Abstract
G Protein-Coupled Receptors (GPCRs) comprise a large family of seven-helix transmembrane proteins which regulate cellular signaling by sensing light, ligands, and binding proteins. The GPCR activation process, however, is not a simple on-off switch; current models suggest a complex conformational landscape in which the active, signaling state includes multiple conformations with similar downstream activity. The present study probes the conformational dynamics of single β2-Adrenergic Receptors (β2ARs) in the solution phase by Anti-Brownian ELectrokinetic (ABEL) trapping. The ABEL trap uses fast electrokinetic feedback in a microfluidic configuration to allow direct observation of a single fluorescently-labeled β2AR for hundreds of milliseconds to seconds. By choosing a reporter dye and labeling site sensitive to ligand binding, we observe a diversity of discrete fluorescence intensity and lifetime levels in single β2ARs, indicating a varying radiative lifetime and a range of discrete conformational states with dwell times of hundreds of milliseconds. We find that binding of agonist increases the dwell times of these states, and furthermore, we observe millisecond fluctuations within states. The intensity autocorrelations of these faster fluctuations are well-described by stretched exponential functions with stretching exponent β ~ 0.5, suggesting protein dynamics over a range of timescales.
INTRODUCTION
G protein-coupled receptors (GPCRs) form a class of over 800 membrane proteins that regulate a large variety of physiological processes by specifically transducing signals (photons, ions, small molecules, peptide and protein hormones) from the outside to the inside of a cell.1, 2 At the molecular level, all GPCRs share a conserved tertiary structure with seven transmembrane α-helical motifs (TMs), the N-terminus outside the cell, and the C-terminus on the cytosolic side of the cell membrane. Agonists bind to the interior of the hydrophobic TMs on the extracellular surface, initiating subtle conformational changes which propagate to the intracellular surface. This transmitted “signal” stimulates binding and activation of G proteins on the intracellular receptor side. G protein activation in turn triggers downstream signaling cascades controlling a broad range of important physiological processes. For this reason, GPCRs have been preferential therapeutic targets for a wide spectrum of diseases.3, 4
Despite their widespread use as drug targets, the detailed molecular mechanisms by which ligand binding modulates GPCR activity remain poorly understood. Like most proteins,5, 6 GPCRs are highly dynamic entities that exist in multiple atomic-scale conformations. They do not act as simple “on-off” switches, but rather like fluctuating integrators of information.4, 7 Even in the ligand-free basal state, they constantly switch between an inactive state (R) and an active state (R*) which can engage the G protein, with each state comprising an ensemble of various conformations.5, 8 Ligand binding modulates receptor function by displacing the conformational equilibrium. Agonists (activating ligands) stabilize conformations which increase G protein-mediated signaling; inverse agonists (inhibiting ligands) stabilize different conformations which decrease the basal level of signaling.2 Structurally very similar ligands can drive the conformational ensemble to diverse equilibria and trigger very different physiological responses.8–11
Recent structural studies of ligand-activated GPCRs have provided some insight into the mechanisms of activation.2, 12–20 An important breakthrough was achieved early this year with the report of the first series of high-resolution crystal structures of ligand-activated GPCRs obtained in the presence of various receptor agonists.21–23 Surprisingly, these studies found that the G protein is required to stabilize the active state: in its absence, the agonist-bound GPCRs crystallized in the inactive R state. Solution studies have however shown the ability of agonists to displace the conformational equilibrium towards the R* state,9, 21, 24–26 suggesting that the active conformations are only of transient nature in the absence of G proteins. Inherently dynamic techniques enabling the capture of transient species are therefore needed in order to gain insight into the active state conformations.
Single-molecule fluorescence spectroscopy has proven to be a particularly valuable tool to study protein dynamics at the molecular level.6, 27–31 By removing the ensemble averaging inherent in bulk experiments, it directly probes the underlying heterogeneity which often arises for biomolecules, for example if the various individual copies of a protein are in different conformations. It further allows unsynchronized or rare events such as transition states to be monitored in addition to the average behavior.32–36
Prolonged observation of single molecules in solution at room temperature in a close-to-native environment has nonetheless been challenging because Brownian motion causes nanoscale objects to diffuse quickly out of the confocal detection volume. Surface immobilization by chemical means has often been used to eliminate this problem, but there remains persistent doubt as to whether surface-attached molecules behave the same as free-solution molecules,37 particularly when the goal is to probe subtle conformational changes and photodynamics. While laser tweezers can trap objects in solution in the 100–1000 nm range,38 the optical trapping force (scaling with volume) is insufficient to trap smaller objects of size <10 nm, including most proteins. On the other hand, the Anti-Brownian ELectrokinetic (ABEL) trap has demonstrated its ability to study single biomolecules free in solution at ambient conditions for prolonged periods of time.39–43 The ABEL trap combines fluorescence-based position estimation with fast electrokinetic feedback to oppose the Brownian motion of a single nanoscale object, hence maintaining its position in the field of view without perturbing its internal degrees of freedom. Current implementations have allowed stable trapping of single-dye-labeled DNA oligomers44 and single organic dyes45 for several seconds, with the trapping time limited only by the photobleaching of the dye. Moreover, as opposed to the case with fluorescence correlation spectroscopy (FCS),46 the molecule is trapped in a region of time-averaged uniform intensity so that useful information can be extracted from the brightness. For example, light-induced conformational changes and fluctuations in the intrinsic chromophore radiative rate for trapped allophycocyanin protein molecules have recently been observed using an ABEL trap; these findings would not have been observable in bulk experiments and differ substantially from previous single-molecule studies in which the same protein had been immobilized in polymers or on a glass surface.42
We describe in this work our use of an ABEL trap to investigate the conformational dynamics of detergent-solubilized single molecules of the β2-adrenergic receptor (β2AR), a canonical GPCR involved in smooth muscle regulation, in the presence and in the absence of the full agonist BI-167107. Detergent-solubilized β2AR labeled with tetramethylrhodamine (TMR) at the end of TM6 (Figure 1) was previously shown to respond in a ligand-specific way to the binding of agonists by undergoing conformational changes which selectively modulate the fluorescence quantum yield of the probe,9 a metric known to depend on the local environment of the fluorophore.47, 48 This picture of multiple distinct substates induced by different ligands has further been confirmed by experiments using plasmon-waveguide resonance.49 An earlier attempt to probe freely-diffusing β2AR obtained indirect evidence for a distribution of more than one conformational state, but was unable to resolve dynamics for each molecule due to the ~ms transit time of each receptor through the confocal probe volume.50 In the present study, however, we perform ABEL trapping of single β2AR molecules for hundreds of milliseconds to seconds, during which time we simultaneously measure both the fluorescence intensity and the excited-state lifetime for every trapped receptor, which change discontinuously with time at the single-receptor level. By combining the observed correlated lifetime-intensity measurements for all molecules, we find a complex conformational landscape as evidenced by a diversity of discrete values for fluorescence intensity and lifetime. These states interconvert on timescales of hundreds of milliseconds, indicating changes in the local protein environment as probed by the fluctuating radiative and nonradiative lifetimes of the TMR reporter dye. We find that agonist binding causes a clear shift in the equilibrium distribution of these states. In addition, agonist binding increases the dwell times within states and reduces the local flexibility of the receptor. We thus obtain the first experimental dynamic insights into conformational fluctuations for single β2AR in solution.
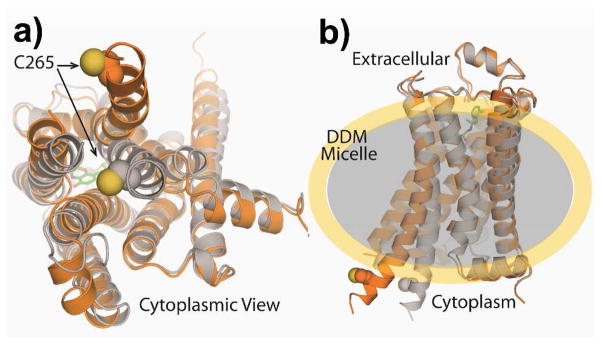
Figure 1.
