain, S., J. G. Dillon, H. L. Gough, and D. A. Stahl. 2003. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69:3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrity, G. M., and J. G. Holt. 2001. Phylum BXI. Chlorobi phy. nov., p. 601-623. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. The Archaea and the deeply branching and phototrophic Bacteria. Springer-Verlag, New York, N.Y.
- 16.Gieskes, J. M., T. Gamo, and H. Brumsack. 1991. Chemical methods for interstitial water analysis aboard JOIDES Resolution. Ocean Drilling Program Technical Note no. 15. Texas A&M University, College Station.
- 17.Götz, D., A. Banta, T. J. Beveridge, A. I. Rushdi, B. R. T. Simoneit, and A.-L. Reysenbach. 2002. Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 52:1349-1359. [DOI] [PubMed] [Google Scholar]
- 18.Harmsen, H. J. M., D. Prieur, and C. Jeanthon. 1997. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl. Environ. Microbiol. 63:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Higashi, Y., M. Sunamura, K. Kitamura, K. Nakamura, Y. Kurusu, J. Ishibashi, T. Urabe, and A. Maruyama. Microbial diversity in hydrothermal surface to sub-surface environments of Suiyo Seamount, Izu-Bonin Arc, using a catheter-type in situ growth chamber. FEMS Microb. Ecol., in press. [DOI] [PubMed]
- 19.Holden, J. F., K. Takai, M. Summit, S. Bolton, J. Zyskowski, and J. A. Baross. 2001. Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern Pacific Ocean. FEMS Microbiol. Ecol. 36:51-60. [DOI] [PubMed] [Google Scholar]
- 20.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 21.Huber, H., M. J. Hohn, K. O. Stetter, and R. Rachel. 2003. The phylum Nanoarchaeota: present knowledge and future perspectives of a unique form of life. Res. Microbiol. 154:165-171. [DOI] [PubMed] [Google Scholar]
- 22.Huber, H., S. Diller, C. Horn, and R. Rachel. 2002. Thermovibrio ruber gen. nov., sp. nov., an extremely thermophilic, chemolithoautotrophic, nitrate-reducing bacterium that forms a deep-branch within the phylum Aquificae. Int. J. Syst. Evol. Microbiol. 52:1859-1865. [DOI] [PubMed] [Google Scholar]
- 23.Jannasch, H. W., C. O. Wirsen, D. C. Nelson, and L. A. Robertson. 1985. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35:422-424. [Google Scholar]
- 24.Jeanthon, C., S. L'Haridon, V. Cueff, A. Banta, A.-L. Reysenbach, and D. Prieur. 2002. Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int. J. Syst. Evol. Microbiol. 52:765-772. [DOI] [PubMed] [Google Scholar]
- 25.Jøorgensen, B. B. 1982. Mineralization of organic matter in the sea bed: the role of sulfate reduction. Nature 296:643-645. [Google Scholar]
- 26.Jøorgensen, B. B., M. F. Isaksen, and H. W. Jannasch. 1992. Bacterial sulfate reduction above 100°C in deep-sea hydrothermal vent sediments. Science 25:1756-1757. [DOI] [PubMed] [Google Scholar]
- 27.Joulian, C., N. B. Ramsing, and K. Ingvorsen. 2001. Congruent phylogenies of most common small-subunit rRNA and dissimilatory sulfite reductase gene sequences retrieved from estuarine sediments. Appl. Environ. Microbiol. 67:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karkhoff-Schweizer, R. R., D. P. Huber, and G. Voordouw. 1995. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karl, D. M. 1995. Ecology of free-hydrothermal vent microbial communities, p. 35-124. In D. M. Karl (ed.), The microbiology of deep-sea hydrothermal vents. CRC Press, Boca Raton, Fla.
- 30.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 33.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa, S., K. Takai, K. Horikoshi, and Y. Sako. 2003. Persephonella hydrogeniphila sp. nov., a novel thermophilic, hydrogen-oxidizing bacterium from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 53:863-869. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa, T., and M. Fukui. 2003. Molecular characterization of community structures and sulfur metabolism within microbial streamers in Japanese hot springs. Appl. Environ. Microbiol. 69:7044-7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa, T., S. Hanada, A. Maruyama, K. Marumo, T. Urabe, and M. Fukui. 2002. Distribution and diversity of thermophilic sulfate-reducing bacteria within a Cu-Pb-Zn mine (Toyoha, Japan). FEMS Microbiol. Ecol. 41:199-209. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa, T., S. Sato, Y. Yamamoto, and M. Fukui. 2002. Successive changes in community structure of an ethylbenzene-degrading sulfate-reducing consortium. Water Res. 36:2813-2823. [DOI] [PubMed] [Google Scholar]
- 40.Nisbet, E. G., J. R. Cann, and C. L. Van Dover. 1995. Origins of photosynthesis. Nature 373:479-480. [Google Scholar]
- 41.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Pérez-Jiménez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35:145-150. [DOI] [PubMed] [Google Scholar]
- 43.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reysenbach, A.-L., and E. Shock. 2002. Merging genomes with geochemistry in hydrothermal ecosystems. Science 296:1077-1082. [DOI] [PubMed] [Google Scholar]
- 46.Ruby, E. G., C. O. Wirsen, and H. W. Jannasch. 1981. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl. Environ. Microbiol. 42:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 48.Slobodkin, A., B. Campbell, S. C. Cary, E. Bonch-Osmolovskaya, and C. Jeanthon. 2001. Evidence for the presence of thermophilic Fe(III)-reducing microorganisms in deep-sea hydrothermal vents at 13°N (East Pacific Rise). FEMS Microbiol. Ecol. 36:235-243. [DOI] [PubMed] [Google Scholar]
- 49.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ɛ-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 50.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teske, A., K. U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Von Damm, K. L., J. M. Edmond, B. Grant, C. I. Measures, B. Walden, and R. F. Weiss. 1985. Chemistry of submarine hydrothermal solutions at 21N, East Pacific Rise. Geochim. Cosmochim. Acta 49:2197-2220. [Google Scholar]
- 55.Von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 56.Wagner, M., A. Roger, J. Flax, G. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 156:81-90. [Google Scholar]
- 58.Watanabe, K., and T. Kajimura. 1994. The hydrothermal mineralization at Suiyo seamount, in the Izu-Ogasawara Arc. Resource Geol. 44:133-140. (In Japanese with English abstract.) [Google Scholar]
- 59.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. IV. Springer-Verlag, New York, N.Y.
- 60.Wirsen, C. O., T. Brinkhoff, J. Kuever, G. Muyzer, S. Molyneaux, and H. W. Jannasch. 1998. Comparison of a new Thiomicrospira strain from the Mid-Atlantic Ridge with known hydrothermal vent isolates. Appl. Environ. Microbiol. 64:4057-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuasa, M., F. Murakami, E. Saito, and K. Watanabe. 1991. Submarine topography of seamounts on the volcanic front of the Izu-Ogasawara, Bonin Arc. Bull. Geol. Surv. Jpn. 42:703-743. [Google Scholar]