Abstract
Adaptations are constructed through the sequential substitution of beneficial mutations by natural selection. However, the rarity of beneficial mutations has precluded efforts to describe even their most basic properties. Do beneficial mutations typically confer small or large fitness gains? Are their fitness effects environment specific, or are they broadly beneficial across a range of environments? To answer these questions, we used two subsets (n = 18 and n = 63) of a large library of mutants carrying antibiotic resistance mutations in the bacterium Pseudomonas fluorescens whose fitness, along with the antibiotic sensitive ancestor, was assayed across 95 novel environments differing in the carbon source available for growth. We explore patterns of genotype-by-environment (G×E) interactions and ecological specialization among the 18 mutants initially found superior to the sensitive ancestor in one environment. We find that G×E is remarkably similar between the two sets of mutants and that beneficial mutants are not typically associated with large costs of adaptation. Fitness effects among beneficial mutants depart from a strict exponential distribution: they assume a variety of shapes that are often roughly L shaped but always right truncated. Distributions of (beneficial) fitness effects predicted by a landscape model assuming multiple traits underlying fitness and a single optimum often provide a good description of the empirical distributions in our data. Simulations of data sets containing a mixture of single and double mutants under this landscape show that inferences about the distribution of fitness effects of beneficial mutants is quite robust to contamination by second-site mutations.
BENEFICIAL mutations provide the raw material for adaptive evolution. Yet little is known about the properties of beneficial mutations because population genetics theory has placed far more emphasis on understanding the more abundant class of deleterious mutations (Eyre-Walker and Keightley 2007). The reason for this derives largely from arguments for the importance of neutrality in molecular evolution, which posit that beneficial mutations should be so rare that they would almost never be seen in nature. The result is a rich body of theory on the importance of deleterious mutations for the evolution of genetic systems such as ploidy, recombination, and life cycles. However, the theory has been comparatively silent on beneficial mutations, the “stuff” of adaptive evolution (Orr 2005).
The realization that a genuinely predictive theory of evolution must be able to accommodate all mutations—whether they be deleterious, neutral, or beneficial—has led to attempts on both the theoretical and empirical fronts to describe the distribution of fitness effects (DFE) among mutations exposed to selection. Although we are still some way from describing the complete DFE among all mutations, theoretical work stemming from the mutational landscape models of Gillespie (1984) suggests that restricting attention to beneficial mutations may provide the beginnings of a general theory of adaptive evolution (Joyce et al. 2008). The essential insight here is that, provided the starting genotype is already fairly well adapted to a given set of conditions, beneficial mutations represent draws from the right-hand tail of a complete DFE. This fact allows us to use extreme value theory (EVT) to describe the distribution of fitness effects among beneficial mutations, even if the complete DFE remains difficult to characterize. The key result is that, regardless of the underlying probability distributions used to model the DFE (the exponential, normal, and gamma distributions are the ones most often employed), the DFE among beneficial mutations has an exponentially distributed right tail, with many mutations of small effect and few of large effect (Orr 2003).
The generality of this prediction can be questioned on at least four counts. First, the theory concerns the spectrum of fitness effects among single mutations, meaning those that are a single nucleotide away from the wild-type sequence. Double mutants are assumed to be rare and effectively ignored. However, direct sequencing of whole genomes from mutation accumulation experiments puts the genome-wide mutation rate, U, approximately two to three orders of magnitude higher than had previously been estimated from phenotypic analyses (reviewed in Halligan and Keightley 2009). In yeast, for example, indirect estimates of U average four mutations in every 1000 genomes replicated, but Lynch et al. (2008) estimated that U = 0.32 using whole-genome resequencing. Double mutants may thus be sufficiently common to impact the DFE among mutations, especially in populations of bacteria or viruses with large population sizes. It is unclear how such contamination of the spectrum of mutants available to selection affects the DFE among all mutations, and beneficial mutations in particular.
Second, the theory, in its current form, does not make any predictions about the joint fitness effects of beneficial mutations across several novel environments. In particular, one would like to know to what extent mutations that improve fitness in one set of conditions also improve it in others. Are the pleiotropic effects of new mutations in different conditions also jointly exponentially distributed? Do beneficial mutations tend to be ecologically specialized—that is, beneficial in a narrow range of environments—or do they tend to maintain high fitness across a broad range of conditions? Knowing the direction and magnitude of the joint fitness effects among beneficial mutations is crucial for understanding the evolution of niche specialization. It is also important in an applied context: when pathogens become resistant to antibiotics or antiviral drugs (that is, gain a beneficial mutation), how large a cost of resistance should we expect to see? The answer matters because the cost of resistance is an important factor governing the prevalence of resistance in the absence of antibiotic (Lipsitch et al. 2000) and so can be used to guide treatment strategies.
Third, an exponential is predicted under EVT only if the underlying DFE of all mutations is in the so-called Gumbel domain of attraction. Beisel et al. (2007) have pointed out that there are actually two other domains of attraction predicted by EVT: the Fréchet domain containing distributions with tails that are heavier than the exponential and the Weibull domain containing distributions that are bounded on the right. Neither of these distributions converge to an exponential, although they can look exponential-like (L shaped). Efforts to identify which distribution best describes empirical data have been undertaken with bacteria (Kassen and Bataillon 2006; Maclean and Buckling 2009; McDonald et al. 2010) and viruses (Rokyta et al. 2008), but the results are conflicting because studies of beneficial mutations comprise very few mutants and attention has been focused on relatively few environments: only four in Kassen and Bataillon (2006) and one in the remainder.
Fourth, recent theoretical work has derived the full DFE among mutations in Fisher’s geometric model (FGM) under the assumption that mutations pleiotropically affect a set of quantitative traits that are under stabilizing selection with fitness declining as a Gaussian function of the distance of a genotype to an optimum. Under this model, the DFE of new mutations is well approximated by a displaced gamma distribution, where the magnitude of the displacement is a measure of how mal-adapted the initial genotype is (Martin and Lenormand 2006). The DFE among beneficial mutations in this model can be markedly different from those in an exponential model, although T converges to an exponential one when fitness is underlain by a large number of independent traits. Martin and Lenormand (2008) have derived a β-approximation for the (scaled) distribution of beneficial mutations under FGM that falls within the Weibull domain of EVT.