Structure of β2AR-TMR in active (orange) and inactive states (gray). Panel (a) shows a cytoplasmic view with the TMR labeling location indicated by space-filling spheres. Panel (b) shows the side view with the full agonist BI-167107 (small green structure on extracellular side) and a sketch of the surrounding DDM detergent micelle (shaded).
METHODS
Sample preparation
β2AR in which four reactive cysteines were mutated (C77V, C327S, C378A, and C406A) was expressed in Sf9 insect cells, solubilized, and purified by affinity chromatography according to previously described procedures.51 Purified receptor was incubated with an equivalent amount of tetramethylrhodamine-5-maleimide (TMR, single isomer, Invitrogen) for 30 minutes at room temperature. The TMR-labeled receptor (β2AR-TMR) was then purified by gel filtration immediately before use, usually resulting in 1–3 μM solutions. Agonist-bound β2AR-TMR was prepared by incubation of 1–3 μM receptor solution with a 10-fold stoichiometric excess of the full agonist BI-16710721 for 2 hours at 4°C immediately prior to experiments. BI-167107 was chosen for its ~nM dissociation constant and extremely slow ~(150 hour)−1 off-rate, enabling extended study in the ABEL trap at single-molecule concentrations. The ligand-free and the agonist-bound receptor stock solutions were used directly for bulk spectroscopy, whereas for trapping experiments, they were diluted by 4–5 orders of magnitude. All samples were handled in PBS buffer (Gibco 10010) at pH 7.4 supplemented with 0.1% (w/w) n-dodecylmaltoside (DDM, Anatrace) and stored on ice in the dark. Experiments were conducted as soon as possible after labeling. Ligand-free and agonist-bound data sets included two independent receptor preparations with multiple ensemble and trapping experiments for each preparation.
Ensemble measurements
All measurements were performed in a 1-cm quartz low-volume cuvette. Absorption spectra were recorded on a Cary 6000i spectrophotometer (Varian), whereas fluorescence excitation and emission spectra were measured on a Fluoromax-4 fluorescence spectrometer (Horiba). Fluorescence quantum yield measurements were performed using rhodamine 6G in ethanol as a standard (Φfl = 0.9452) and changes in index of refraction were taken into account when necessary.
Excited-state lifetimes were acquired with the time-correlated single-photon counting (TCSPC) technique. Excitation at 515 nm was achieved using the frequency-doubled output of a 1030 nm mode-locked femtosecond fiber laser (Mercury 2000-200-MOD, Polaronyx). The average power was ~0.5 mW at 43 MHz repetition rate with <1 ps pulses. Fluorescence was collected at 90 degrees and passed through a 550-nm long-pass filter (HQ550LP, Chroma) and an analyzer set at the magic angle with respect to the vertical excitation polarization. A Si single-photon detection module with fast timing resolution (PDM-5CTC, Micro Photon Devices) ensured efficient detection. Acquisition was performed with a TCSPC module (PicoHarp-300, Picoquant) connected to a computer with an overall instrument response function (IRF) of ca. 400 ps (full width at half maximum, FWHM). For time-resolved fluorescence polarization anisotropy experiments, two fluorescence decay traces were recorded with the analyzer set at parallel (VV) and perpendicular (VH) orientation with respect to the excitation polarization. The anisotropy decay r(t) was computed using the standard formula53
The polarization correction factor g, quantifying the difference in detection efficiency between parallel and perpendicular polarizations, was measured to be 1.0 for our setup. Decays included at least 106 total photons with negligible laser-induced background.
ABEL trap experiments
ABEL trap microfluidic cells were assembled by bonding a molded polydimethylsiloxane (PDMS) chamber to a glass coverslip as previously detailed.54 Before injection into the ABEL trap, diluted samples were combined with the anti-adsorption polymer POP6 without denaturant (Applied Biosystems) at 10% (v/v) to prevent the receptors from sticking to the coverslip surface. The presence of the polymer did not affect the photophysics of β2AR-TMR.
ABEL trapping experiments were performed on an Olympus IX71 inverted microscope equipped with a high-NA oil objective (1.35 NA, 60x, Olympus). The 515-nm pulsed excitation beam (same as described in ensemble TCSPC measurements, but with average power 40 μW before objective) was sent through an orthogonal pair of acousto-optic modulators (46080-3-LTD, NEOS) to deflect the beam along a circular path with angular frequency 26 kHz. The circularly-revolving beam was then steered into the microscope and imaged to the focal plane with beam radius 0.7 μm and revolution radius 0.3 μm, yielding a time-averaged uniform intensity profile. Fluorescence was detected with a fiber-coupled avalanche photodiode (SPCM-AQ4C, Perkin Elmer) after filtering through a 545-nm long-pass filter and a 610-nm short-pass filter (HQ545LP, E610SP; Chroma). In such a configuration, illuminated receptors emit fluorescence photons whose detection times depend on the position of the revolving beam; a lock-in scheme implemented on a field-programmable gate array (7833-R, National Instruments) estimated the angular position of the receptor.55 Based on the angular estimate, feedback voltages of up to ±30 V were applied to the solution with a home-built amplifier using a set of four platinum electrodes to oppose the Brownian motion of the receptor, trapping it until the TMR dye photobleached. The static and uniform electric fields induced by these relatively small voltages across a distance of ~10 μm have been shown to have no effect on single-molecule photophysics.42
Data Analysis
All data analysis was performed in MATLAB (The Mathworks Inc.). For ABEL trap data, the time-tagged photon stream was binned into 10 ms bins to create a time trace of the intensity. We selected trapped receptors by applying an automatic threshold. For a receptor to be included in the analysis, we required (1) that the intensity of each 10 ms bin comprising it be at least 2 standard deviations above the mean background level, (2) that the burst duration be at least 30 ms, and (3) that the burst contain at least 200 photons above background. In order to accurately set the background level, we hand-selected ~5 s of background photons from the time trace in each 300 s interval of acquired data. These photons determined both the mean background level for the burst selection procedure and the time-resolved background decay for lifetime determination detailed below.
As many bursts in our data set show obvious digital steps from one intensity state to another, we applied the change-point determination algorithm developed by Watkins and Yang56 to identify different states. The change-point algorithm was configured to include only states lasting at least 30 ms, and the resulting set of states was further filtered to calculate dwell times and fluctuation timescales. To determine the fluorescence lifetime of TMR in each state and in each 10 ms bin, the fitting procedure of Zander et al. was applied.57 Briefly, a trial single exponential decay with decay constant τ was convolved with the measured IRF (0.4 ns FWHM) of the detection system and then added to the measured background decay (see Supplementary Information). This trial decay function was then optimized according to the maximum likelihood estimation (MLE) algorithm to reproduce the data57 with the estimated lifetime being the optimal decay constant τ. The color-shift delay between the measured IRF and the experimental decay was included as a free parameter in the fit. The lifetime decay of each 10 ms bin contains a smaller number of photons (typically hundreds rather than thousands) and so the traces are noisier, leading to higher uncertainties in the fitting parameters, but the gain in time resolution allowed us to detect substantial shifts in lifetime even at relatively constant intensity (Figure 3d). Lifetimes from ensemble measurements were extracted according to the same MLE-based method. The far higher signal-to-background ratio made full background decay correction unnecessary, but a time-independent offset term was included to adjust for dark counts.
