Abstract
The absence of alcoholic fermentation makes pyruvate decarboxylase-negative (Pdc−) strains of Saccharomyces cerevisiae an interesting platform for further metabolic engineering of central metabolism. However, Pdc− S. cerevisiae strains have two growth defects: (i) growth on synthetic medium in glucose-limited chemostat cultures requires the addition of small amounts of ethanol or acetate and (ii) even in the presence of a C2 compound, these strains cannot grow in batch cultures on synthetic medium with glucose. We used two subsequent phenotypic selection strategies to obtain a Pdc− strain without these growth defects. An acetate-independent Pdc− mutant was obtained via (otherwise) glucose-limited chemostat cultivation by progressively lowering the acetate content in the feed. Transcriptome analysis did not reveal the mechanisms behind the C2 independence. Further selection for glucose tolerance in shake flasks resulted in a Pdc− S. cerevisiae mutant (TAM) that could grow in batch cultures (μmax = 0.20 h−1) on synthetic medium, with glucose as the sole carbon source. Although the exact molecular mechanisms underlying the glucose-tolerant phenotype were not resolved, transcriptome analysis of the TAM strain revealed increased transcript levels of many glucose-repressible genes relative to the isogenic wild type in nitrogen-limited chemostat cultures with excess glucose. In pH-controlled aerobic batch cultures, the TAM strain produced large amounts of pyruvate. By repeated glucose feeding, a pyruvate concentration of 135 g liter−1 was obtained, with a specific pyruvate production rate of 6 to 7 mmol g of biomass−1 h−1 during the exponential-growth phase and an overall yield of 0.54 g of pyruvate g of glucose−1.
Traditionally, Saccharomyces cerevisiae has been used to rapidly ferment sugars to ethanol and carbon dioxide. More recently, developments in molecular biology have led to the application of S. cerevisiae as a host for therapeutic protein production (13) and for the production of chemicals with commercial value via metabolic engineering (28, 29, 31). In view of the process economy of manufacturing bulk products, the yield of the desired product should be maximized. In the case of yeasts as production organisms, this necessitates the redirection of carbon fluxes away from alcoholic fermentation towards the desired product (1, 4, 7, 8, 19).
Pyruvate decarboxylase (EC 4.1.1.1) is located at the branch point between fermentative and respiratory sugar catabolism and catalyzes the first step in the fermentative branch. S. cerevisiae contains three structural genes (PDC1, PDC5, and PDC6) that encode active pyruvate decarboxylase isoenzymes (18). Pyruvate decarboxylase was long considered to be a strictly catabolic enzyme, but recently a biosynthetic function of the enzyme was discovered (8). Growth of pyruvate decarboxylase-negative (Pdc−) S. cerevisiae in aerobic glucose-limited chemostat cultures on synthetic media required a supply of acetate or ethanol corresponding to ca. 5% of the carbon fed to the cultures (6, 8). This requirement for a C2 compound probably reflects an essential function of pyruvate decarboxylase in the synthesis of cytosolic acetyl-coenzyme A (CoA) (Fig. 1), which is required for lysine and fatty acid synthesis (6).
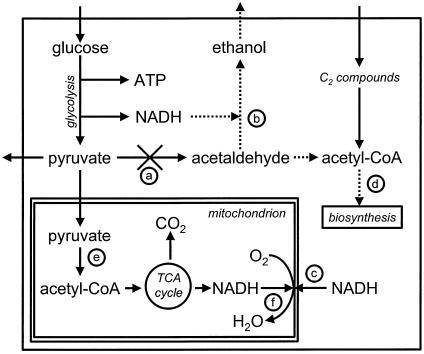
FIG. 1.
Schematic representation of the metabolism of pyruvate decarboxylase-negative S. cerevisiae growing on glucose. By deletion of all genes encoding pyruvate decarboxylase (reaction a), two important processes (dotted lines) are impaired as follows. First, reoxidation of cytosolic NADH via alcohol dehydrogenase (reaction b) is blocked. Cytosolic NADH must therefore be oxidized by the mitochondria via external NADH dehydrogenase (reaction c) or redox shuttle systems. Second, the formation of cytosolic acetyl-CoA from acetaldehyde is blocked. Instead, the C2 compounds required for cytosolic acetyl-CoA for lysine and fatty acid biosynthesis (reaction d) must be taken up from the environment. When oxygen consumption exceeds the amount of oxygen necessary for oxidation of glucose to pyruvate, mitochondrial oxidation of pyruvate, via pyruvate dehydrogenase (reaction e) and the tricarboxylic acid cycle (TCA cycle), can occur, resulting in CO2 formation and the oxidation of NADH via internal NADH dehydrogenase (reaction f).
The overproduction of threonine aldolase, catalyzing the cleavage of threonine to glycine and acetaldehyde, can circumvent the essential biosynthetic role of pyruvate decarboxylase (46). Even when the C2 requirement of Pdc− strains is met by overexpression of threonine aldolase or by inclusion of ethanol or acetate in the medium, Pdc− strains can only grow on glucose when the glucose supply is growth limiting. When Pdc− strains are exposed to the glucose concentrations normally applied in batch cultures, they excrete significant amounts of pyruvate but are completely unable to grow (9). This glucose sensitivity is a general characteristic of Pdc− strains (6, 46).
The exact cause of the glucose sensitivity of Pdc− strains remains unknown. In the absence of alcoholic fermentation, which is blocked in Pdc− S. cerevisiae, cells rely on respiration (Fig. 1) for the reoxidation of cytosolic NADH (33). However, respiration of wild-type S. cerevisiae in batch cultures on glucose is repressed but not blocked, judging from the significant oxygen consumption rate under these conditions (1, 4). It is therefore unlikely that glucose repression of respiration is the sole cause of the glucose sensitivity of Pdc− S. cerevisiae.