Here we address each of these issues empirically by using a subset of a large library of single-step resistance mutants in P. fluorescens previously used by Kassen and Bataillon (2006). Specifically, we are interested in the following four questions: (1) What are the effects of beneficial mutations on the degree of niche specialization? This question is one component of the larger issue concerning the extent to which single mutations contribute to G×E interaction (Remold and Lenski 2001). (2) How general is the prediction of an exponential DFE? We test this prediction directly. (3) Does a simple fitness landscape model such as FGM provide an adequate description of the DFE among all mutations, including beneficial ones? (4) How robust are the predictions of FGM to the inclusion of at least some genotypes that carry multiple mutations? To answer these questions, we make use of two subsets of mutants reported in Kassen and Bataillon (2006)—those that were previously identified as being beneficial (hereafter referred to as the “top 18” mutants) in the same environment in which resistance was selected and a collection of 63 mutants collected at random without regard to their fitness under the original test conditions (hereafter referred to as the “random 63”). For both subsets, we assay the fitness of these mutants, together with the ancestral strain, across a wide range of novel environments, each differing in the carbon substrate available for growth.
Materials and Methods
Mutant strains
The protocol for mutant collection is given in Kassen and Bataillon (2006). Briefly, a collection of 673 independently derived strains containing mutations conferring resistance to the quinolone antibiotic nalidixic acid was obtained by screening >2000 populations of the wild-type strain SBW25 of P. fluorescens in a conventional fluctuation-style assay. This method permits the collection of novel genotypes derived from independent mutational events occurring naturally during population expansion without regard to their fitness effects in the assay environments. The primary targets of chromosomal quinolone resistance in Gram-negative bacteria are the DNA gyrases, GyrA and GyrB, where point mutations in a highly conserved region (the quinolone resistance determining region, or QRDR) within gyrA and gyrB are especially common, and topoisomerase IV, porins, or efflux systems (Bagel et al. 1999; Jalal et al. 2000; Kohler and Pechere 2001). Although we have not undertaken an exhaustive analysis to identify all possible sites conferring resistance in our collection, we have found five distinct nonsynonymous mutations in 15 of the 18 strains sequenced from a 500-bp region of the QRDR of gyrA (Table 1). Four mutations are substitutions in either amino acids 83 (Thr) or 87 (Asp), which are among the most common sites associated with resistance to quinolones in clinical isolates of Pseudomonas aeruginosa (Wong and Kassen 2011), and one is at position 81 (Gly). Additional sequencing and phenotypic assays (see below and Table 1) reveal at least 15 unique genotypes in our top 18 collection.
Table 1. DNA sequence variation among the top 18 mutants.
| Strain | Fitness rank in LB | gyrAa,b | Second-site mutationsc | Fold increase in minimum inhibitory concentration (MIC) to nalidixic acid | Difference in log(MIC) following addition of efflux inhibitor d,e | Minimum number of mutations | Genotype classf |
|---|---|---|---|---|---|---|---|
| SBW25 | 19 | — | — | 1 | 0.798 ± 0.177 | 0 | Wild type |
| 3-1-B1 | 4 | — | NA | NA | NA | 1 | A |
| 1-5-D8 | 7 | — | mexCD regulator: Leu134Phe (C514G) | 47.4 | 1.457 ± 0.129** | 1 | B |
| 2-10-G10 | 15 | — | NA | NA | NA | 1 | A |
| 1-7-F5 | 2 | Asp87Ala (GAC260GCC) | NA | 47.4 | 1.545 ± 0.116** | 1 | C |
| 1-4-B9 | 1 | Asp87Gly (GAC260GGC) | mexAB regulator: GC ins stop codon at AA 44 | 47.4 | 0.088 ± 0.173** | 2 | D |
| 1-8-G4 | 11 | Asp87Gly (GAC260GGC) | NA | 25.2 | 1.007 ± 0.128 | 1 | E |
| 1-6-F8 | 12 | Asp87Gly (GAC260GGC) | NA | 25.2 | 0.968 ± 0.173 | 1 | E |
| 2-9-H4 | 3 | Asp87Tyr (GAC259TAC) | — | 25.2 | 0.304 ± 0.128* | 1 | F |
| 1-1-H10 | 6 | Asp87Tyr (GAC259TAC) | mexEF regulator: Ser88Asn (G863A) | 101.0 | 0.389 ± 0.112* | 2 | G |
| 2-4-A12 | 8 | Asp87Tyr (GAC259TAC) | — | 101.0 | 0 ± 0** | 2 | H |
| 2-8-F7 | 10 | Asp87Tyr (GAC259TAC) | NA | 21.0 | 0.576 ± 0.128 | 1 | I |
| 1-10-H10 | 13 | Asp87Tyr (GAC259TAC) | NA | 101.0 | 0 ± 0** | 2 | H |
| 1-1-G1 | 17 | Asp87Tyr (GAC259TAC) | NA | 25.2 | 0.176 ± 0.245* | 1 | J |
| 2-8-F8 | 5 | Gly81Cys (GGC241TGC) | — | 56.8 | 1.537 ± 0.208* | 1 | K |
| 3-1-A6 | 9 | Thr83Ile (ACT248ATT) | NA | 31.6 | 0 ± 0.164** | 1 | L |
| 1-6-A9 | 14 | Thr83Ile (ACT248ATT) | NA | 19.9 | 0.401 ± 0.208* | 1 | M |
| 1-8-A1 | 16 | Thr83Ile (ACT248ATT) | NA | 101.0 | 0.908 ± 0.210 | 1 | N |
| 2-7-C3 | 18 | Thr83Ile (ACT248ATT) | mexAB regulator: Arg90His (G269A) | 56.8 | 0 ± 0** | 2 | O |
—, identical to wild type (SBW25).
NA, not sequenced or data not available.
Sequences assayed were the QRDR of gyrB, parC, and parE, as well as the DNA-binding sites of the putative efflux pump regulators mexAB, mexCD, mexEF, and a generic mex-family regulator; see Materials and Methods for details.
Values shown are mean difference in log(MIC) without and with the efflux inhibitor, PAbN ± 95% confidence intervals; see Materials and Methods for details.
Significant from SBW25 at *P < 0.05. Significant from SBW25 following Bonferroni correction for 16 tests such that **P < 0.003.