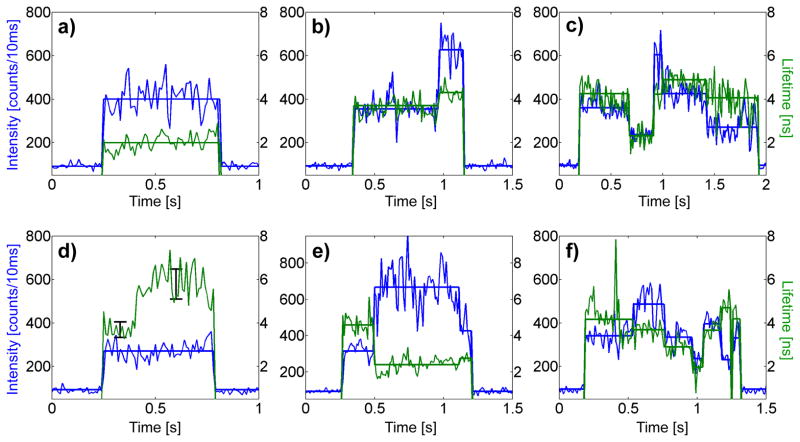
Figure 3.
Examples of intensity-lifetime traces with 10 ms bin size from single trapped ligand-free receptors. Real-time intensity (blue) and fluorescence lifetime (green) are shown with calculated intensity change-point states overlaid (thick lines). Transitions among a range of intensity-lifetime states are observed, in which intensity may be correlated (b,c), uncorrelated (d), or anti-correlated (e) with lifetime. Because trace (d) shows a large, uncorrelated change in lifetime for constant intensity, we replace the thick line for mean lifetime with 1σ error bars calculated from the 10 ms binned lifetime.
Autocorrelations were calculated by combining the time-tagged photons in each state into 50 μs bins. This shorter bin time was chosen for higher time resolution, although time resolution is limited by the revolution period of the excitation beam, (26 kHz)−1 = 38 μs and the detected photon count rate. Estimates of the autocorrelation in the experimentally-relevant 100 μs to 1 s time window are insensitive to bin time. The beam revolution produces, as expected, a sharp oscillating component in the autocorrelation (see Supplementary Information), but only autocorrelation lags greater than 200 μs, after the oscillation has died out, were included in the fitting and analysis. For additional controls, see Supplementary Information. Autocorrelation fluctuation timescales and stretching exponents as well as mean dwell times were extracted by standard nonlinear least squares fitting with all parameters bounded by 0 and infinity.
RESULTS
Labeling and Bulk Spectroscopy
We labeled purified receptors with the environment-sensitive dye tetramethylrhodamine (TMR) at the base of the sixth transmembrane helix (Figure 1), a position known from previous studies to be well-suited to sense conformational changes during receptor activation without interfering with receptor function.9, 21, 26, 50 The two projections in Figure 1 show the cytoplasmic view (a) and the side view (b) of the receptor with the TMR labeling location indicated by space-filling spheres. The active form of the receptor is shown in orange and the inactive form in gray with a shaded ellipsoid around the equatorial region suggestive of the surrounding micelle. We performed bulk absorption, fluorescence, fluorescence quantum yield, fluorescence lifetime, and time-resolved fluorescence polarization anisotropy decay measurements on both ligand-free β2AR-TMR and β2AR-TMR incubated with the full agonist BI-167107 in order to quantify the bulk-level values of these parameters.
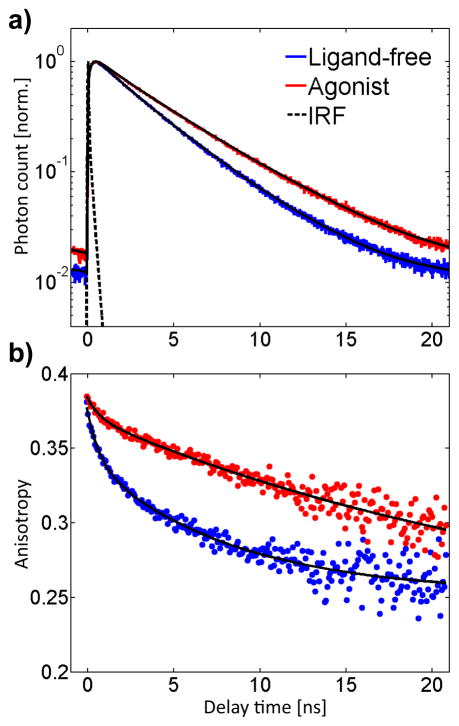
No significant change in the absorption spectrum and only a minor red-shift (4 nm) in the fluorescence spectrum were observed upon agonist binding (Figure S1), and the fluorescence quantum yield of the single TMR label increased from 0.33 to 0.38 (Table 1). In correlation with the rise in fluorescence quantum yield, the fluorescence lifetime also increased in the presence of agonist (Figure 2a). Significantly, the probe on both ligand-free and agonist-bound receptors displays biexponential fluorescence lifetime decays (Table 1), as often observed for fluorescent dyes interacting with proteins,58, 59 indicating heterogeneity in sample or conformation. Ligand-free receptors have 3.6 ns (90%) and 1.6 ns components, whereas agonist-bound receptors have 4.5 ns (95%) and 1.6 ns decay components. The lifetime of TMR is known to increase with decreasing medium polarity,47, 48 likely indicating that TMR explores a less polar microenvironment in the agonist-bound receptor, as already suggested by the hypsochromic shift of the fluorescence. The 1.6 ns component common to both samples could indicate a shared conformation in equilibrium with a more dynamic state whose sensed lifetime increases from 3.6 to 4.5 ns upon activation. However, biexponential fluorescence decays do not necessarily imply the existence of two distinct emitting populations of receptor; they could also originate from a distribution of populations with distinct fluorescence decay times.60, 61 Such closely-spaced lifetime states or distributions of lifetimes cannot be resolved from ensemble-averaged measurements.
Table 1.
Fluorescence quantum yield Φfl, excited-state lifetime decay components τ1 and τ2 (and amplitudes), average lifetime τav, and radiative lifetime τrad for β2AR-TMR in the absence and in the presence of BI-167107, all from bulk measurements.
| Ligand | Φfl | τ1 (ns) | τ2 (ns) | τav (ns) | τrad (ns) |
|---|---|---|---|---|---|
| Ligand-free | 0.33 ± 0.02 | 1.6 (10%) | 3.6 (90%) | 3.4 | 10.3 |
| BI-167107 | 0.38 ± 0.02 | 1.6 (5%) | 4.5 (95%) | 4.4 | 11.6 |
Figure 2.
Bulk fluorescence lifetime (a, 8 ps bins) and time-resolved fluorescence polarization anisotropy decays (b, 80 ps bins) of TMR on ligand-free and agonist-bound receptors. The agonist-bound receptors display a longer fluorescence lifetime and higher anisotropy of the TMR reporter dye.
Bulk measurements of the decay of the fluorescence polarization anisotropy further indicate that the TMR probe senses β2AR conformational changes, as the decay rate substantially decreases for agonist-bound receptors compared to ligand-free receptors (Figure 2b). Again, the traces are best reproduced by two exponential terms for both samples. Ligand-free receptors exhibit rotational relaxation times of 8.2 ns (74%) and 0.7 ns, whereas the anisotropy of agonist-bound receptors decays with 30 ns (93%) and 0.7 ns time constants. Biexponential fluorescence polarization anisotropy decays are often observed when dyes are attached to macromolecules and generally analyzed using the “wobbling-in-a-cone” model (Table S1).62, 63 A fast time scale (0.7–0.8 ns) is used to describe the local wobbling motion of the probe while a slower time scale (8.2–30 ns) describes tumbling of the macromolecule or of segments of it. In the case of β2AR-TMR (Table S1), the fraction of fluorescence polarization anisotropy decay associated with TMR wobbling (26%) drastically decreases in the presence of agonist (7%), suggesting that the probe is on average highly confined in agonist-bound conformations. On the other hand, the 30 ns time scale (this value has large uncertainty given the available experimental time window compared to the excited-state lifetime of the probe) associated with β2AR tumbling is consistent with the size of a receptor micelle (expected value ~50 ns). This time scale is not directly observed with ligand-free β2AR-TMR, which indicates that the 8.2 ns component measured in this case is likely the convolution of two motions, one arising from the motion of a local receptor segment such as the third intracellular loop or TM6, and one from the overall receptor tumbling. This motion attributed to local segment motion becomes totally suppressed in the presence of BI-167107. Our results thus suggest that agonist binding locally stabilizes the receptor into less dynamic and more restricted conformations. These bulk experiments motivate detailed single-molecule investigation of the receptor conformational states and dynamics.