From the time of their invention, chemostats have been associated with selection of spontaneous mutants (25, 26). The first chemostat studies had already described the selection of an Escherichia coli strain with a higher affinity for the growth-limiting nutrient (26). Subsequent review articles cited a variety of other examples of selection in chemostats (5, 37) and elaborated on the theory of selection during chemostat cultivation (17). Similarly, extended cultivation of microorganisms in shake flasks can be used to select for spontaneous mutants that grow under conditions in which the original strain would not grow (10, 37).
The first goal of this study was to apply selection pressure in batch and chemostat cultures to obtain a Pdc− S. cerevisiae strain capable of growth in batch cultures on synthetic medium containing high concentrations of glucose as the sole carbon and energy source. Prolonged chemostat cultivation on glucose with progressively decreasing acetate feeds was used to select for C2-independent Pdc− S. cerevisiae. A subsequent round of selection was performed in batch cultures to select for C2-independent Pdc− S. cerevisiae that could grow on high concentrations of glucose. A second goal was to physiologically characterize the selected strain and to gain insight into the molecular mechanisms underlying the selected phenotype. To this end, biomass and product yields were analyzed in batch and chemostat cultures and genome-wide transcriptome analysis was performed, using nitrogen-limited chemostat cultures grown under conditions of excess glucose.
MATERIALS AND METHODS
Strains and maintenance.
All S. cerevisiae strains used for this study (Table 1) were derived from the congenic CEN.PK family (41). Stock cultures were prepared from shake flask or chemostat cultures by the addition of 20% (vol/vol) glycerol to cultures and storage of 2-ml aliquots in sterile vials at −80°C.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| CEN.PK 113-7D | MATaURA3 PDC1 PDC5 PDC6 |
| CEN.PK 182 | MATapdc1(−6,−2)::loxP pdc5(−6,−2)::loxP pdc6(−6,−2)::loxP |
| CEN.PK 111-61A | MATα ura3-52 leu2-112 his3-Δ1 |
| RWB837 | MATapdc1(−6,−2)::loxP pdc5(−6,−2)::loxP pdc6(−6,−2)::loxP ura3-52 |
| RWB837* | MATapdc1(−6,−2)::loxP pdc5(−6,−2)::loxP pdc6(−6,−2)::loxP ura3-52, selected for C2 independence in glucose-limited chemostats |
| TAM | MATapdc1(−6,−2)::loxP pdc5(−6,−2)::loxP pdc6(−6,−2)::loxP ura3-52, selected for C2 independence in glucose-limited chemostats and glucose-tolerant growth in batch culture |
Strain construction.
RWB837 was obtained from a cross between CEN.PK182 and CEN.PK111-61A (constructed by P. Kötter, Frankfurt, Germany, and obtained from Jefferson C. Lievense, Tate and Lyle North America). The resulting diploid was sporulated and the asci were heated for 15 min at 56°C. This random dissection mix was then plated on YP medium, with 0.2% acetate as the carbon source. The resulting colonies were tested for growth on YP medium with glucose or ethanol. Colonies that could not grow on glucose were subsequently checked by PCR for the presence of a disrupted PDC6 gene and for mating type. Determination of the auxotrophic markers present, in this case ura3-52, then gave RWB837. The final strain (designated TAM), selected as described below, was transformed with YEplac195 (14) according to the high-efficiency protocol described by Gietz and Woods (15), resulting in the prototrophic (ura+) TAM+YEplac195 strain.
Chemostat cultivation.
Aerobic carbon-limited or nitrogen-limited chemostat cultivation reactions were performed as described previously (45). To complement auxotrophy, 0.15 g of uracil liter−1 (32) was added to the media. The synthetic medium for the glucose-limited chemostat cultures contained 250 mM substrate carbon. When acetate was present, this was added in addition to the 250 mM carbon from glucose, at concentrations corresponding to 0 to 5% acetate on a substrate-carbon basis. For nitrogen-limited cultures, the glucose concentration in the synthetic medium was adjusted during a trial run to result in a residual glucose concentration in the culture broth of approximately 100 mM. Afterwards, reproducible duplicate cultures were obtained at this glucose concentration.
Shake flask cultivation.
The 500-ml shake flasks, containing 100 ml of synthetic medium (49), were incubated at 30°C in a rotary shaker (200 rpm). To rescue auxotrophy, 0.15 g of uracil liter−1 (32) was added to the media. Precultures of RWB837 were grown in 2% ethanol. For all other shake flask cultures, glucose was used as the carbon source, with concentrations ranging from 2 to 10% (wt/vol). The selected strain was routinely checked for uracil auxotrophy to verify culture purity.
Fermenter batch cultivation.
Aerobic batch cultivation was performed at 30°C in 2-liter fermenters (Applikon, Schiedam, The Netherlands) with a working volume of 1 liter. The pH was controlled at 5.0 via automated addition of 10 M KOH (Applikon ADI 1030 biocontroller). The dissolved-oxygen concentration was maintained above 10% air saturation at all times by adjusting the stirrer speed between 800 and 1,000 rpm and the airflow between 0.50 and 0.75 liter min−1. A synthetic medium with twice the concentrations described by Verduyn et al. (49) was used for cultures. The initial glucose concentration was 100 g liter−1. During the repeated batch cultivation, 100 g of nonsterile solid glucose was added twice, at 32 and 48 h after inoculation. Antifoam (BDH) product was added to the fermenters when required. Culture purity was checked microscopically at the end of the fermentation and no contaminants were observed.
Microarray analysis.
Sampling of cells from chemostats, probe preparation, and hybridization to Affymetrix GeneChip microarrays were performed as described previously (30). The results were derived from two independent replicate cultures for the selected Pdc− strain and from three independent replicate cultures for the wild type.
Microarray data acquisition and analysis.
Acquisition and quantification of array images and data filtering were performed with the Affymetrix software packages Microarray Suite, v. 5.0, MicroDB, v. 3.0, and Data Mining Tool, v. 3.0. For further statistical analysis, Microsoft Excel running the Significance Analysis of Microarrays (v. 1.12) add-in was used, with a delta value that corresponded to the minimum expected median false-positive rate and a minimum change of twofold (40). In our experience, these criteria establish a data set that is able to be reproduced by an independent laboratory (30).