Genotypes sharing the same letter are putatively identical on the basis of direct sequencing and the efflux inhibitor assay. Wild type is the genome of the ancestral, nalidixic acid-sensitive SBW25 strain from which all strains are derived.
We assessed the prevalence of second-site mutations in our top 18 mutant collection using two approaches. First, we sequenced the QRDRs of gyrB, parC, and parE and the putative efflux pump regulatory sites mexAB, mexCD, mexEF, and an additional unidentified mex-family regulator. Putative efflux pump transcriptional regulators were identified as transcription factors situated immediately upstream of mex-type multidrug efflux pump operons. As efflux pump systems in P. fluorescens are poorly characterized, we pinpointed orthologs of the P. aeruginosa MexAB-OprM, MexCD-OprJ, and MexEF-OprN efflux pumps in the P. fluorescens SBW25 genome using BLAST analysis. Primers used for sequencing are available from the authors upon request. Second, we assayed for efflux pump-mediated resistance by examining the difference in minimum inhibitory concentration (MIC) to nalidixic acid in the presence and absence of Phe-Arg-β-napthylamide (PAbN). PAbN, also referred to in the literature as MC-207-110, was first identified as an inhibitor of three resistance-nodule-division multidrug efflux pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN) in P. aeruginosa (Lomovskaya et al. 2001). PAbN enhances the effect of a number of antibiotics including fluoroquinolones, chloramphenicol, oxazolidinones, and rifampicin. It is believed that PAbN acts as a competitive inhibitor of antibiotics by binding to antibiotic-binding sites within membrane transporter proteins, thus impairing antibiotic efflux (Lomovskaya and Bostian 2006). We estimated the minimum number of mutations for a given strain by first grouping mutants according to their gyrA genotype and then by examining each of these for the presence of second-site mutations at all non-gyrA targets. For those strains where no second-site mutations were found, we then compared the PAbN phenotype to that of the wild type using two-tailed t-tests. A significant difference in MIC in the presence and absence of PAbN relative to the wild type was taken as evidence of an additional mutation affecting resistance.
Fitness assays
All strains, including the nalidixic acid-sensitive ancestor, SBW25, were first streaked to single colonies from frozen cultures and then grown overnight at 28° in liquid media containing 5 ml of Luria–Bertrani [LB; 10 gL-1 (grams per liter) bacto-tryptone, 5 gL-1 yeast extract, 10 gL-1 NaCl] medium. Resistant strains were grown in the presence of antibiotic (100 μl⋅ml−1 nalidixic acid). Aliquots of 50 μl from overnight cultures were then starved for 2 hr in 50 ml of M9 minimum salts (1 gL-1 NH4Cl, 3 gL-1 KH2PO4, 0.5 gL-1 NaCl, 6.8 gL-1 Na2HPO4, supplemented with 15 mgL-1 CaCl2 and 0.5 gL-1 MgSO4) while being shaken at 150 × g at 28°. A total of 150 μl from the starved cultures was then inoculated into each well of commercially available microwell plates containing 95 different carbon substrates (Biolog), and the optical density (OD) was read at 630 nm with an automated microwell plate reader. Cultures were allowed to grow, unshaken, at 28° for 48 hr and the OD was read again. Fitness was estimated as the difference between the final and initial OD readings (ΔOD). All assays were conducted in triplicate, giving a total of (19 strains × 3 replicates) + (64 × 3 replicates) = 249 Biolog plates for both experiments.
PCR amplification and sequence determination
Primers were designed to amplify the fragments of gyrA positions 1–791, gyrB positions 1268–2000, parC positions 1–797, and parE positions 1120–1905 containing the P. fluorescens QRDR region, and primers for the putative efflux pump regulators were designed to amplify the entire gene. The PCR amplification protocol composed of denaturation for 5 min at 95° and then 35 cycles of denaturation for 30 sec at 95°, annealing for 30 sec at a temperature between 55° and 65° that was optimized for each gene, elongation for 1 min at 72°, and a final elongation step for 10 min at 72°. The reactions were carried out in a 50-µl volume with 2.5 units of Taq DNA polymerase (Invitrogen). The PCR-amplified DNA was sent to the Genome Québec Innovation Centre at McGill University for sequencing on the forward strand, and mutations were identified using BLAST analysis against the SWB25 genome (GenBank accession no. AM181176).
Efflux pump inhibition assay
MIC analysis was performed in cation-adjusted Mueller–Hinton broth in 96-well microtiter plates following a microbroth dilution method adapted from the National Committee for Clinical Laboratory Standards protocol (Schwalbe et al. 2007). An inoculum density of ∼5 × 105 cfu/ml was employed in all experiments, and nalidixic acid MIC values for the wild-type and mutant strains in the presence or absence of 5 µg/ml of the efflux pump inhibitor PAbN were determined in two replicates following a 24-hr incubation period at 28°.
Niche specialization among the top 18 mutants
We assayed the degree of niche specialization in two ways. First, we counted the number of environments on which each beneficial mutant grew relative to the ancestral strain. Second, for each strain we calculated its fitness effect, which was the difference in fitness between a mutant strain and the ancestor, in each environment and then took the average fitness effect for each strain across all environments. We omitted from this analysis any environments where the mutant strain grew but the ancestor did not.
Genotype-by-environment interaction
We performed a two-way ANOVA with strains and environment as random main effects. From this we calculated the variance components associated with each main effect and their interaction by equating expected with observed mean squares using the lmer function in the lme4 package in R (R Development Core Team 2009). These calculations were done by including only environments in which a strain showed evidence of growth assessed as a mean OD that was greater than twice the standard deviation of OD measurements taken from the blank well of a BIOLOG plate. The sensitive ancestor, SBW25, was omitted from this analysis. We further decomposed the G×E interaction variance, VGE, using Robertson’s (1959) equation for the G×E between any pair of environments: VGE = 1/2 (SG1 – SG2)2 + SG1⋅SG2 (1 − rG1G2), where SGi is the genetic standard deviation in environment i, and rG1G2 is the cross-environment genetic correlation. In this model, (SG1 – SG2)2 expresses the squared difference in the amount of genetic variation between the two environments. We regressed VGE against the squared difference in average performance between a pair of environments, which represents macroenvironmental variance expressed by the same collection of genotypes tested in a pair of environments. We assessed the significance of the slope of the regression in each case using a randomization procedure (1000 randomizations were used) written in R (R Development Core Team 2009).
