Abstract
Mounting evidence points to differences in gene regulation as a major source of phenotypic variation. MicroRNA-mediated post-transcriptional regulation has emerged recently as a key factor controlling gene activity during development. MicroRNA genes are abundant in genomes, acting as managers of gene expression by directing translational repression. Thus, understanding the role of microRNA sequence variation within populations is essential for fully dissecting the origin and maintenance of phenotypic diversity in nature. In this study, we investigate allelic variation at microRNA loci in the nematode Caenorhabditis briggsae, a close relative of C. elegans. Phylogeographic structure in C. briggsae partitions most strains from around the globe into a “temperate” or a “tropical” clade, with a few strains having divergent, geographically restricted genotypes. Remarkably, strains that follow this latitudinal dichotomy also differ in temperature-associated fitness. With this phylogeographic pattern in mind, we examined polymorphisms in 18 miRNAs in a global sample of C. briggsae isolates and tested whether newly isolated strains conform to this phylogeography. Surprisingly, nucleotide diversity is relatively high in this class of gene that generally experiences strong purifying selection. In particular, we find that miRNAs in C. briggsae are substantially more polymorphic than in Arabidopsis thaliana, despite similar background levels of neutral site diversity between the two species. We find that some mutations suggest functional divergence on the basis of requirements for target site recognition and computational prediction of the effects of the polymorphisms on RNA folding. These findings demonstrate the potential for miRNA polymorphisms to contribute to phenotypic variation within a species. Sequences were deposited in GenBank under accession nos. JN251323–JN251744.
GROWING empirical evidence points to altered gene regulation as a major cause of phenotypic variation (Wray 2007; Carroll 2008). For instance, in nematodes, morphological differences in the excretory duct cell between Caenorhabditis elegans and C. briggsae are due in part to the divergence of cis-regulatory sequences (Wang and Chamberlin 2002). Similarly, divergent regulatory sequences may lead to differential chemosensory receptor expression patterns in nematode neurons (Nokes et al. 2009). Moreover, the high level of transcription factor sequence variation both between Caenorhabditis species and segregating within populations highlights the potential significance of transcriptional regulation evolution in this genus (Haerty et al. 2008; Jovelin 2009; Jovelin et al. 2009).
However, gene expression regulation is not limited to transcriptional regulation. Changes in regulation can occur at many levels, including chromatin remodeling and accessibility, alternative splicing, transcription factor and RNA stability, and post-translational modifications (Alonso and Wilkins 2005). In addition, post-transcriptional gene regulation mediated by small RNAs has emerged recently as a key factor controlling gene activity (Bartel and Chen 2004). One class of small RNA genes, the microRNAs (miRNAs), plays critical roles in development by fine tuning gene expression and by buffering regulatory networks against noise and environmental perturbations (Wu et al. 2009; Herranz and Cohen 2010). MiRNAs are abundant small regulatory RNAs (∼22 bp) that bind to the 3′-UTR of mRNAs to direct translational repression or, less frequently, mRNA cleavage, when overall sequence complementarity is high (Ambros 2004; Bartel 2004; Plasterk 2006). First identified in the nematode C. elegans (Lee et al. 1993), miRNAs are widespread among animals, plants, fungi, and viruses, with 15,172 miRNA genes from 142 species now annotated in miRBase release 16 (Kozomara and Griffiths-Jones 2011). Primary miRNA transcripts (pri-miRNA) are cleaved in the nucleus by the Drosha RNase III endonuclease, liberating a miRNA precursor (pre-miRNA) that is exported to the cytoplasm by the export receptor exportin-5. Pre-miRNAs have a stereotypical stem-loop hairpin structure that is critical for processing by the enzyme Dicer, then generating an RNA heteroduplex comprising the mature miRNA (miR) and its complementary sequence (miR*). The mature miRNA is then loaded into the RNA-induced silencing complex (RISC) where it can bind to its target mRNAs by sequence complementarity of a 7-bp long motif, the seed sequence (Bartel 2004).
In the nematode C. briggsae, a close relative to C. elegans, nucleotide variation is strongly partitioned according to geographical origin, with the majority of strains collected from nature falling into one of the two “temperate” and “tropical” clades and a minority of strains exhibiting genetically divergent multilocus haplotypes that do not follow this latitudinal differentiation (Graustein et al. 2002; Cutter et al. 2006; Dolgin et al. 2008; Howe and Denver 2008; Cutter et al. 2010; Raboin et al. 2010). C. elegans and C. briggsae both are cosmopolitan species and found commonly on rotting vegetation, suggesting that, contrary to a long-standing paradigm, their natural habitat is not soil, but environments rich in bacteria, on which they feed (Felix and Braendle 2010). Temperature is an important ecological variable for many ectothermic organisms, including nematodes (see Anderson et al. 2007). In particular, fitness-associated responses to temperature vary across genetic backgrounds in C. briggsae, depending strongly on the geographical origin of the strains. Temperate-latitude isolates have greater fecundity at low temperatures and reduced fecundity at high temperatures than do isolates from the tropical phylogeographic clade, perhaps reflecting local adaptation to climatic temperature profiles (Prasad et al. 2010).