Diversity of Intensity and Lifetime States of Single Receptors
We use the ABEL trap to observe single β2AR-TMR molecules in solution for prolonged periods of time.39 The design of the ABEL trap is such that a trapped TMR-labeled receptor experiences a spatially-uniform excitation profile on a time scale slower than ~200 μs, and for this reason, any observed fluorescence intensity fluctuations can be attributed to intrinsic fluctuations of the TMR-receptor system. In addition, pulsed excitation combined with time-tagging of each detected photon allowed us to calculate both the mean excited-state lifetime of TMR for each trapped receptor, as well as a running estimate of the lifetime for every 10 ms bin. Measuring excited-state lifetime concurrently with fluorescence intensity for each single molecule provides not only correlated values but also an added experimental dimension, enabling, for example, direct access to the intrinsic radiative lifetime. Such correlated measurements cannot be obtained from ensemble studies.
Figure 3 shows six trapped ligand-free receptors exhibiting a representative range of behaviors. Fluorescence intensity (blue) and fluorescence lifetime (green) are plotted on the same time axis with 10 ms bins, where the initial increase represents the initial entry of a single molecule into the trap. The most striking feature of the data is the presence of discrete intensity steps. Given these evident digital transitions, we applied the change-point finding algorithm of Watkins and Yang56 to statistically identify the time points (“change-points”) during which the intensity of a trapped receptor changes significantly. Hereafter we use the word “state” to describe the microscopic conformation of the single receptor in between such change-points, as reported by the mean fluorescence intensity, mean lifetime, and mean fluctuation timescales (see below) of the fluorescence photons from the TMR reporter dye. Our implementation of the change-point algorithm provides an automatic way to identify and catalog receptor states consistent with those that might be chosen by eye. The 10 ms binned data indicates calculated intensity (blue) and lifetime (green) in every 10 ms interval, whereas the heavy, straight lines indicate change-point mean intensity states and the much more accurate mean lifetime value calculated from pooling together and fitting the decay from all photons in the state.
The largest fraction (~90%) of trapped receptors remain in a single intensity state until the TMR dye photobleaches (Figure 3a). The remaining ~10% of receptors show state-to-state dynamics with a range of different behaviors. Figure 3b shows a clear step upward in intensity, combined with a small correlated increase in lifetime. In Figure 3c, the trapped receptor exhibits multiple intensity steps with closely correlated changes in lifetime. Other receptors, however, displayed large changes in probe lifetime even at constant intensity (Figure 3d). Instances of clear anti-correlation between intensity and probe lifetime were also observed (Figure 3e). Unusually, these behaviors indicate a sudden, substantial change in the radiative lifetime of the probe. In some cases, single receptors undergoing lifetime transitions both correlated and anti-correlated with intensity could be monitored (Figure 3f). Taken together, the different single-molecule examples in Figure 3 show that the various different receptors sample a range of discrete intensity-lifetime states. It would be ideal to be able to observe one single receptor for far longer times to see if each receptor can sample all possible states. Since this is not possible due to eventual photobleaching of the probe, we make the reasonable assumption that each receptor is only sampling a portion of the states available.
We observed qualitatively similar behavior for both ligand-free (3962 receptors trapped and observed) and agonist-bound receptors (4312 receptors trapped and observed); by eye, individual trapped receptors from both groups cannot be distinguished. Quantitative analysis of the correlated fluorescence intensity-lifetime values observed for all molecules, however, revealed substantial differences. Figure 4 represents a two-dimensional density plot (histogram) of the observed distribution of intensity-lifetime states for both ligand-free and agonist-bound receptors, with representative fitted excited-state lifetime decays from two specific locations in the plot. In agreement with bulk measurements, the agonist-bound receptors display higher mean intensity and higher mean lifetime compared to the ligand-free receptors. The peak of the distribution is likewise at higher intensity and lifetime for the agonist-bound receptors. The ligand-free receptors show a broad distribution of states from low intensity and low lifetime to high intensity and high lifetime, including a peak near 170 counts per bin and 4 ns. The agonist-bound receptors, however, explore far fewer states at low intensity and low lifetime, although the rough linear trend of positive correlation remains visible. The peak region of the agonist-bound distribution is narrower in lifetime, but with greater spread along the intensity axis, evident in the oblong peak shape. To probe changes in radiative rate, we note that the intensity I and the excited-state lifetime τ can be written as usual as
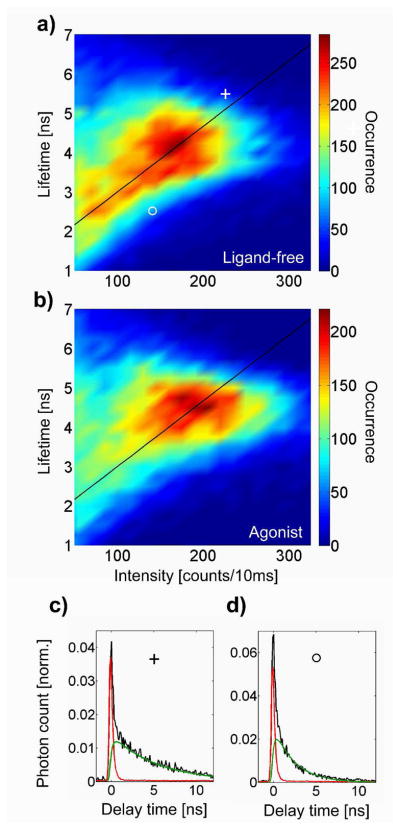
Figure 4.
Peak-normalized density plots of the intensity-lifetime states for the full population of ligand-free (a) and agonist-bound single receptors (b). The color bar indicates the number of states observed in each (15 cts/10 ms) × (0.25 ns) bin. Agonist-bound receptors show a tighter distribution of states with higher peak intensity and lifetime compared to ligand-free receptors. The diagonal line is a guide to the eye, highlighting states of constant radiative lifetime. In (c) and (d) we show typical lifetime decays (black) from states at coordinates ○ and +, which include the fitted lifetime component (green) together with the background component (red).
where krad is the intrinsic radiative rate and knrad is the nonradiative (quenching) rate. Dividing these equations yields the radiative rate alone, up to a constant which depends on the detection efficiency of the setup. We also assume that the absorption spectrum, and therefore the excitation rate, remains constant for the TMR-receptor system, as bulk absorption spectra of ligand-free and agonist-bound receptors are extremely similar (data not shown). Thus, as a guide to the eye, we plot a diagonal line to highlight states of constant radiative lifetime, τrad = krad−1 ∝ τ/I. States at different coordinates along the line experience varying degrees of non-radiative quenching with correlated lifetime and intensity, whereas states far from the line exhibit a different radiative lifetime. Distributions for both samples have detailed structure and high asymmetry, and do not show obvious clustering into well-resolved peaks, making them inconsistent with a single ligand-free (inactive) or a single agonist-bound (active) state. Indeed, the range of intensity-lifetime states along the line of constant radiative lifetime provides evidence for a range of discrete conformational substates in which the TMR probe experiences slightly different microenvironments.