Analysis of the DFE among beneficial mutants
Irrespective of statistical significance within each environment, we deem as “beneficial” all mutant genotypes that had a higher mean fitness than the ancestral genotype. We first analyzed our data sets under the assumption that all mutants contain a single mutation affecting fitness. We then examined the robustness of our inferences about the DFE in the presence of contamination by second-site mutations (see Monte Carlo simulation of DFE expected in empirical data sets). Instead of relying on the likelihood ratio test (LRT) developed by Kassen and Bataillon (2006), we used a likelihood framework developed by Beisel et al. (2007) that allows testing of the exponential against other domains of attraction predicted by EVT. In particular, we obtained the empirical DFE in each BIOLOG environment by scaling mutation effects relative to the smallest beneficial mutations instead of the ancestral type. This shifting procedure is expected to make our analysis much more robust to missed beneficial mutations and only “costs” 1 d.f. by decreasing the sample size by 1 in each environment. Within each environment, we obtain the likelihood of our data (our observation consisting of the shifted beneficial mutation effects) under a generalized Pareto model with two parameters (shape and scale). A formal test for the hypothesis that the DFE of beneficial mutations is exponential is conducted using a LRT comparing the likelihood of the full model with that of a reduced model (scale parameter κ set to zero in the exponential case). A P-value for the LRT is obtained by parametric bootstrapping (10,000 boostrap samples from an exponential with the fitted scale were used). The analysis of the data was done in R [R Development Core Team (2009)].
Given that each test is performed on a small sample size, we conducted a pooled test across environments where at least six beneficial mutations were detected. Briefly, within each environment i, a P-value pi is collected for the LRT described above. Under the null hypothesis that the DFE of beneficial mutation is exponential in all k environments, the pi’s are uniformly distributed between 0 and 1, and the statistic L = −2 Σi ln(pi), with i = 1..k, is distributed as a χ2 with 2k d.f.
Finally, we consider the predictions about the DFE among beneficial mutations using FGM under the assumption that the wild type is close to an optimum (Martin and Lenormand 2008). We first rescaled the data using the largest beneficial mutant as a proxy for the distance to the optimum and then fit a constrained β-distribution Be[a = 1, b] to this rescaled data. We compare the fit of the constrained β to a fit with two free parameters (the shape parameter a is also free to vary) using a LRT. Note that this procedure should not be interpreted as a test of the fit of an explicit landscape model against any other model such as those that assume only EVT conditions. This is because both models make the same underlying assumption that the wild-type genotype is initially well adapted and, moreover, that the two models converge when the number of independent characters determining fitness is large.
Monte Carlo simulation of DFE expected in empirical data sets
We examined how the presence of second-site mutations affected our ability to infer the DFE among beneficial mutations by first simulating the DFE expected under ideal conditions (all mutants are exactly one mutational step away from the wild type) and under “less than ideal,” but more realistic, conditions where the collection of mutants comprises a mixture of true single-step mutations and those carrying extra mutations influencing fitness. To simulate the DFE, we used a fitness model based on FGM. The rationale for this choice is twofold. First, FGM generates explicit predictions about epistasis and the DFE among mutants carrying an arbitrarily large number of mutations (Martin et al. 2007), something that the mutational landscape model cannot easily do. Second, because we have fit our data to a landscape model generated using predictions obtained under ideal conditions, it provides a natural means of assessing the degree to which contamination by double mutants changes the DFE.
Simulations followed the parameterization of FGM described in Martin et al. (2007), and we examined different scenarios by varying the effective number of dimensions of the landscape (m), the mean fitness effect of a single mutation, E(s), and the initial distance to the optimum (so). To obtain the DFE of all mutants, we simulated the fitness of 100,000 independent mutants carrying 1 + c mutations affecting fitness, where c is a stochastic number of “contaminating” mutations. Following the description of our procedure for isolating mutants, we assumed that c was Poisson distributed with a mean λ. Thus, λ = 0 generates the case of pure single-step mutants while λ = 0.5 represents the less than ideal condition of allowing for double mutants at a rate that roughly matches, and may even be higher than, the proportion of mutants carrying more than a single fitness affecting a mutation uncovered in our samples (see below).
We then studied how the shape of the DFE, κ, associated with beneficial mutations was affected by the presence of multiple mutations. We focused on κ as it is of central interest in characterizing beneficial mutations. We examined how κ changes with different combinations of the parameters determining the DFE [m, so, E(s), λ] by sampling 10–20 mutants from each of 1000 simulated data sets for each distribution and re-estimating the shape parameter κ.
Results and Discussion
DNA sequence variation among top 18 mutants
We used both sequencing and a phenotypic assay in an effort to identify the genetic targets of quinolone resistance in our top 18 mutants. Our results are reported in Table 1. As is often observed in clinical strains of Gram-negative bacteria resistant to quinolones, all but three strains contained mutations in the QRDR of gyrA. Within this site we found five distinct mutations, four of which occur at two amino acid sites (83 and 87) known to be associated with binding of GyrA to DNA (Wong and Kassen 2011). No further mutations were found at the sites often associated with the primary incidences of quinolone resistance—gyrB, parC, or parE—among the subset of the top 18 chosen for further sequencing. We did, however, uncover four additional unique mutations in putative regulators of efflux pumps, three of which occurred alongside a gyrA mutation. These results, combined with our phenotypic assay, suggest that we have at least 15 unique strains and five double mutants in our collection. Note that this assay is conservative because we have examined only those sites that are known or thought to be involved in conferring resistance to quinolones. Recent efforts to identify alternative genetic targets of resistance to the fluoroquinolone ciprofloxacin in P. aeruginosa, a close relative of P. fluorescens, suggest that there may be >40 additional genes capable of conferring a more than twofold reduction in susceptibility as measured by MIC (Breidenstein et al. 2008). Given this wide range of putative resistance targets, we suspect that the majority, if not all, of our strains are genetically unique as required by theory.