Before comparison, all arrays were globally scaled to a target value of 150, using the average signal from all gene features, with Microarray Suite, v. 5.0. For the 9,335 transcript features on the YG-S98 arrays, a filter was applied to extract 6,383 yeast open reading frames, of which 6,084 were different genes. This discrepancy was due to several genes being represented more than once when suboptimal probe sets were used in the array design. Since the 900 transcripts with the lowest transcription level could not be reliably measured, their level was set to a value of 12 for the comparison analyses.
Promoter analyses were performed with the web-based software Regulatory Sequence Analysis Tools (42), as described previously (2).
Analytical procedures.
Dry weight determination, glucose, acetate, and metabolite analysis, off-gas analysis, and pyruvate decarboxylase and threonine aldolase assays were performed as described previously (46). The protein content of whole cells was determined by a modified biuret method (48).
RESULTS
Selection of C2-independent Pdc− S. cerevisiae in chemostat cultures.
For this study, the power of chemostat cultivation as a tool for the selection of microorganisms (5, 17) was used in an attempt to eliminate the C2 compound requirement of Pdc− S. cerevisiae (6, 8). A pdc1,5,6Δ ura3Δ S. cerevisiae strain (RWB837) was used for selection. The ura3Δ auxotrophic marker was used to facilitate controls for culture purity. First, a steady state of Pdc− S. cerevisiae on a mixture of 5% acetate and 95% (on the basis of carbon) glucose was established. The metabolism of this culture was fully respiratory, as indicated by a respiratory quotient of just over one carbon dioxide molecule produced per oxygen molecule consumed. The biomass yield on carbon was 14.6 g of biomass mol of carbon−1, and all glucose and acetate were consumed. The acetate content of the synthetic medium was then reduced in five consecutive steps, from 5% of the total carbon content to zero. Each step lasted five volume changes. During this slow transition, RWB837 adapted to growth in aerobic carbon-limited chemostat cultures, with glucose as the sole carbon source, at a dilution rate of 0.10 h−1. The biomass yield on substrate (14.7 g of biomass mol of carbon−1), oxygen consumption rate, and carbon dioxide production rate (both around 2.9 mmol g of biomass−1 h−1) of this glucose-limited culture indicated respiratory carbon metabolism of the C2-independent Pdc− S. cerevisiae culture, as was observed with wild-type S. cerevisiae under these conditions (43).
Transcriptome analysis of the C2-independent Pdc− S. cerevisiae strain.
Transcriptome analysis of the glucose-limited chemostat culture of the C2-independent S. cerevisiae strain was performed to study the genetic changes responsible for C2 independence. The C2-independent Pdc− strain was compared to glucose-limited chemostat cultures of the wild type (30). Of the genes with a known function, only 18 were upregulated and only 16 were downregulated in the selected strain. These upregulated genes included 11 that were involved in meiosis or sporulation (HOP2, IME2, REC102, REC104, RED1, SLZ1, SPO13, SPO16, SPR1, YER179W, and ZIP1). The other seven upregulated genes were CAR1, ECM1, HXT3, HXT4, IRE1, NUF1, and NUF2. The downregulated genes included four expected genes (PDC1, PDC5, PDC6, and URA3) and, in addition, ALP1, AQY1, GND2, FUI1, HSP30, HXT5, MEP2, MLS1, PDR12, PHO4, SSA3, and SSA4. None of these genes had a clear link to the C2 independence of the selected mutant. Transcript levels of the GLY1 gene, overexpression of which alleviates the C2 requirement of Pdc− S. cerevisiae (46), were not significantly changed in the selected strain.
Selection for glucose tolerance in shake flask cultures.
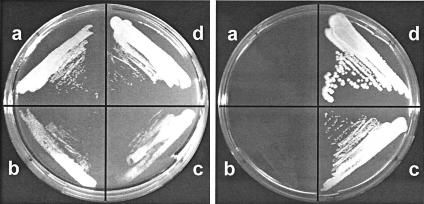
After selection for C2 independence, a small aliquot of the chemostat culture was transferred to a shake flask with synthetic medium containing uracil and 20 g of glucose liter−1. As was expected from previous results, neither the original Pdc− S. cerevisiae nor the C2-independent Pdc− strain grew on agar plates with synthetic medium, uracil, and 2% glucose (Fig. 2, right panel), whereas both strains did grow on agar plates with synthetic medium, uracil, and 2% ethanol (Fig. 2, left panels). In agreement with this, no growth was observed during the first 7 days of the initial shake flask culture of the C2-independent Pdc− strain on 2% glucose. Prolonged cultivation of the C2-independent Pdc− strain, however, resulted in significant biomass formation, indicating an accumulation of spontaneous glucose-tolerant mutants. The observed growth rate was well below 0.01 h−1. After growth had ceased, which occurred at a relatively low biomass density due to acidification of the culture by pyruvic acid accumulation, 1 ml of the culture was transferred to a new shake flask with identical synthetic medium.
FIG. 2.
Growth of three Pdc− S. cerevisiae strains and wild-type S. cerevisiae on synthetic medium agar plates with ethanol (left plate) or glucose (right plate) as the carbon source. Both plates were supplemented with uracil to alleviate the auxotrophy of the Pdc− S. cerevisiae strains. Ethanol plates were incubated for 7 days, and glucose plates were incubated for 3 days. Strains: a, RWB837 (Pdc− S. cerevisiae); b, RWB837* (selected C2-independent S. cerevisiae); c, TAM (selected C2-independent and glucose-tolerant Pdc− S. cerevisiae); d, CEN.PK 113-7D (wild type).
The process of serial transfer was repeated 27 times in total. The specific growth rate of the Pdc− strain after the sixth transfer was already 0.10 h−1 on 20 g of glucose liter−1. After 14 shake flask cultivations and an obtained specific growth rate of 0.18 h−1, the glucose content of the medium was raised to 32, 54, 69, and 100 g liter−1 in consecutive cultures. At 100 g of glucose liter−1, the finally obtained C2-independent, glucose-tolerant Pdc− S. cerevisiae culture grew at a specific growth rate of 0.20 h−1.