Timescales of Conformational Dynamics
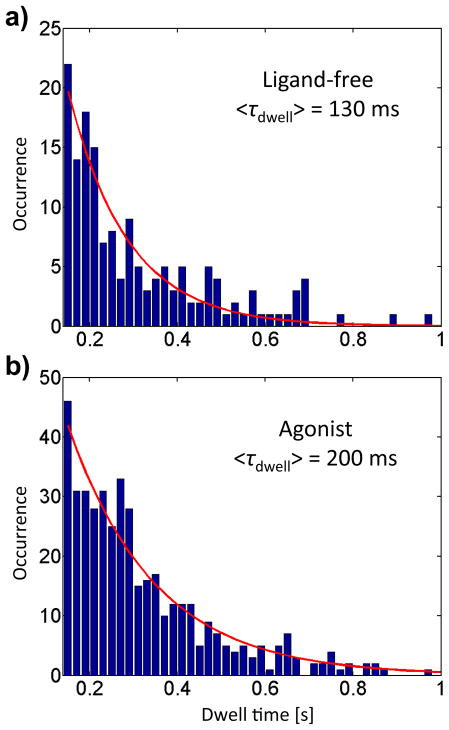
In addition to probing the equilibrium distribution of fluorescence intensity-lifetime states, the prolonged observation time provided by ABEL trapping allowed us likewise to investigate the dynamics. In Figure 5, we examine the distribution of state dwell times, defined as the residence time of the receptor in a given state before it transitions to a different state. An essential caveat is that transitions among receptor states occur independently of trapping; residence in a state does not begin when the receptor is trapped, nor does it end when the TMR probe photobleaches. Thus, to obtain an unbiased dwell time distribution, we included only dwell times of the intermediate time states (not first or last) of trapped receptors traversing at least three states. Furthermore, because the accuracy of the change-point algorithm increases for longer-lived states due to more data, we have highest confidence in long dwell times.56 On the other hand, shorter states are more numerous, allowing more robust statistics and fitting. As a compromise, we took into account only states lasting at least 150 ms. The agonist-bound receptors show a clear increase in mean dwell time (200 ms, extracted from a single-exponential fit) compared with the ligand-free receptors (130 ms). Equivalently, transitions between states occur less often, on average, in agonist-bound receptors.
Figure 5.
Histograms of state dwell times for the full population of ligand-free (a) and agonist-bound receptors (b). The mean dwell times are estimated from a single exponential fit with time constant τdwell. Agonist-bound receptors show longer state dwell times than ligand-free receptors.
Although our use of the change-point algorithm accurately identifies large changes in intensity, it does not simultaneously capture smaller, faster fluctuations. The state in Figure 3a and the second state in Figure 3e, for example, display large fluctuations within single identified states. Indeed, more than 90% of states in both samples show fluctuations in excess of shot noise (data not shown). To characterize these faster fluctuations, we computed the intensity autocorrelation of the photons comprising each state.
The intensity autocorrelation G(τAC) of a time-varying intensity trace essentially describes, on average, how much the intensity changes between time t and time t+τAC, as a function of the lag time τAC. High values of G(τAC) indicate small change (high correlation) whereas low values of G(τAC) indicate significant change (low correlation). Most dynamic physical systems show a smooth decay between high correlation at short times and low correlation at long times; examination of this decay provides access to the underlying dynamical timescale. For each state, we calculate the intensity autocorrelation
where τAC is the autocorrelation time, <…> denotes averaging over the intensity trace, and δI(t) = I(t) − <I(t)> is the mean-subtracted intensity. Intensity autocorrelation finds wide application in FCS,46 where it is used to quantify, among other quantities, the mean transit time of a fluorescent object through a confocal excitation volume of known size, allowing extraction of the diffusion coefficient of the object. More generally, autocorrelation can be used to quantify conformational dynamics,32, 64 or indeed any time-dependent fluctuation, but in typical FCS experiments, the dynamics from many single molecules are averaged, as opposed to the case here. An on-off telegraph signal with mean relaxation time τ̄AC will have theoretical intensity autocorrelation G(τAC) ∝ exp(−τ̄AC/τ̄AC) and as such the single exponential form is often used to estimate the mean relaxation timescale <τAC> of a fluctuation process.64 Note that in this instance, <…> indicates averaging over the autocorrelation decay, not averaging over the intensity trace.
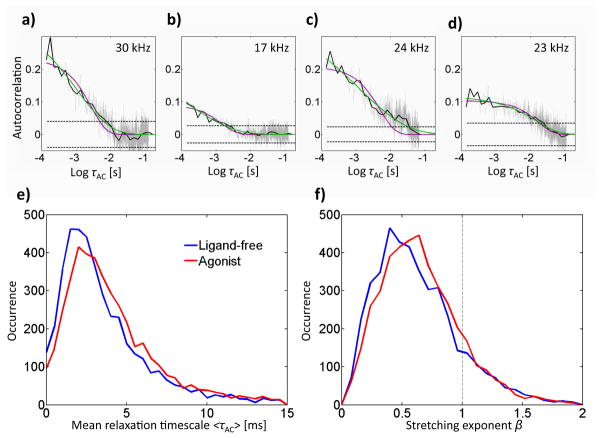
Figure 6 shows four representative autocorrelations obtained from four ligand-free receptor states. In order to accurately estimate the autocorrelation on ~ms timescales (see below), only states of at least 200 ms duration were included in the autocorrelation analysis. In each case, the average photon detection count rate was sufficient to detect fluctuations on the timescale plotted. We plot the intensity autocorrelation (light gray), calculated up to τAC = tmax/4, where tmax is the duration of the state, as recommended by Box et al.65 The uncertainty in the calculated autocorrelation increases at higher time lags τAC as a result of poorer sampling; to counteract this effect, we performed all fits on the logarithmically-binned autocorrelation (solid black line) to more robustly sample the longer time lags. We also plot confidence intervals about zero (dashed line, 95% confidence) to confirm the existence of positive autocorrelation above noise.65–67 According to this test, greater than 97% of states longer than 200 ms displayed significant autocorrelation; the remainder were excluded from further analysis.
Figure 6.
Intensity autocorrelation of individual intensity-lifetime states longer than 200 ms. We show example autocorrelations (a,b,c,d) calculated with linear bins (gray), logarithmic bins (black), and we include 95% confidence bounds on zero (dashed) and the mean signal photon count rate of the state (upper right). The autocorrelation (a) is from the state in Fig. 3(a), and (b) is from the state in Fig. 3(d). We use a stretched exponential fit (green) rather than a single exponential fit (purple) because it more accurately describes the data. We show histograms of mean decay times (e) and stretching exponents (f) for ligand-free and agonist-bound receptors.