That being said, we have also clearly violated the assumption that all mutants in our collection are a single-nucleotide step away from the wild type, with at least 28% of our strains carrying second-site mutations. This is likely to be something of an underestimate because, by design, we have searched for double mutants only at those sites known to be clinically important for conferring resistance to quinolones and fluoroquinolones in related species. If genomic mutation rates are indeed higher than previously thought, we may have missed second-site mutations occurring elsewhere in the genome. The observation of double mutants is not unusual, at least among clinical isolates of P. aeruginosa. Wong and Kassen (2011) showed that clinical isolates of fluoroquinolone-resistant P. aeruginosa often contain multiple mutations to both gyrases and efflux pumps and that the identity of the efflux pump mutation depends on whether the infection is acute or chronic. Although it is unlikely that double mutants arise simultaneously in clinical strains, the fact that they do happen at all suggests that having more than one resistance-associated mutation confers a fitness advantage. Our results (Table 1) indicate that double mutants in our collection have MICs to nalidixic acid that are nearly 10× higher than single mutants (Welch’s one-tailed two sample, t10.6 = 3.66, P = 0.002). However, double mutants did not tend to have higher fitness ranks in LB than single mutants (Welch’s one-tailed two sample, t6 = 0.128, P = 0.549), nor was there any detectable relationship between the MIC conferred and the fitness rank in LB among strains in general (slope ± standard error = −0.006 ± 0.030, F1,14 = 0.046, P = 0.832). Taken together, these results suggest that, although the second-site mutations that we identified tend to substantially decrease susceptibility to nalidixic acid, they contribute little to the variation in fitness in the absence of a drug. Below we consider in more detail how the occurrence of such double mutants might impact inferences about the DFE among mutations.
Most beneficial mutants do not pay a substantial cost of adaptation in new environments
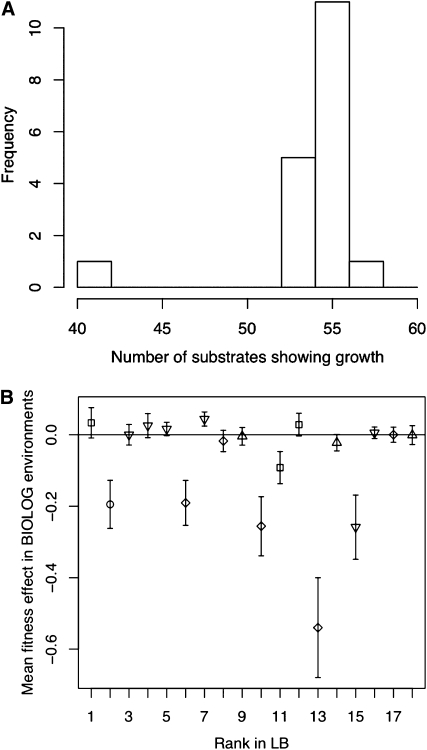
We used several indicators to quantify the amount of ecological specialization associated with the set of 18 mutants deemed beneficial in the original LB environment. First, we characterized the niche breadth of a genotype as the number of carbon sources that it effectively utilizes. The ancestral type grew on 55 of the 95 carbon sources available. With the exception of one mutant genotype (which lost the ability to grow in >10 environments relative to wild type), all other beneficial mutants had a niche breadth similar to wild type (Figure 1A). Second, we calculated a mean fitness effect, relative to the wild type, of each mutant across all environments (where growth was available). We found no relationship between the mean fitness effect of a mutant across environment and the fitness rank of the mutant in permissive LB media, the media in which beneficial effects were assessed in Kassen and Bataillon (2006) (Figure 1B). Taken together, these results suggest that a mutant beneficial in one environment is not substantially deleterious in other environments. Thus, specialization underlain by a cost of adaptation (i.e., the loss of fitness in the original environment relative to the ancestor) is unlikely to evolve in a single mutational step; rather, it will require the substitution of multiple beneficial mutations.
Figure 1 .
Patterns of ecological specialization of the 18 beneficial mutations. (A) Distribution of number of environments showing growth in the top 18 mutants. Note that the niche breadth of the ancestral type is at the mode of the distribution (i.e., could grow in 55 environments). (B) Mean fitness effects (averaged across all carbon sources) of the top 18 mutants. Rank is based on the performance in the initial environment (LB). Symbols designate known (to date) mutations in the 500-bp region of the gyrA QRDR: □, Asp87Gly; ○, Asp87Ala; ▿, WT; ⋄, Asp87Tyr; ▵, Thr83Ile.
Patterns of G×E interaction among single-step mutants do not differ significantly between beneficial mutants and a random sample of mutants
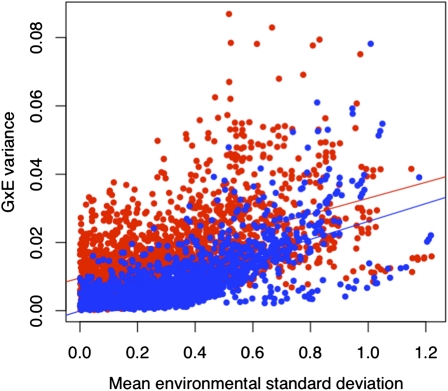
We contrasted patterns of G×E interactions in the set of 18 mutants previously deemed beneficial in LB medium and a set of 63 mutants selected at random from the library of resistant strains. Notably, the random 63 mutants displayed a wide range of fitness values that included both deleterious and beneficial effects in LB. If there is something unusual or distinct about the fitness effects of these beneficial mutations in our top 18 collection in novel environments, then we should see this reflected in the pattern of G×E variance between the two sets of strains. Overall, the fraction of genetic variance attributable to G×E (VGE) effects was similar between the two sets: VGE accounted for 21 and 32%, respectively, of the total genetic variance in the top 18 and the random 63 data sets. The top 18 set therefore does not exhibit any extraordinary amount of VGE relative to the subset random 63. This was confirmed by a second analysis where we considered pairs of environments and examined how VGE changed with the macroenvironmental variance between each pair of environments. In both cases, VGE increases as the macroenvironmental variance between a pair of environments increases, as has been observed previously (Kassen and Bell 2000), but there is little difference in the slope of this relationship for each subset (Figure 2). Together, this suggests that beneficial mutants are neither more canalized nor more reactive than a random sample of comparable mutants when exposed to the range of novel environments used here.
Figure 2 .