The culture, possibly consisting of a mixture of different spontaneous mutants, was streaked onto agar plates containing synthetic medium, glucose, and uracil. Four of the resulting colonies were tested for growth in shake flasks on glucose, and no significant differences in specific growth rate were observed. One of these cultures was chosen for further study, and this C2-independent glucose-tolerant Pdc− S. cerevisiae strain will be referred to as TAM in this and future work.
The differences in growth among the original Pdc− strain (RWB837), the C2-independent Pdc− strain, the TAM strain, and the isogenic wild-type strain on synthetic medium, with glucose or ethanol as the sole carbon source, were clearly demonstrated by agar plate growth, as depicted in Fig. 2. Although the TAM strain displayed a 3-day-longer lag phase, all four strains grew on plates with ethanol as the carbon source. As described above, when glucose was the carbon source, the original Pdc− strain (RWB837) and the C2-independent Pdc− strain did not grow. Consistent with the growth in shake flasks with glucose, the selected TAM strain, and of course the wild type, proliferated well on the agar plates with glucose (Fig. 2).
Pyruvate production by the selected TAM strain in fermenter cultures.
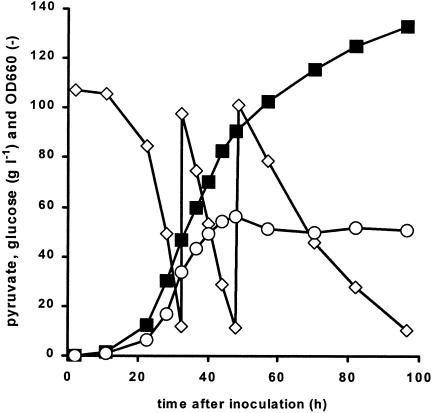
During selection for glucose tolerance in shake flasks, rapid acidification of the culture due to pyruvate excretion was observed. To study the growth and pyruvate production of the TAM strain under controlled conditions, aerobic batch cultivations with 100 g of glucose liter−1 were performed in fermenters at a constant pH of 5.0. During the exponential-growth phase (Fig. 3), the maximum specific growth rate of the TAM strain was 0.20 h−1, which was equal to the maximum specific growth rate in shake flasks. Consistent with the observations with shake flasks, large amounts of pyruvate were produced in fermenter cultures. The rate of pyruvate production during the exponential-growth phase was 6 to 7 mmol g of biomass−1 h−1. In the first 40 h of this batch, starting with a low biomass concentration (optical density at 660 nm [OD660], 0.1), a pyruvate concentration of 50 g liter−1 was obtained, with a yield of 0.55 g of pyruvate g of glucose−1.
FIG. 3.
Growth and pyruvate production during an aerobic repeated batch culture on glucose of the selected TAM strain. The results shown are from one representative batch experiment. Biomass and pyruvate concentrations in independent replicate experiments varied by <5%. Closed squares, pyruvate concentration; open symbols, glucose concentration (diamonds) or OD660 (circles).
For further assessment of the potential of the TAM strain to produce pyruvate, the fermentation was continued as a repeated batch by the addition of solid glucose to the fermenter (Fig. 3). During this repeated batch phase, the specific growth rate gradually decreased and growth ultimately ceased, probably due to nutrient limitations in the medium. The pyruvate concentration in the supernatant exceeded 100 g liter−1 within 60 h of inoculation of the fermenter. The final concentration of pyruvate obtained after 100 h was 135 g liter−1, with an overall yield of 0.54 g of pyruvate g of glucose−1.
Glucose-limited chemostat cultivation of the TAM strain.
The obtained maximum specific growth rate of the TAM strain in batch culture with glucose was 0.20 h−1. Under the same conditions, wild-type S. cerevisiae CEN.PK 113-7D grew with a higher maximum specific growth rate of 0.37 h−1 (data not shown). For further study of this deviation in growth, glucose-limited chemostat cultures of the TAM strain were performed at increasing dilution rates. At a dilution rate of 0.10 h−1, glucose dissimilation by the TAM strain was fully respiratory, without the accumulation of metabolites. Except for a lower biomass yield of the selected strain (0.43 g of biomass g of glucose−1, compared to 0.48 g of biomass g of glucose−1 for the wild type), physiological parameters were comparable to those of the wild type (43). At a dilution rate of 0.15 h−1, the biomass yield of the TAM strain had increased to 0.47 g of biomass g of glucose−1, which is still lower than the 0.50 g of biomass g of glucose−1 for the wild type at this dilution rate. The TAM strain was capable of growth at a dilution rate of 0.20 h−1 in glucose-limited chemostat cultures, but this growth was accompanied by a variable pyruvate production rate (0.25 to 0.45 mmol of pyruvate g of biomass−1 h−1). At a dilution rate of 0.23 h−1, the TAM strain washed out of the chemostats, indicating a maximum specific growth rate of this strain between 0.20 and 0.23 h−1 in glucose-limited chemostat cultures. After this prolonged glucose-limited chemostat cultivation, an aliquot of the culture was transferred to a shake flask with 100 g of glucose liter−1. With this shake flask, rapid growth was observed, indicating that the culture had maintained its glucose-tolerant phenotype.
Nitrogen-limited chemostat cultivation of the TAM strain.
Comparison of the selected TAM strain with wild-type S. cerevisiae CEN.PK 113-7D is best performed at high glucose concentrations and in the absence of C2 compounds in the medium. Nitrogen-limited chemostat cultivation at the same dilution rate, with glucose as the sole carbon source, combines these conditions with the advantages of chemostat cultivation for reproducible physiological studies. The glucose concentrations in the synthetic medium were chosen such that approximately the same residual glucose concentration was obtained in the cultivations of both strains (Table 2).
TABLE 2.