RNA folding and stability are temperature dependent, suggesting that selection could favor alleles that confer a stable secondary structure over a broad range of temperatures or, alternatively, alleles that form temperature-specific hairpins. For instance, in A. thaliana, two MIR824 alleles, one with a temperature-sensitive structure and one with a thermoresistant structure, are maintained at intermediate frequency by balancing selection (de Meaux et al. 2008). Thus, distinct climatic regimes experienced by different populations might impose divergent selective pressure on miRNA folding structures, providing either a target for preserved function of gene regulation or a means of facilitating local adaptation.
Motivated by the phylogeographic divergence and temperature-dependent phenotypic differences in C. briggsae, in this study, we investigate nucleotide variation at 18 miRNA loci in a global sample of C. briggsae isolates that covers the range of known diversity in this species. These miRNA genes represent 12% of the C. briggsae miRNAs (de Wit et al. 2009), for which we (i) quantify the level of segregating variation and investigate selective pressures acting in present-day populations, (ii) test for divergent alleles between tropical and temperate strains, and (iii) test for divergent miRNA forms in the strains derived from unique geographic locations. We also examine 19 newly collected strains from 13 locations from around the globe, relative to previously sampled strains. We find that despite purifying selection to maintain the miRNA hairpin secondary structure, some of the strains from unique geographic locations carry alleles with mutations indicative of functional divergence.
Materials and Methods
C. briggsae strains, PCR amplification, and sequencing
We examined nucleotide diversity at miRNA loci for 25 to 48 strains, including 19 new isolates from 13 locations (Table 1). The full list of strains used to examine miRNA nucleotide variation is given in Supporting Information, Table S1. In addition, we investigated the relationships of 17 of these newly collected strains (Table 1 and Figure S1) relative to a worldwide sample of 117 strains that were used in previous studies on C. briggsae diversity, based on a standard set of six loci (Cutter et al. 2006, 2010; Dolgin et al. 2008). Newly isolated nematode strains were gifts from Marie-Anne Felix (“JU” strain prefix), Matthew Rockman (QG), Christian Braendle (NIC), Guo-Xiu Wang (VX), and Erik Anderson and Leonid Kruglyak (QX) (Table 1). The new strains were collected mostly from rotting fruits (Table 1), identified through mating tests, and founded from a single hermaphrodite individual to establish selfing strains. All strains were maintained using standard C. elegans protocols (Brenner 1974).
Table 1 . List of the newly sequenced strains used in this study with their geographical origin.
| Latitude | Longitude | Date | |||
|---|---|---|---|---|---|
| Strain | (+N, −S) | (+E, −W) | Locality | (M/Y) | Substrate |
| JU1884 | 15.56 | −88.64 | Near Cerro San Gil, Guatemala | 12/09 | Rotting fruit |
| JU1885 | 7.30 | 80.64 | Near Kandy, Sri Lanka | 12/09 | Rotting fruit |
| JU1907 | −15.51 | 145.61 | Queensland, Australia | 2/10 | Rotting fruit |
| JU1914 | −27.46 | 152.97 | Queensland, Australia | 3/10 | Rotting fruit |
| NIC5 | 47.21 | 7.53 | Solothurn, Switzerland | 7/08 | Rotting fruit |
| NIC17 | −21.02 | 55.54 | La Réunion, France | 11/09 | Fruit |
| NIC19 | 25.16 | 121.55 | Yangmingshan, Taiwan | 11/09 | Fruit |
| NIC88 | 4.08 | 52.68 | French Guiana, France | 11/09 | Fruit |
| NIC106 | 4.09 | 52.67 | French Guiana, France | 11/09 | Fruit |
| NIC107 | 4.09 | 52.67 | French Guiana, France | 11/09 | Fruit |
| NIC108 | 4.09 | 52.67 | French Guiana, France | 11/09 | Fruit |
| NIC115 | 16.95 | −61.57 | Guadeloupe, France | 3/10 | Rotting fruit |
| QG106 | 40.73 | −74.07 | Jersey City, New Jersey | 7/09 | Rotten tomato |
| QG117 | 40.73 | −74.07 | Jersey City, New Jersey | 7/09 | Rotten tomato |
| QG119 | 40.73 | −74.07 | Jersey City, New Jersey | 7/09 | Rotten tomato |
| QX1545 | 39.97 | −82.99 | Columbus, Ohio | 8/09 | Rotting vegetable |
| QX1547 | 35.97 | −83.92 | Knoxville, Tennessee | 8/09 | Rotting fruit |
| VX0033 | 30.61 | 114.32 | Wuhan, Hubei, China | 7/08 | Snail |
| VX0034 | 31.64 | 113.82 | Guangshui, Hubei, China | 7/08 | Rotting bamboo leaves |
See Table S1 for the full list of strains used for investigation of miRNA allelic variation.