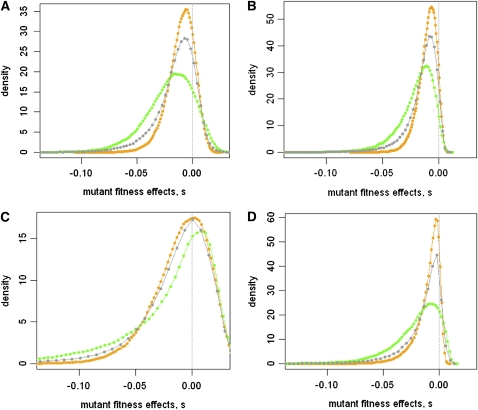
Distribution of fitness effects among pure vs. contaminated mutant genotypes. All distributions are based on the simulation of 100,000 independent mutant genotypes under the assumption of FGM parameterized as described in Materials and Methods. Note that for graphical convenience we do not display these empirical distributions as histograms but report here smoothed distributions. Orange: DFE of pure single-step mutant genotypes (ideal conditions) Gray: DFE of mutant genotypes carrying one mutation affecting fitness plus a Poisson-distributed number of extra mutations (rate of contamination, λ = 0.5). This generates a mixture of pure single-step mutants contaminated with genotypes carrying extra mutations affecting fitness. (A) m = 5; E(s) = 0.01; so = 0.05. (B) m = 5; E(s) = 0.01; so = 0.01. (C) m = 2; E(s) = 0.01; so = 0.05; Poisson rate of contamination, λ = 0.5. (D) m = 2; E(s) = 0.01; so = 0.01; Poisson rate of contamination, λ = 0.5.
It is notable that the magnitude of VGE is on the same order as that found using a small set of random insertion mutants in Escherichia coli (Remold and Lenski 2001) or in agronomic studies using a set of genotypes that are potentially a lot more divergent genetically (Bell 1997). We have at present no compelling explanation for this finding, except to say that this reinforces the conclusion that there is little that is distinctive about the top 18 mutants that were beneficial in LB.
DFE of beneficial mutants is not universally exponential across environments
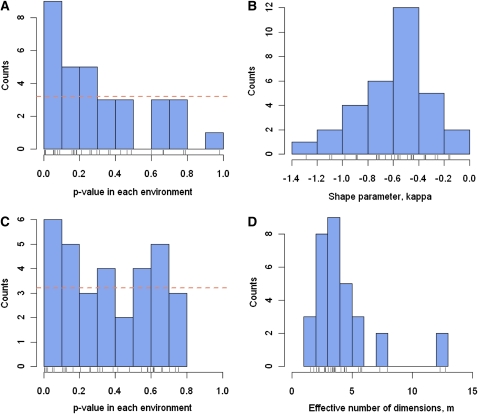
In 32 of 95 environments, at least six mutants among the top 18 were found to be better than SBW25, irrespective of formal tests of significance. We asked whether, in these environments, the exponential provided an adequate fit to the empirical distribution of fitness effects among beneficial mutants. We tested whether the shape parameter (κ) of a generalized Pareto distribution (GPD) estimated from the data differed significantly from that expected under an exponential distribution (κ = 0). We estimated the shape parameter using both the maximum likelihood (ML) estimator of Beisel et al. (2007) on (rescaled) data and an alternative estimator proposed by Rokyta et al. (2008). In all environments, estimates for the shape parameter of the GPD distribution are negative and occasionally smaller than −1 (Figure 3B), suggesting that a right-truncated distribution (whether J or L shaped) provides a better a fit of the data than an exponential distribution. Due to low sample size in each environment (from n = 6 to n = 13), the null hypothesis of an exponential distribution could rarely be rejected (of 32 tests, only 5 tests had a P-value <5% before correction for multiple testing). However, combining tests across 32 environments strongly rejects the global null hypothesis of an exponential distribution across all environments (P < 0.001; see also Figure 3A for distribution of P-values). These results are consistent with re-analyses of two viral data sets (Rokyta et al. 2008). Taken together, these results suggest that the DFE among beneficial mutants is not exponential but is, rather, located in the so-called Weibull domain of the GPD (shape parameter κ < 0) where the distributions are right truncated. It is further notable that two recent studies on the DFE of new mutations in P. fluorescens (McDonald et al. 2010) and P. aeruginosa (Maclean and Buckling 2009) also strongly reject the exponential, although in these cases the wild-type strain was initially very maladapted, and so EVT arguments are, a priori, expected to perform poorly.
Figure 3 .
Meta-analysis of the DFE across environments. (A) Distribution of P-values associated with the test of the departure from an exponential DFE in each environment. The horizontal line denotes the expected counts of P-values in each bin on the basis of the global null hypothesis that DFE is exponential in every environment. (B) Distribution of the shape parameter, κ, of the DFE of beneficial mutations in each environment. κ was estimated in each environment using an estimator proposed by Rokyta et al. (2008) that is more accurate than maximum-likelihood estimates when the shape parameter is negative. (C) Distribution of P-values associated with LRT for the departure from the null hypothesis of a constrained β-distribution for DFE in each environment. The horizontal line denotes the expected counts of P-values in each bin based on the global null hypothesis that DFE is a constrained β, Be[1,b], in every environment. (D) Distribution of the effective number of independent phenotypic traits, m, describing Fisher’s geometric landscape in each environment. Estimates of m are based on the theoretical relationship between m and the shape parameter (κ) of the DFE of beneficial mutations (κ).
Can a simple fitness landscape model predict the DFE of beneficial mutations?
Martin and Lenormand (2008) derived a β-approximation for the DFE of beneficial mutations using an explicit fitness landscape model featuring a single smooth fitness peak. Fitting their model requires that we know the distance of the initial genotype to the fitness optimum. Using the fitness effect of the largest mutant as a proxy for the distance to the optimum [a similar approximation is effectively used by Rokyta et al. (2008) to derive their alternative estimator for the shape parameter of the GPD], we first rescaled our data by this value and then fit the rescaled data to a β-distribution constrained to be Be[1, b]. Comparing the fit of this model to a nonconstrained β allows us to ask directly whether the fitness landscape model provides an adequate fit to the data in each environment.