Physiology of strain TAM (C2-independent, glucose-tolerant Pdc− S. cerevisiae) and the isogenic wild-type strain CEN.PK 113-7D in aerobic nitrogen-limited chemostat culture at a dilution rate of 0.10 h−1a
| Characteristicb | Wild type | TAM |
|---|---|---|
| Reservoir glucose concentration (g liter−1) | 58.8 ± 0.1 | 35.1 ± 0.1 |
| Residual glucose concentration (g liter−1) | 16.7 ± 0.7 | 20.4 ± 0.1 |
| Ysx (gbiomass gglucose−1) | 0.09 ± 0.00 | 0.21 ± 0.00 |
| Ynx (gbiomass gN−1) | 18.8 ± 0.1 | 14.7 ± 0.1 |
| Protein content (gprotein gbiomass−1) | 0.29 ± 0.01 | 0.33 ± 0.01 |
| Respiratory quotient | 4.5 ± 0.2 | 0.70 ± 0.01 |
| qglucose (mmol gbiomass−1 h−1) | 5.8 ± 0.1 | 2.6 ± 0.1 |
| qethanol (mmol gbiomass−1 h−1) | 8.0 ± 0.1 | <0.01 |
| qpyruvate (mmol gbiomass−1 h−1) | 0.1 ± 0.0 | 2.8 ± 0.0 |
| qglycerol (mmol gbiomass−1 h−1) | 0.08 ± 0.00 | <0.01 |
| qacetate (mmol gbiomass−1 h−1) | 0.06 ± 0.02 | <0.01 |
| qCO2 (mmol gbiomass−1 h−1) | 12.1 ± 0.2 | 2.8 ± 0.0 |
| qO2 (mmol gbiomass−1 h−1) | 2.7 ± 0.1 | 4.0 ± 0.1 |
| Recovery of consumed carbon (%) | 94.0 ± 1.0 | 97.4 ± 0.7 |
| Total carbon recovery (%) | 95.6 ± 0.7 | 98.6 ± 0.4 |
The wild-type data were obtained from the same cultures as those used by Boer et al. (2).
Averages and mean deviations were obtained from duplicate (TAM) and triplicate (wild type) experiments with independent steady-state cultures. Calculations of carbon recovery were based on a 48% (wt/vol) carbon content of the biomass. Ysx and Ynx are the biomass yields on glucose and nitrogen, respectively. Specific production or consumption rates are indicated with a q.
The wild type showed alcoholic fermentation, as is characteristic for S. cerevisiae under conditions of excess glucose. This resulted in a low biomass yield on glucose (0.09 g of biomass g of glucose−1), an ethanol production rate of 8.0 mmol g of biomass−1 h−1, and a respiratory quotient of 4.5 mmol of carbon dioxide produced per mmol of oxygen consumed. The protein content (0.29 g of protein g of biomass−1) and the biomass yield on nitrogen (18.8 g of biomass g of nitrogen−1) were in good agreement with previously published values (44, 45) for nitrogen-limited chemostat cultures of wild-type strain CEN.PK 113-7D.
Under the same conditions, the TAM strain, which fully depends on respiration in the absence of alcoholic fermentation, had a higher biomass yield on glucose (0.21 g of biomass g of glucose−1) and produced pyruvate as the only major by-product, at a rate of 2.8 mmol g of biomass−1 h−1 (Table 2). The oxygen consumption rate was 4.0 mmol g of biomass−1 h−1, compared to 2.7 mmol g of biomass−1 h−1 for the wild type. The respiratory oxidation of the NADH formed during pyruvate formation lowered the respiratory quotient to 0.70 mmol of carbon dioxide produced per mmol of oxygen consumed. The protein content of the biomass was slightly higher for the TAM strain (0.33 g of protein g of biomass−1) than for the wild type (0.29 g of protein g of biomass−1). This higher protein content of the cells partially explains the significantly lower yield on nitrogen of the TAM strain (14.7 g of biomass g of nitrogen−1) than of the wild type (18.8 g of biomass g of nitrogen−1).
Transcriptome analysis of the TAM strain.
Central in transcriptome analysis is the choice of adequate culture conditions for the comparison. In the case of the selected TAM strain, the absence of C2 compounds and the presence of high levels of glucose in the broth are typical for uncovering its phenotype. To combine the benefit of chemostat cultures in microarray studies (30) and the requirement for excess glucose, the nitrogen-limited chemostat cultures of the TAM strain and the isogenic wild-type strain CEN.PK 113-7D were chosen for the transcriptome analysis.
The comparison of the nitrogen-limited chemostat cultures revealed 305 genes of which the mRNA levels were significantly changed and at least twofold higher in the TAM strain than in the wild type. The mRNA abundance of 168 genes was significantly changed and at least twofold lower in the TAM strain than in the wild type. In total, these changed genes comprise almost 8% of the total S. cerevisiae genome. Of these changed genes, 273 (58%) have an unknown function, which is higher than the percentage of not fully annotated genes in the whole S. cerevisiae genome (47%).
Sequence analysis of the upstream regions of genes that are upregulated in the selected strain showed an overrepresentation of possible Mig1p-binding sites among these genes, indicating an (partial) alleviation of Mig1p-mediated repression. Although the transcript level of the primarily posttranscriptionally (12) regulated MIG1 was not changed, the transcript level of its close homologue MIG2 was downregulated almost 11-fold. Many genes required for growth on carbon sources other than glucose were upregulated in the TAM strain. This included genes involved in gluconeogenesis and ethanol utilization (ACS1, ADH2, ADR1, CAT8, FBP1, and SIP4), fatty acid metabolism (CAT2, CRC1, ECI1, FAA2, FOX2, PEX11, POT1, POT1, and YAT2), galactose metabolism (GAL2, GAL3, and GAL4), maltose metabolism (MPH2, MPH3, and YFL052W), and pyruvate and lactate metabolism (DLD1 and JEN1).