Single exponential decays fail to adequately describe most autocorrelations (Figure 6a–d). A better fit to the data is provided by the stretched exponential form
where 0 < β < 1 is the stretching exponent and τ̄AC is the characteristic timescale.68 The stretched exponential form can be interpreted as a superposition of multiple exponentials and thus describes a distribution of timescales, increasingly broadened as β decreases from 1. In contrast to the single exponential form, for which the characteristic time τ̄AC, a parameter of the fit, is equal to the mean relaxation timescale <τAC >, the mean relaxation timescale of a stretched exponential is obtained from , where Γ(x) denotes the gamma function. It is the single-molecule distribution of this (more physical) quantity for unbound and agonist-bound receptors which is represented in Figure 4e. Both distributions are similarly broad and asymmetric, peaking around 2 ms, although agonist-bound receptors show a slight shift to longer timescales. This timescale is too slow to arise from motion of the probe around its linker in the nanosecond regime (Figure 2b), and we thus attribute these observed intensity fluctuations to intrinsic fluctuations of the receptor. The distributions of the stretching exponent β in Figure 4g are again similar for both samples, peaking around β ~ 0.5, although the agonist-bound receptors show a slight shift toward higher β, indicating a tighter distribution of timescales (closer to single exponential at β=1). The broadness of the β distribution is due to the difficulty of estimating stretched exponential parameters from short time series. Indeed, Lu et al. find substantial broadening in β distributions for single-molecule intensity traces, although for time series at least 100 times longer than the characteristic autocorrelation timescale, the peak β provides an accurate estimate for the true β.69 Given our fluctuation timescale of ~2 ms and state duration of at least 200 ms, we expect an accurate estimate of β from its peak value. We confirmed that no correlation exists between dwell time and mean relaxation time for either sample, thus ruling out the possibility that relaxation timescales may be influenced by artifacts of sampling. Indeed, we find that peak timescales obtained from the stretched exponential fits are the same as those obtained from single exponential fits (data not shown). The fact that the vast majority of extracted β values are decidedly less than 1, however, is strong evidence that the stretched exponential form provides a more accurate description of the data.
DISCUSSION
By labeling the end of TM6 with the environment-sensitive fluorescent probe TMR, we were able to experimentally follow conformational dynamics of β2AR, a model GPCR, in the absence and in the presence of the full agonist BI-167107. The cytoplasmic end of TM6 is particularly well-suited to sense conformational changes as this segment undergoes a large outward movement of 11 Å upon transition of the receptor from an inactive to an active state21 and TMR was shown in this and previous studies to selectively report on agonist-induced conformational transitions by having its fluorescence quantum yield, fluorescence lifetime, or fluorescence polarization anisotropy decay modulated.9, 70, 71 Our results suggest that the probe experiences a less polar, conformationally more restricted environment in the agonist-bound form of the receptor. The mechanism by which TMR senses changes in its environment is related to polarity, the fluorescence quantum yield and lifetime increasing with decreasing polarity.47, 48 The longest reported lifetime for TMR in apolar media is 3.7 ns, a value close to the mean lifetime of ligand-free β2AR-TMR measured here (3.4 ns). We observe, however, an even longer mean lifetime of 4.4 ns for agonist-bound β2AR-TMR, incompatible with a solely polarization-related mechanism. Radiative lifetime fluctuation arising from modulation of the refractive index and hyperpolarizability of the environment of the dye due to nanoconfinement could account for this observation and act as a second fluorescence contrast mechanism.72 The very slow decay of the fluorescence polarization anisotropy of agonist-bound β2AR-TMR due to quasi-absence of TMR wobbling motion is consistent with nanoconfinement of the probe. This mechanism would also support the changes in radiative lifetime witnessed both in ensemble (Table 1) and in single-molecule (Figure 3) measurements, as neither changes in the orientation of the dipole moment of the probe nor spectral shifts can account for this observation in the solution phase.73
Although we do not have a crystal structure showing the actual position of the fluorescent probe relative to the atomic positions of the GPCR, we can speculate on the origins of the fluorescence changes observed in this work. In the inactive form (gray in Figure 1), the labeling position is nestled among the helices of the protein, while in the active form (orange in Figure 1), the labeled helix extends outward and rotates. Although this position may appear more hydrophilic, the increased rigidity and hydrophobicity we observe for agonist-bound GPCR may be due to the ability of the probe on its long linker to interact more strongly with the nearby detergent molecules. Rearrangement of the micelle itself is expected to take place on timescales of tens of nanoseconds or faster and cannot account for that effect.74–76
The account of receptor dynamics which emerges from single-molecule experiments confirms the complexity of the conformational landscape of GPCRs and, consistent with other observations, rules out any simple two-state activation model.7 The numerous distinct states that ligand-free β2AR explores (Figure 4a) reflect the mechanism underlying the constitutive activity of the receptor; even in the absence of agonist, β2AR switches between several inactive and active conformations. The striking feature of Figure 4a, analogous to a state map obtained along one conformational coordinate (i.e., that probed by TMR), however, is its lack of clear local structure which could allow conformations to be grouped together and discrete intermediates in the activation pathway of the receptor to be defined. Our data rather suggests the existence of a large number of discrete states along the activation pathway. Agonist binding displaces the equilibrium toward active conformations (Figure 4b), but, as suggested by molecular dynamics simulations,22 this event alone is not sufficient to lock the receptor into active states, even if it seems to lead to a partial stiffening of the protein as suggested by the longer average dwell time (Figure 5) and the fluorescence polarization anisotropy data (Figure 2b). Although agonist binding lowers the difference in energy between inactive and active conformations, inactive conformations remain energetically favored in the absence of G protein,22 hence the similarities in the behaviors observed on the single-receptor level in the presence and in the absence of agonist. The dynamic nature of β2AR is reinforced by the millisecond-time scale fluctuations recorded within fluorescence intensity and lifetime states. The fact that a distribution of timescales is needed to accurately describe these fluctuations is consistent with a complex and dynamic conformational landscape encompassing numerous distinct states.77
CONCLUSION
In this work we present the first single-molecule study of β2AR conformational dynamics in solution. By permitting observation of single receptors for prolonged times, the ABEL trap allows us to access not only the distributions of conformational states, but also their interconversion rates and fast fluctuations. The novel single-molecule results of this work are the following: direct experimental observation of both discrete, stepwise as well as fluctuating conformational dynamics as reported by intensity and lifetime of TMR in single receptors (Figure 3); the first two-dimensional “map” of the shift in conformational equilibria in the ensemble caused by agonist binding, represented in correlated intensity-lifetime space (Figure 4); and measurement of timescales of interconversion of these states over three orders of magnitude, milliseconds to seconds, which have proved inaccessible to other techniques (Figures 5,6). These results suggest a rugged, dynamic energy landscape consistent with the highly-evolved GPCR cellular regulation mechanisms known from structural and biochemical assays. On the other hand, evidence for multiple interconverting conformational substates is only a statement of the problem; more work is needed to resolve discrete atomic-scale conformations and to integrate them into a more complete dynamical model of G protein activation. Our results additionally suggest that fluctuations in radiative rate could serve as a novel probe of protein dielectric microenvironment.
Supplementary Material
Acknowledgments
We gratefully acknowledge Quan Wang for critical support and FPGA implementation, Randall Goldsmith and Yan Jiang for helpful discussions and MATLAB scripts, Adam Cohen for initial trap design and soft lithography, and Stefanie Nishimura (Life Technologies) for the antiadsorption polymer POP6 without denaturant. This work was supported in part by Grant No. 1R21-RR023149-03 from the National Center for Research Resources (W.E.M.), an NSF Graduate Fellowship (S.B.), a Swiss National Science Foundation post-doctoral fellowship (A.F.), and by National Institutes of Health Grants NS028471 and GM083118 and the Mathers Foundation (X.J.Y. and B.K.K.).
Footnotes
Author Contributions
S.B, A.F., B.K.K., and W.E.M. conceived and designed experiments; S.B. performed ABEL trap experiments; A.F. performed bulk experiments; X.J.Y. and B.K.K. expressed, purified, and labeled receptors; S.B., A.F., and W.E.M. analyzed data and wrote the paper.