Keeping in mind that our procedure for rescaling the data to obtain an estimate for distance to the optimum is rather crude, the constrained β predicted under a fitness landscape model does seem to provide an acceptable fit in many environments: the constrained β-model is rejected in favor of the nonconstrained β-model in only 5 of the 32 environments (using a 5% level without correction for multiple tests). Note again, however, that, although the empirical distribution of P-values associated with the test above is fairly flat (Figure 3C), a combined test pooling P-values across all 32 environments again rejects the composite null hypothesis that data in all environments is compatible with the constrained β (P < 0.01). Further work is needed to infer more rigorously the parameters of a fitness landscape from empirical data. One can also, in principle, use the shape parameter of the empirical DFE to make an approximate inference about m, the number of effectively independent traits underlying the fitness landscape in each environment. Here our analysis suggests that m is not very large, being as small to 2–4 in the majority of environments (Figure 3D). This squares nicely with theory by Martin and Lenormand (2008) showing that, under the assumption of their fitness landscape model, the DFE among beneficial mutations will be well approximated by an exponential only if a relatively large number of independent traits underlie fitness.
Inferences about the DFE among beneficial mutations are robust to contamination with multiple mutations
Our results may be criticized because our top 18 data set violates the assumption that all mutants are a single mutational step away from the wild type. This may not be so surprising in retrospect if, as seems to be the case, genomic mutation rates are two to three times higher than previously thought. However, it does call for caution when interpreting the results of experiments such as ours that have used fluctuation-style assays to “trap” mutations of interest in target genes, as these cannot always be assumed to be single mutants (see especially Maclean and Buckling 2009).
How robust are our inferences about the DFE and contamination by double mutants? Intuition suggests that the realized DFE of a collection of single and double mutants can be interpreted as the DFE among true single-step mutations with the addition of draws from the DFE of mutants carrying two mutations or more. Stochastic simulations under FGM (the fitness landscape model used above to fit our data) confirm this intuition (Figure 4; see also Martin et al. 2007). For rates of contamination compatible with our data, the DFEs containing true single-step mutations and those contaminated by multiple mutations are actually quite similar, with estimates of κ (which characterizes the DFE of beneficial mutations) from small data sets (10–20 mutants) being very similar between ideal and contaminated data sets (Table S1). Thus empirical tests of theory appear to be fairly robust even when violating the assumption that all mutants are a single mutational step from the wild type, which will likely be the case when trapping large numbers of mutants with the type of assays described above. Finally, there is little reason to think that the presence of double mutants in our data set severely biases our most important conclusion—that the DFE among beneficial mutants is not exponential but located in the Weibull domain of the GPD (shape parameter κ < 0) as the sign of κ is always negative even if the values themselves are slightly different.
Figure 4 .
Comparison of the effect of environmental contrast on the amount of variance due to G×E, VGE, in both data sets. The macroenvironmental contrast is calculated as the squared difference in mean performance in each environment (averaged across the set of strains).
Conclusion
The development of a predictive theory of adaptive evolution has benefited from the analysis of both sequence- and phenotype-based models. Empirical evaluation of these models has been hampered, however, because beneficial mutations are so rare that sample sizes tend to be small and the inferential strength of these tests remains modest at best.
To begin to redress this lack of data, we have assayed the fitness of a sample of mutants that had previously been found to be beneficial in a single environment across a large array of new environments. This procedure allows us both to describe the pattern of niche breadth—a key ecological characteristic for understanding the evolution of specialization and the maintenance of diversity—and to provide a more comprehensive test of the theoretical predictions regarding the DFE among beneficial mutations.
In terms of niche characteristics, our main finding is that the mutants that we had previously identified as beneficial in one environment are not severely compromised in fitness in a wide range of alternative environments. Indeed, there is little evidence for strong costs of adaptation relative to the antibiotic-sensitive wild type in different environments; most mutants deemed beneficial in LB (the environment in which they were originally selected) remain beneficial, or at least neutral, across a wide range of environments. A similar result has been observed for mutants conferring a fitness advantage in a minimal glucose environment in E. coli (Ostrowski et al. 2005), suggesting that our results are not unique for being resistance mutations. Moreover, the quantity of VGE in this mutant collection does not appear to be unusual in any way, either when compared to a larger random sample of resistant mutants from the same library that includes many deleterious strains or when compared to similar analyses performed on widely different organisms (Bell 1997).
These results have at least two important implications. First, the evolution of specialization underlain by costs of adaptation is unlikely to occur through the substitution of a single mutation. Costs of adaptation, when they are found, are likely to be built up through the substitution of multiple mutations contributing to adaptation. Perhaps this result is not surprising, but it does provide an explanation for why strong fitness trade-offs and large costs of adaptation do not readily evolve in most laboratory selection experiments lasting between a few hundred and a thousand generations (reviewed in Kassen 2002). The timescale of these experiments is simply too short for large costs of adaptation to evolve. Second, to the extent that antibiotic resistance is attributable to single or double mutants that arise rapidly under antibiotic selection, our efforts to eliminate or control resistance must do more than simply halt the use of the offending antibiotic. Costs of resistance, which are a special case of costs of adaptation, are not likely to be of sufficient magnitude to eliminate resistant strains through competition with a sensitive wild type alone. Alternative strategies that use antibiotic cycling or multidrug cocktails are likely to prove much more effective.
What of the DFE among beneficial mutations? By assaying the fitness of a collection of mutants across many different environments, we have obtained a large collection of DFEs among beneficial mutants. This approach allows us to sidestep two problems at once. On the one hand, it alleviates the pervasive issue of small sample sizes. Instead, we now have many DFEs, even if each one may still be based on small sample sizes within a given environment. At the same time, this approach means that we do not have to be overly concerned with what the “natural” environment is for our mutants. Rather, we can ask the more general question, how variable is the DFE among beneficial mutants across environments, given that the wild type is fairly well adapted?