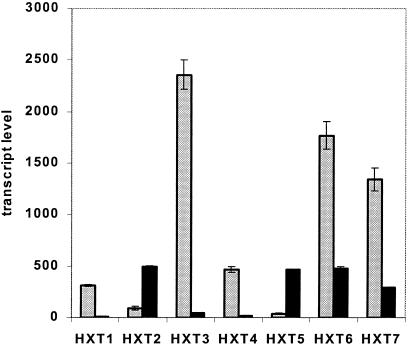
A striking observation was the change in expression of the genes coding for the hexose transporters. Despite the high glucose concentrations under nitrogen limitation, the low-affinity transporters (HXT1 and HXT3) were downregulated 50-fold in the TAM strain compared to in the wild type (Fig. 4). The known high-affinity transporters (HXT6 and HXT7) were also downregulated (fourfold) in this strain (Fig. 4). As a result, the summed transcript abundance of all HXT genes represented on the arrays (HXT1 to HXT10, HXT12, HXT14, and HXT16) was four times lower in the TAM strain under nitrogen limitation than in the wild type. In the glucose-responsive regulatory network of the HXT genes (35), the only significant transcriptional change was the 12-fold downregulation of STD1, a glucose-concentration-dependent modulator of expression, in the TAM strain compared to the wild type.
FIG. 4.
Transcript-level comparison of the main hexose transporter genes (HXT1 to -7) between the selected TAM strain and its isogenic wild type in nitrogen-limited chemostat cultures with glucose as the carbon source. The wild-type data were obtained from the same cultures used by Boer et al. (2). The data represented were obtained from independent duplicate (TAM) or triplicate (wild type) chemostat cultivations. Gray bars, wild type; black bars, TAM strain. Error bars represent standard deviations.
Since the TAM strain was not only glucose tolerant, but also independent of C2 compounds for growth on glucose, it would be interesting to know to what extent transcriptional changes are responsible for the obtained C2 independence. The transcript abundance of GLY1, a previously demonstrated source of cytosolic C2 compounds (46), was upregulated 2.5-fold in the TAM strain. However, the threonine aldolase activity in the TAM strain was still below the detection limit of 0.005 U mg of protein−1. The derepressed genes of fatty acid metabolism can also be interpreted as a modification in the metabolism of cytosolic C2 compounds. In addition, two genes (CAR1 and CAR2) for arginine metabolism, which includes reactions involving acetyl-CoA, were upregulated sixfold in the TAM strain. Thus, no immediate and complete explanation for the C2 independence of this strain could be established via transcriptome analysis.
A surprisingly large group of upregulated genes in the selected Pdc− strain were involved in mating (3 genes), meiosis (17 genes), and sporulation (8 genes). Since both the original strain (RWB837) and the selected Pdc− strain are confirmed haploids, the mechanism behind and the origin of the expression of these genes, including the early meiotic transcription factor IME1, remain unknown. Sequence analysis of the upstream regions of the upregulated genes in the selected Pdc− strain also clearly showed an overrepresentation of the binding sites for UME6 and IME1, both involved in the regulation of meiosis.
DISCUSSION
Selection of C2-independent Pdc− S. cerevisiae mutants.
Progressive reduction of the acetate feed during prolonged chemostat cultivation resulted in selection of a Pdc− strain that did not require the addition of C2 compounds to the growth environment (Fig. 1). Transcriptome analysis did not reveal the mechanism underlying the physiological changes in this strain. However, based on physiology, some possible sources of cytosolic acetyl-CoA can be excluded.
Massive overexpression of GLY1, encoding threonine aldolase, has been shown to circumvent the essential biosynthetic role of pyruvate decarboxylase in Pdc− S. cerevisiae (45). Despite the higher GLY1 transcript levels in the TAM strain, the low affinity of Gly1p for threonine (Km, 55 mM [22]) and the relatively low intracellular threonine concentration (5 to 10 mM [16, 23]), combined with the low in vitro activity (<0.005 U mg of protein−1), make it unlikely that threonine aldolase is responsible for its C2-independent phenotype. In addition, the absence from S. cerevisiae of de novo carnitine biosynthesis (47) and of ATP-citrate lyase (34) eliminates the carnitine shuttle and citrate efflux from the mitochondria as possible sources of cytosolic acetyl-CoA. In vitro pyruvate decarboxylase activity was not observed, nor was acetate or ethanol detected under any of the tested conditions.
Selection of glucose-tolerant Pdc− S. cerevisiae mutants.
The exact reason for the inability of Pdc− strains to grow on high concentrations of glucose, even in the presence of C2 compounds or with a threonine aldolase overproduction construct, is still not known. Respiration is obviously essential for the reoxidation of cytosolic NADH formed by glycolysis (Fig. 1). However, the significant respiration rate of wild-type S. cerevisiae (1, 4, 20) under conditions of excess glucose suggests that oxidative regeneration of cytosolic NADH, and therefore ATP synthesis via glycolysis and oxidative phosphorylation, should also be possible in Pdc− S. cerevisiae under these conditions. Furthermore, any surplus respiratory capacity can be used for oxidative dissimilation by the mitochondria. What then causes the inability of Pdc− S. cerevisiae to grow under conditions of excess glucose?
This study does not answer the question satisfactorily. However, transcriptome analysis indicated a partial relief of repression of genes, with a possible Mig1p-binding box in the upstream sequence and a fourfold decrease in transcript levels of genes involved in glucose uptake. Combined deletion of MIG1 and MIG2 has been reported to increase the respiratory capacity and specific growth rate of S. cerevisiae in aerobic glucose-grown cultures (19). In addition, it has been shown by a study of phosphoglycerate mutase-negative S. cerevisiae that partial alleviation of carbon catabolite repression in a suppressor mutant coincided with a decreased glucose uptake capacity (11). Interestingly, transcript levels of HAP4, encoding a known positive transcriptional regulator of genes involved in respiratory metabolism (27, 36, 39), and HXK2, encoding a central protein in glucose repression (3), were not affected in the selected strain.