Supporting Information Available
The Supporting Information provides details of absorption and fluorescence spectroscopy for quantum yield determination, single-molecule lifetime fitting, autocorrelation analysis and controls, and bulk anisotropy measurements. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-Transmembrane Receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The Structure and Function of G-Protein-Coupled Receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenakin T. Efficacy at G-Protein-Coupled Receptors. Nat Rev Drug Discov. 2002;1:103–110. doi: 10.1038/nrd722. [DOI] [PubMed] [Google Scholar]
- 4.Kenakin TP. Cellular Assays as Portals to Seven-Transmembrane Receptor-Based Drug Discovery. Nat Rev Drug Discov. 2009;8:617–626. doi: 10.1038/nrd2838. [DOI] [PubMed] [Google Scholar]
- 5.Frauenfelder H, Parak F, Young RD. Conformational Substates in Proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 6.Henzler-Wildman K, Kern D. Dynamic Personalities of Proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 7.Park PSH, Lodowski DT, Palczewski K. Activation of G Protein-Coupled Receptors: Beyond Two-State Models and Tertiary Conformational Changes. Annu Rev Pharmacol Toxicol. 2008;48:107–141. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilardaga J, Steinmeyer R, Harms GS, Lohse MJ. Molecular Basis of Inverse Agonism in a G Protein-Coupled Receptor. Nat Chem Biol. 2005;1:25–28. doi: 10.1038/nchembio705. [DOI] [PubMed] [Google Scholar]
- 9.Swaminath G, Xiang Y, Lee TW, Steenhuis JJ, Parnot C, Kobilka BK. Sequential Binding of Agonists to the Beta-2-Adrenoreceptor. J Biol Chem. 2004;279:686–691. doi: 10.1074/jbc.M310888200. [DOI] [PubMed] [Google Scholar]
- 10.Gaertner H, Cerini F, Escola J, Kuenzi G, Melotti A, Offord R, Rossitto-Borlat I, Nedellec R, Salkowitz J, Gorochov G, Mosier D, Hartley O. Highly Potent, Fully Recombinant Anti-HIV Chemokines: Reengineering a Low-Cost Microbicide. Proc Natl Acad Sci U S A. 2008;105:17706–17711. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilardaga J, Bünemann M, Krasel C, Castro M, Lohse MJ. Measurement of the Millisecond Activation Switch of G Protein-Coupled Receptors in Living Cells. Nature Biotech. 2003;21:807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 12.Bokoch MP, et al. Ligand-Specific Regulation of the Extracellular Surface of a G-Protein-Coupled Receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sprang SR. Cell Signalling: Binding the Receptor at both Ends. Nature. 2011;469:172–3. doi: 10.1038/469172a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen SGF, Choi H, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal Structure of the Human Beta 2 Adrenergic G-Protein-Coupled Receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 15.Cherezov V, et al. High-Resolution Crystal Structure of an Engineered Human Beta 2-Adrenergic G Protein-Coupled Receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum DM, et al. GPCR Engineering Yields High-Resolution Structural Insights into Beta 2-Adrenergic Receptor Function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 17.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a Beta 1-Adrenergic G-Protein-Coupled Receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaakola V, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2. 6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien EYT, Liu W, Zhao QA, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu BL, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen SGF, et al. Structure of a Nanobody-Stabilized Active State of the Beta 2 Adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbaum DM, et al. Structure and Function of an Irreversible Agonist-Beta 2 Adrenoceptor Complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AGW, Schertler GFX, Tate CG. The Structural Basis for Agonist and Partial Agonist Action on a Beta 1-Adrenergic Receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanpain C, Vanderwinden J, Cihak J, Wittamer V, Le Poul E, Issafras H, Stangassinger M, Vassart G, Marullo S, Schlöndorff D, Parmentier M, Mack M. Mulitple Active States Oligomerization of CCR5 Revealed by Functional Properties of Monoclonal Antibodies. Mol Biol Cell. 2002;13:723–737. doi: 10.1091/mbc.01-03-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghanouni P, Steenhuis JJ, Farrens DL, Kobilka BK. Agonist-Induced Conformational Changes in the G-Protein-Coupling Domain of the Beta-2-Adrenergic Receptor. Proc Natl Acad Sci U S A. 2001;98:5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao XJ, Ruiz GV, Whorton MR, Rasmussen SGF, DeVree BT, Deupi X, Sunahara RK, Kobilka B. The Effect of Ligand Efficacy on the Formation and Stability of a GPCR-G Protein Complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. S9501/1–S9501/3. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss S. Fluorescence Spectroscopy of Single Biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 28.Moerner WE. New Directions in Single-Molecule Imaging and Analysis. Proc Natl Acad Sci U S A. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying LM. Single Molecule Biology: Coming of Age. Mol Biosyst. 2007;3:377–380. doi: 10.1039/b702845h. [DOI] [PubMed] [Google Scholar]
- 30.Blank K, Rocha S, de Cremer G, Roeffaers MBJ, Uji-i H, Hofkens J. Watching Individual Enzymes at Work. Springer Ser Chem Phys. 2010;96:495–511. [Google Scholar]
- 31.Michalet X, Weiss S, Jager M. Single-Molecule Fluorescence Studies of Protein Folding and Conformational Dynamics. Chem Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Luo G, Karnchanaphanurach P, Louie T, Rech I, Cova S, Xun L, Xie XS. Protein Conformational Dynamics Probed by Single-Molecule Electron Transfer. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 33.Li CB, Yang H, Kornatsuzaki T. Multiscale Complex Network of Protein Conformational Fluctuations in Single-Molecule Time Series. Proc Natl Acad Sci U S A. 2008;105:536–541. doi: 10.1073/pnas.0707378105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorokina M, Koh H, Patel SS, Ha T. Fluoresecent Lifetime Trajectories of a Single Fluorophore Reveal Reaction Intermediates during Transcription Initiation. J Am Chem Soc. 2009;131:9630–9631. doi: 10.1021/ja902861f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, Langowski J, Seidel CAM. Nucleosome Disassembly Intermediates Characterized by Single-Molecule FRET. Proc Natl Acad Sci U S A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoso Y, Joyce CM, Potapova O, Le Reste L, Hohlbein J, Torella JP, Grindley NDF, Kapanidis AN. Conformational Transitions in DNA Polymerase I Revealed by Single-Molecule FRET. Proc Natl Acad Sci U S A. 2010;107:715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasnik I, McKinney SA, Ha T. Surfaces and Orientations: Much to FRET about? Acc Chem Res. 2005;38:542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 38.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a Single-Beam Gradient Force Optical Trap for Dielectric Particles. Opt Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 39.Cohen AE, Moerner WE. Suppressing Brownian Motion of Individual Biomolecules in Solution. Proc Natl Acad Sci U S A. 2006;103:4362–4365. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen AE, Moerner WE. Principal-Components Analysis of Shape Fluctuations of Single DNA Molecules. Proc Natl Acad Sci U S A. 2007;104:12622–12627. doi: 10.1073/pnas.0610396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen AE, Moerner WE. Controlling Brownian Motion of Single Protein Molecules and Single Fluorophores in Aqueous Buffer. Opt Express. 2008;16:6941–6956. doi: 10.1364/oe.16.006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldsmith RH, Moerner WE. Watching Conformational- and Photodynamics of Single Fluorescent Proteins in Solution. Nat Chem. 2010;2:179–186. doi: 10.1038/nchem.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields AP, Cohen AE. Anti-Brownian Traps for Studies on Single Molecules. Methods Enzymol. 2010;475:149–174. doi: 10.1016/S0076-6879(10)75007-2. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Moerner WE. An Adaptive Anti-Brownian ELectrokinetic (ABEL) Trap with Real-Time Information on Single-Molecule Diffusivity and Mobility. appearing ACS Nano. 2011 doi: 10.1021/nn2014968. [DOI] [PubMed] [Google Scholar]