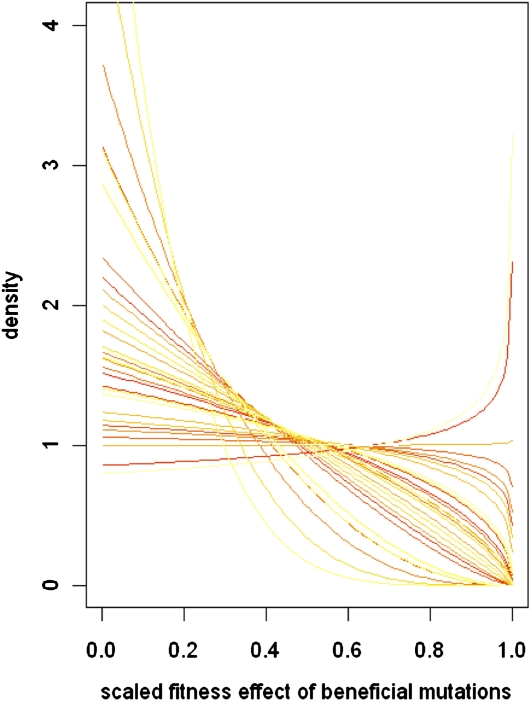
The empirical picture emerging from our study is quite clear: the DFE among beneficial mutants is one that is characterized by many mutations of small effect and few of large effect. This, again, is not surprising. But, importantly for theory, this distribution is located within the Weibull domain of attraction, not the Gumbel domain as has previously been thought. It is important to note that a wide range of κ-values within the Weibull can give rise to decreasing distributions that can, at first glance at least, resemble an exponential (Figure 5). The broad similarity among these alternative distributions notwithstanding, our meta-analysis confirms the results of previous work in bacteria (Maclean and Buckling 2009; McDonald et al. 2010), phage (Rokyta et al. 2008; Miller et al. 2011), and filamentous fungus (Schoustra et al. 2009) that the DFE among beneficial mutations is not exponential but is instead characterized by having a truncated right tail. Moreover, these conclusions are not severely compromised by the fact that our mutant collection contained some strains that carried two mutations. Our simulations of the DFE under a fitness landscape model suggest that estimation of the shape parameter for the DFE among beneficial mutants is fairly robust to violating the assumption that all mutations are a single step away from the wild type. An alternative approach might be to follow that of Martin and Lenormand (2008) and use a β-distribution to characterize the scaled DFE among beneficial mutations. Our results suggest that this approach may work quite well. Future work should focus on theory to quantify the joint DFE of new mutations in multiple environments and the development of a theoretical framework to understand the DFE in populations that are both initially well or poorly adapted.
Figure 5 .
Diversity of shapes for the scaled DFE of beneficial mutations. Each curve represents the constrained β-distribution best fitting the scaled empirical DFE of beneficial mutants in each environment (n = 32). All DFE predicted under the FGM model have a right tail that is truncated but, depending on the underlying number of dimensions, m, the distributions assume a variety of shapes. Distributions with m > 2 are essentially L shaped. When m = 2, distributions are flat (uniform). DFE can even be J shaped (when m < 2).
Acknowledgments
We thank G. Martin for sharing unpublished R code for efficient simulation of Fisher’s geometric landscape, M. Al-Azzabi for technical help, two anonymous reviewers, and Jim Bull for comments. T.B. was supported for part of this work by a Steno fellowship awarded by the Danish Council for Independent Research in the Natural Sciences. R.K. was supported by grants from the Natural Sciences and Engineering Research Council of Canada. T.B. and R.K. also acknowledge the French Embassy in Ottawa for additional funding.
Literature Cited
- Bagel S., Hullen V., Wiedemann B., Heisig P., 1999. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43: 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C. J., Rokyta D. R., Wichman H. A., Joyce P., 2007. Testing the extreme value domain of attraction for distributions of beneficial fitness effects. Genetics 176: 2441–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., 1997. Selection: The Mechanism of Evolution. Chapman & Hall, London/New York [Google Scholar]
- Breidenstein E. B., Khaira B. K., Wiegand I., Overhage J., Hancock R. E., 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52: 4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618 [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129 [DOI] [PubMed] [Google Scholar]
- Halligan D., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172 [Google Scholar]
- Jalal S., Ciofu O., Hoiby N., Gotoh N., Wretlind B., 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44: 710–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce P., Rokyta D. R., Beisel C. J., Orr H. A., 2008. A general extreme value theory model for the adaptation of DNA sequences under strong selection and weak mutation. Genetics 180: 1627–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., 2002. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15: 173–190 [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488 [DOI] [PubMed] [Google Scholar]
- Kassen R., Bell G., 2000. The ecology and genetics of fitness in Chlamydomonas. X. The relationship between genetic correlation and genetic distance. Evolution 54: 425–432 [DOI] [PubMed] [Google Scholar]
- Kohler T., Pechere C., 2001. In vitro selection of antibiotic resistance in Pseudomonas aeruginosa. Clin. Microbiol. Infect. 7(Suppl. 5): 7–10 [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Bergstrom C. T., Levin B. R., 2000. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 97: 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Warren M. S., Lee A., Galazzo J., Fronko R., et al. , 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Bostian K. A., 2006. Practical applications and feasibility of efflux pump inhibitors in the clinic: a vision for applied use. Biochem. Pharmacol. 71: 910–918 [DOI] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean R. C., Buckling A., 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5: e1000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 60: 893–907 [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2008. The distribution of beneficial and fixed mutation fitness effects close to an optimum. Genetics 179: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Elena S. F., Lenormand T., 2007. Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nat. Genet. 39: 555–560 [DOI] [PubMed] [Google Scholar]
- McDonald M. J., Cooper T. F., Beaumont H. J., Rainey P. B., 2010. The distribution of fitness effects of new beneficial mutations in Pseudomonas fluorescens. Biol. Lett. 7: 98–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. R., Joyce P., Wichman H. A., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski E. A., Rozen D. E., Lenski R. E., 2005. Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution 59: 2343–2352 [PubMed] [Google Scholar]
- Orr H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6: 119–127 [DOI] [PubMed] [Google Scholar]
- Remold S. K., Lenski R. E., 2001. Contribution of individual random mutations to genotype-by-environment interactions in Escherichia coli. Proc. Natl. Acad. Sci. USA 98: 11388–11393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A., 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15: 469–485 [Google Scholar]
- Rokyta D. R., Beisel C. J., Joyce P., Ferris M. T., Burch C. L., et al. , 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2009. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna [Google Scholar]
- Schoustra S. E., Bataillon T., Gifford D. R., Kassen R., 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol. 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbe R., Steele-Moore L., Goodwin A., 2007. Antimicrobial Susceptibility Testing Protocols. CRC Press, New York [Google Scholar]
- Wong A., Kassen R., 2011. Parallel evolution and local differentiation in quinoloneresistance in Pseudomonas aeruginosa. Microbiology 157: 937–944 [DOI] [PubMed] [Google Scholar]