Recently, Boer et al. (2) presented a four-way transcriptome analysis of glucose-, nitrogen-, phosphate-, and sulfur-limited chemostat cultivations of wild-type S. cerevisiae CEN.PK 113-7D. Their analysis yielded a subset of genes that were uniquely up (164 genes) or down (62 genes) regulated in aerobic glucose-limited chemostat cultivations. Fifty-seven percent of these genes were also found to be up (94 genes) or down (35 genes) regulated in the nitrogen-limited chemostat cultures of the TAM strain relative to nitrogen-limited chemostat cultures of the isogenic wild type. This resemblance to glucose-limited wild-type cultures, the upregulation of genes with a possible Mig1p-binding box, and the fourfold decrease in HXT transcript levels all support the hypothesis that a release of glucose catabolite repression contributes to the glucose tolerance of the selected Pdc− strain. Testing of this hypothesis by thorough analysis of Pdc− strains, with additional deletions in transcriptional regulators involved in glucose catabolite repression, is currently in progress.
DNA microarrays as a diagnostic tool for yeast strain improvement.
To maximize the quality of the transcriptome data, the TAM strain and an isogenic reference strain were grown in replicate chemostat cultures under identical, carefully defined conditions. Although a substantial number of genes yielded a significantly altered transcript level, this did not give clear insight into the molecular mechanisms responsible for C2 independence. In the case of glucose tolerance, only a possible correlation with glucose catabolite repression was observed. This demonstrates that the application of high-information-density analytical tools will not always yield clear-cut answers to physiological questions. DNA microarrays, however valuable for identifying changes in transcript level, cannot identify many other relevant changes, such as point mutations or changes in posttranscriptional regulation. The absence of a clearly established biological function for many of the differentially transcribed genes further complicates interpretation of transcriptome analysis. Taking into account that similar constraints exist for proteome and metabolome analysis, we have to conclude that the full analysis of complex phenotypes will continue to require time- and labor-intensive genetic dissection of the genotypes of selected strains.
Metabolic fluxes in the TAM strain and wild-type S. cerevisiae.
During carbon-limited growth at a dilution rate of 0.10 h−1, both the TAM strain and wild-type S. cerevisiae displayed full respiratory glucose metabolism, although the biomass yield of the TAM strain was almost 10% lower. In nitrogen-limited chemostat cultures at a dilution rate of 0.10 h−1, a more suitable environment for investigating the selected glucose-tolerant phenotype, the oxygen consumption rate of the TAM strain (4.0 mmol g of biomass−1 h−1) was higher than that of the wild type (2.7 mmol g of biomass−1 h−1) (Table 2). Interestingly, the increase in oxygen consumption rate (4.0 − 2.7 = 1.3 mmol g of biomass−1 h−1) almost equals that required to regenerate the cytosolic NADH formed during pyruvate production (0.5 × 2.8 = 1.4 mmol g of biomass−1 h−1). The rate of oxidative pyruvate dissimilation by the mitochondria is apparently identical in nitrogen-limited chemostat cultures of the TAM strain and of wild-type S. cerevisiae.
Production of pyruvate by the selected TAM strain.
The excretion of pyruvate by S. cerevisiae with reduced or absent pyruvate decarboxylase activity has been observed before (9, 38). The TAM strain, however, displays rapid growth on synthetic medium with glucose as the sole carbon source, whereas other Pdc− S. cerevisiae strains completely fail to grow under these conditions. The TAM strain has the additional benefit of the complete absence of ethanol as a by-product and, in contrast to many other pyruvate-producing microorganisms, does not require the addition or omission of specific compounds in the medium (21). The final pyruvate concentration of 135 g liter−1 (Fig. 3) is almost twofold higher than the highest concentration previously obtained by fermentation (24). The high specific rate of pyruvate production (6 to 7 mmol of pyruvate g of biomass−1 h−1) resulted in 100 g of pyruvate liter−1, starting with a low density inoculum (OD660 of 0.1), in <60 h. The amount of pyruvate obtained was 0.54 g of pyruvate g of glucose−1 for both the repeated batch and the nitrogen-limited chemostat cultures of the TAM strain. This yield is lower than the highest reported yield in the public domain (0.68 g of pyruvate g of glucose−1) (24). Engineering of respiratory pyruvate degradation, either by decreasing internal mitochondrial respiration or by decreasing the activity of the pyruvate dehydrogenase complex, might result in improvement of the pyruvate yield on glucose by the TAM strain. Even allowing for its suboptimal pyruvate yield, the high final pyruvate concentration (135 g liter−1) and pyruvate production rate (6 to 7 mmol g of biomass−1 h−1) already make the TAM strain a serious contender for industrial pyruvate production via microbial fermentation (21).
Acknowledgments
This work was supported by Tate and Lyle North America. The research group of J.T.P. is part of the Kluyver Centre for Genomics of Industrial Fermentation, which is supported by The Netherlands Genomics Initiative.
We thank Jefferson C. Lievense and Chi-Li Liu for stimulating discussions and for critically reading the manuscript and our colleagues Marijke Luttik and Viktor Boer for experimental assistance.