- 45.Fields AP, Cohen AE. Electrokinetic Trapping at the One Nanometer Limit. Proc Nat Acad Sci (USA) 2011;108 doi: 10.1073/pnas.1103554108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigler R, Elson E, Schaefer FP, Toennies JP, Zinth W, editors. Springer Series in Chemical Physics. Vol. 65. Springer; Berlin: 2001. Fluorescence Correlation Spectroscopy; p. 487. [Google Scholar]
- 47.Eggeling C, Fries JR, Brand L, Gunther R, Seidel CAM. Monitoring Conformational Dynamics of a Single Moleucle by Selective Fluorescence Spectroscopy. Proc Natl Acad Sci U S A. 1998;95:1556–1561. doi: 10.1073/pnas.95.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel M, Rettig W, Sens R, Drexhage KH. Structural Relaxation of Rhodamine Dyes with Different N-Substitution Patterns - a Study of Fluorescence Decay Times and Quantum Yields. Chem Phys Lett. 1988;147:452–460. [Google Scholar]
- 49.Devanathan S, Yao Z, Salamon Z, Kobilka BK, Tollin G. Plasmon-Waveguide Resonance Studies of Ligan Binding to the Human Beta-2-Adrenergic Receptor. Biochemistry. 2004;43:3280–3288. doi: 10.1021/bi035825a. [DOI] [PubMed] [Google Scholar]
- 50.Peleg G, Ghanouni P, Kobilka BK, Zare RN. Single-Molecule Spectroscopy of the Beta-2 Adrenergic Receptor: Observation of Conformational Substates in a Membrane Protein. Proc Natl Acad Sci U S A. 2001;98:8469–8474. doi: 10.1073/pnas.151239698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao XJ, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B. Coupling Ligand Structure to Specific Conformational Switches in the Beta(2)-Adrenoceptor. Nat Chem Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 52.Fischer M, Georges J. Fluorescence Quantum Yield of Rhodamine 6G in Ethanol as a Function of Concentration using Thermal Lens Spectrometry. Chem Phys Lett. 1996;260:115–118. [Google Scholar]
- 53.Lakowicz JR. Principles of Fluorescence Spectroscopy; Kluwer Academic; New York: 1999. [Google Scholar]
- 54.Cohen AE, Moerner WE. The Anti-Brownian Electrophoretic Trap (ABEL Trap): Fabrication and Software. Proc SPIE. 2005;5699:296–305. [Google Scholar]
- 55.Jiang Y, Wang Q, Cohen AE, Douglas N, Frydman J, Moerner WE. Hardware-Based Anti-Brownian Electrokinetic Trap (ABEL Trap) for Single Molecules: Control Loop Simulations and Application to ATP Binding Stoichiometry in Multi-Subunit Enzymes. Proc Soc Photo Opt Instrum Eng. 2008;7038:703807. doi: 10.1117/12.798093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watkins LP, Yang H. Detection of Intensity Change Points in Time-Resolved Single-Molecule Measurements. J Phys Chem B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 57.Zander C, Sauer M, Drexhage KH, Ko D, Schulz A, Wolfrum J, Brand L, Eggeling C, Seidel CAM. Detection Characterization of Single Molecules in Aqueous Solution. Appl Phy B. 1996;63:517–523. [Google Scholar]
- 58.Baptista MS, Indig GL. Effect of BSA Binding on Photophysical and Photochemical Properties of Triarylmethane Dyes. J Phys Chem B. 1998;102:4678–4688. [Google Scholar]
- 59.Fürstenberg A, Kel O, Gradinaru J, Ward TR, Emery D, Bollot G, Mareda J, Vauthey E. Site-Dependent Excited-State Dynamics of a Fluorescent Probe Bound to Avidin and Streptavidin. Chem Phys Chem. 2009;10:1517–1532. doi: 10.1002/cphc.200900132. [DOI] [PubMed] [Google Scholar]
- 60.Wlodarczyk J, Kierdaszuk B. Interpretation of Fluorescence Decays using a Power-Like Model. Biophys J. 2003;85:589–598. doi: 10.1016/S0006-3495(03)74503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siemiarczuk A, Wagner BD, Ware WR. Comparison of the Maximum Entropy and Exponential Series Methods for the Recovery of Distributions of Lifetimes from Fluorescence Lifetime Data. J Phys Chem. 1990;94:1661–6. [Google Scholar]
- 62.Kinosita K, Jr, Kawato S, Ikegami A. A Theory of Fluorescence Polarization Decay in Membranes. Biophys J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipari G, Szabo A. Effect of Librational Motion on Fluorescence Depolarization and Nuclear Magnetic Resonance Relaxation in Macromolecules and Membranes. Biophys J. 1980;30:489–506. doi: 10.1016/S0006-3495(80)85109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwille P, Kummer S, Heikal AA, Moerner WE, Webb WW. Fluorescence Correlation Spectroscopy Reveals Fast Optical Excitation-Driven Intermolecular Dynamics of Yellow Fluorescent Proteins. Proc Natl Acad Sci U S A. 2000;97:151–156. doi: 10.1073/pnas.97.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Box GEP, Jenkins GM. Time Series Analysis forecasting and control; Holden-Day, Inc; San Francisco: 1970. [Google Scholar]
- 66.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/Off Blinking and Switching Behavior of Single Green Fluorescent Protein Molecules. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 67.Hanson J, Yang H. A General Statistical Test for Correlations in a Finite-Length Time Series. J Chem Phys. 2008;128:214101. doi: 10.1063/1.2931943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung SH, Stevens JR. Time-Dependent Correlation and the Evaluation of the Stretched Exponential Or Kohlrausch-Williams-Watts Function. Amer J Phys. 1991;59:1024–1030. [Google Scholar]
- 69.Lu C, VandenBout DA. Effect of Finite Trajectory Length on the Correlation Function Analysis of Single Molecule Data. J Chem Phys. 2006;125:124701-1–124701-9. doi: 10.1063/1.2352748. [DOI] [PubMed] [Google Scholar]
- 70.Neumann L, Wohland TJWR, Zare RN, Kobilka BK. Functional Immobilization of a Ligand-Activated G-Protein-Coupled Receptor. Chem Bio Chem. 2002;3:993–998. doi: 10.1002/1439-7633(20021004)3:10<993::AID-CBIC993>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 71.Swaminath G, Deupi X, Lee TW, Zhu W, Thian FS, Kobilka TS, Kobilka B. Probing the Beta(2) Adrenoceptor Binding Site with Catechol Reveals Differences in Binding and Activation by Agonists and Partial Agonists. J Biol Chem. 2005;280:22165–22171. doi: 10.1074/jbc.M502352200. [DOI] [PubMed] [Google Scholar]
- 72.Nau WM, Hennig A, Koner AL. Squeezing Fluorescent Dyes into Nanoscale Containers-the Supramolecular Approach to Radiative Decay Engineering. Springer Ser Fluoresc. 2008;4:185–211. [Google Scholar]
- 73.Stracke F, Blum C, Becker S, Mullen K, Meixner AJ. Correlation of Emission Intensity and Spectral Diffusion in Room Temperature Single-Molecule Spectroscopy. Chem Phys Chem. 2005;6:1242–1246. doi: 10.1002/cphc.200400436. [DOI] [PubMed] [Google Scholar]
- 74.Watanabe K, Klein ML. Shape Fluctuations in Ionic Micelles. J Phys Chem. 1989;93:6897–6901. [Google Scholar]
- 75.Halle B, Landgren M, Jonsson B. The Shape of Ionic Micelles. Journal de Physique. 1988;49:1235–1259. [Google Scholar]
- 76.Bond PJ, Cuthbertson JM, Deol SS, Sansom MSP. MD Simulations of Spontaneous Membrane protein/detergent Micelle Formation. J Am Chem Soc. 2004;126:15948–15949. doi: 10.1021/ja044819e. [DOI] [PubMed] [Google Scholar]
- 77.Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE. Identification of Two Distinct Inactive Conformations of the Beta(2)-Adrenergic Receptor Reconciles Structural and Biochemical Observations. Proc Natl Acad Sci U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.