REFERENCES
- 1.Blom, J., M. J. Teixeira de Mattos, and L. A. Grivell. 2000. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl. Environ. Microbiol. 66:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. W. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 3.Diderich, J. A., L. M. Raamsdonk, A. L. Kruckeberg, J. A. Berden, and K. van Dam. 2001. Physiological properties of Saccharomyces cerevisiae from which hexokinase II has been deleted. Appl. Environ. Microbiol. 67:1587-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diderich, J. A., L. M. Raamsdonk, A. Kuiper, A. L. Kruckeberg, M. J. Teixeira de Mattos, and K. van Dam. 2002. Effects of a hexokinase II deletion on the dynamics of glycolysis in continuous cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 2:165-172. [DOI] [PubMed] [Google Scholar]
- 5.Dykhuizen, D. E., and D. L. Hartl. 1983. Selection in chemostats. Microbiol. Rev. 47:150-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flikweert, M. T., M. de Swaaf, J. P. van Dijken, and J. T. Pronk. 1999. Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol. Lett. 174:73-79. [DOI] [PubMed] [Google Scholar]
- 7.Flikweert, M. T., M. Kuyper, A. J. A. van Maris, P. Kötter, J. P. van Dijken, and J. T. Pronk. 1999. Steady-state and transient-state analysis of growth and metabolite production in a Saccharomyces cerevisiae strain with reduced pyruvate-decarboxylase activity. Biotechnol. Bioeng. 66:42-50. [DOI] [PubMed] [Google Scholar]
- 8.Flikweert, M. T., L. van der Zanden, W. M. T. M. Janssen, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1996. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12:247-257. [DOI] [PubMed] [Google Scholar]
- 9.Flikweert, M. T., J. P. van Dijken, and J. T. Pronk. 1997. Metabolic response of pyruvate-decarboxylase-negative Saccharomyces cerevisiae to glucose excess. Appl. Environ. Microbiol. 63:3399-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, P. L. 2000. Adaptive mutation: implications for evolution. BioEssays 22:1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamo, F.-J., M. J. Lafuente, and C. Gancedo. 1994. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 176:7423-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellissen, G., K. Melber, Z. A. Janowicz, U. M. Dahlems, U. Weydemann, M. Piontek, A. W. Strasser, and C. P. Hollenberg. 1992. Heterologous protein production in yeast. Antonie Leeuwenhoek 62:79-93. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Hans, M. A., E. Heinzle, and C. Wittmann. 2001. Quantification of intracellular amino acids in batch cultures of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 26:776-779. [DOI] [PubMed] [Google Scholar]
- 17.Harder, W., J. G. Kuenen, and A. Matin. 1977. Microbial selection in continuous culture. J. Appl. Bacteriol. 43:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Hohmann, S. 1991. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J. Bacteriol. 173:7963-7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein, C. J. L., J. J. Rasmussen, B. Rønnow, L. Olsson, and J. Nielsen. 1999. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J. Biotechnol. 68:197-212. [DOI] [PubMed] [Google Scholar]
- 20.Larsson, C., U. von Stockar, I. Marison, and L. Gustafsson. 1993. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J. Bacteriol. 175:4809-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., J. Chen, and S. Y. Lun. 2001. Biotechnological production of pyruvic acid. Appl. Microbiol. Biotechnol. 57:451-459. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J.-Q., S. Nagata, T. Dairi, H. Misono, S. Shimizu, and H. Yamada. 1997. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific l-threonine aldolase that catalyzes cleavage of l-allo-threonine and l-threonine to glycine. Eur. J. Biochem. 245:289-293. [DOI] [PubMed] [Google Scholar]
- 23.Messenguy, F., D. Colin, and J. P. ten Have. 1980. Regulation of compartmentation of amino acid pools in Saccharomyces cerevisiae and its effects on metabolic control. Eur. J. Biochem. 180:439-447. [DOI] [PubMed] [Google Scholar]
- 24.Miyata, R., and T. Yonehara. 1999. Breeding of high-pyruvate-producing Torulopsis glabrata with acquired reduced pyruvate decarboxylase. J. Biosci. Bioeng. 88:173-178. [DOI] [PubMed] [Google Scholar]
- 25.Novick, A., and L. Szilard. 1950. Description of the chemostat. Science 112:715-716. [DOI] [PubMed] [Google Scholar]
- 26.Novick, A., and L. Szilard. 1950. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA 36:708-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olesen, J., S. Hahn, and L. Guarente. 1987. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell 51:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Ostergaard, S., L. Olsson, and J. Nielsen. 2000. Metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 64:34-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overkamp, K. M., B. M. Bakker, P. Kötter, M. Luttik, J. P. van Dijken, and J. T. Pronk. 2002. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68:2814-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piper, M. D. W., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses: an interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 31.Porro, D., L. Brambilla, B. M. Ranzi, E. Martegani, and L. Alberghina. 1995. Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol. Prog. 11:294-298. [DOI] [PubMed] [Google Scholar]
- 32.Pronk, J. T. 2002. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 68:2095-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pronk, J. T., H. Y. Steensma, and J. P. van Dijken. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607-1633. [DOI] [PubMed] [Google Scholar]
- 34.Ratledge, C., and C. T. Evans. 1989. Lipids and their metabolism, p. 367-455. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 3. Academic Press, San Diego, Calif.
- 35.Rolland, F., J. Winderickx, and J. M. Thevelein. 2002. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2:183-201. [DOI] [PubMed] [Google Scholar]
- 36.Rosenkrantz, M., C. S. Kell, E. A. Pennell, and L. J. Devenish. 1989. The HAP2, 3, 4 transcriptional activator is required for derepression of the yeast citrate synthase gene, CIT1. Mol. Microbiol. 13:119-131. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 73:130-169. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, H. D., and F. K. Zimmermann. 1982. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J. Bacteriol. 151:1146-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, J. C., and L. Guarente. 1991. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 11:4934-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. F. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindeløv, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 42.van Helden, J., B. André, and J. Collado-Vides. 2000. A web site for the computational analysis of yeast regulatory sequences. Yeast 16:177-187. [DOI] [PubMed] [Google Scholar]
- 43.van Hoek, P., M. T. Flikweert, Q. J. M. van der Aart, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1998. Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branchpoint in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64:2133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoek, P., J. P. van Dijken, and J. T. Pronk. 2000. Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb. Technol. 26:724-736. [DOI] [PubMed] [Google Scholar]
- 45.van Maris, A. J. A., B. M. Bakker, M. Brandt, A. Boorsma, M. J. Teixeira de Mattos, A. R. Grivell, J. T. Pronk, and J. Blom. 2001. Modulating the distribution of fluxes among respiration and fermentation by overexpression of HAP4 in Saccharomyces cerevisiae. FEMS Yeast Res. 1:139-149. [DOI] [PubMed] [Google Scholar]
- 46.van Maris, A. J. A., M. A. H. Luttik, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2003. Overproduction of threonine aldolase circumvents the biosynthetic role of pyruvate decarboxylase in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2094-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Roermund, C. W. T., E. H. Hettema, M. van den Berg, H. F. Tabak, and R. J. A. Wanders. 1999. Molecular characterization of carnitine dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2. EMBO J. 18:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:395-403. [DOI] [PubMed] [Google Scholar]
- 49.Verduyn, C., T. P. L. Zomerdijk, J. P. van Dijken, and W. A. Scheffers. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]