Gene fragments of ∼700 bp, containing mostly intronic sequence and corresponding to the six loci used in previous studies of C. briggsae diversity, were amplified to examine the relationship of the newly collected strains to the global distribution of C. briggsae. PCR and sequencing of these six loci used published primers (Cutter et al. 2006). A total of 18 miRNAs corresponding to 12% of the C. briggsae miRNA complement (de Wit et al. 2009) also were amplified and sequenced, along with flanking sequence. MiRNA-specific primers were designed from the C. briggsae genomic sequence (Table S2) (Stein et al. 2003). For each strain, DNA was isolated from large populations of worms using the DNeasy Blood and Tissue kit (Qiagen). Amplifications were processed in 30-μl reaction volumes with 1.5 μl DMSO, 3 μl dNTPs (6.6 mm), 3 μl 10X Buffer (Fermentas), 2.4 μl MgCl2, 0.36 μl of each primer (50 μm), 0.18 μl of Taq polymerase (Fermentas), and 2 μl of genomic DNA. Cycling conditions were: 95° for 4 min followed by 35 cycles of 95° for 1 min, 55° or 58° for 1 min, and 72° for 1 min. Amplification products were sequenced at the University of Arizona Genetics Core sequencing facility. Primers were designed such that forward and reverse sequences strongly overlap, resulting in all loci being sequenced on both strands. All polymorphisms were visually verified in sequence chromatograms. In addition, we independently reamplified and resequenced all 18 miRNAs in a subset of four strains (NIC19, JU1341, JU1348, and BW287) to confirm that PCR or sequencing errors were not the cause of observed polymorphisms. This redundant sequencing confirmed all observed allelic variation for all miRNAs within these four strains, indicating that sequencing errors do not present spurious miRNA alleles into our dataset. Primer sequences were manually deleted from each locus prior to analysis. Sequences were deposited in GenBank under accession nos. JN251323–JN251744.
Sequence analyses
Orthologs of the six protein-coding genes and the 18 miRNAs were identified in the Caenorhabditis sp. 9 genome assembly using the BLASTN program (http://genome.wustl.edu/tools/blast) (Altschul et al. 1990). Each miRNA with ∼600 bp of flanking sequence was used in the BLAST search to ensure identification of the C. sp. 9 ortholog and to avoid retrieving nonorthologous miRNAs with similar sequence. C. sp. 9 was used as an outgroup because C. sp. 9 and C. briggsae are closely related and do not exhibit saturation at synonymous sites (Cutter et al. 2010; Woodruff et al. 2010; Jovelin and Phillips 2011).
We constructed multiple-sequence alignments by eye for each gene using BioEdit (Hall 1999). Sequences from the six loci used in studies of C. briggsae diversity were concatenated and combined with published sequences (Cutter et al. 2006, 2010; Dolgin et al. 2008). We then inferred relationships among C. briggsae strains from the concatenated sequences using unrooted neighbor networks generated with a Jukes–antor distance in SplitsTree 4.10 (Huson and Bryant 2006). Neighbor networks are particularly suited for analyzing relationshipsC among taxa for which reticulated evolution such as recombination events may be nonnegligible (Huson and Scornavacca 2011).
We combined our polymorphism data from the six nuclear loci with published data from 23 other loci (Cutter and Choi 2010) to compare nucleotide variation at miRNA and non-miRNA loci. Nucleotide diversity (π) (Nei 1987) was measured for each locus using DnaSP 5.10 (Librado and Rozas 2009). We determined the ancestral/derived state of each polymorphism found in the C. briggsae miRNAs by comparing to the C. sp. 9 miRNA orthologs and examined the frequency of the derived polymorphism in the global sample of C. briggsae alleles.
We then examined the structural context of each miRNA polymorphism, first by using the stem-loop structure of the AF16 reference strain allele (de Wit et al. 2009). We also investigated how temperature might affect miRNA folding patterns by examining the minimum free energy (MFE) structure of each miRNA allele at different temperatures, using the Vienna RNA server (Gruber et al. 2008). We examined the effect of polymorphism on miRNA folding at two extreme temperatures, 14° and 30°, in addition to an intermediate temperature of 20°. The effect of temperature on self-fecundity is heritable in C. briggsae and discriminates strains on the basis of their geographical origin. Lifetime fecundity is higher for temperate strains at low temperatures and higher for tropical strains at higher temperature (Prasad et al. 2010), prompting us to hypothesize that temperature-dependent RNA folding by miRNAs might correlate with phenotypic differentiation.
Finally, we investigated selective constraints on miRNAs by examining single nucleotide polymorphism (SNP) density and nucleotide divergence on the full pre-miRNAs, mature sequences (miRs), and flanking sequence. Flanking sequences include non-miRNA sequence upstream and downstream of miRNAs as well as intergenic sequence for miRNAs found in clusters. We included indels in the SNP density calculation, with indels larger than 1 bp being treated as a single polymorphism, and applied a correction for the difference in sample size among the miRNAs investigated, analogous to Watterson’s sample size correction (Watterson 1975). The SNP density per base pair is the ratio of the number of SNPs to the total length of sequence examined, divided by log[n − 1], where n is the sample size. Divergence was measured between C. briggsae AF16 and C. sp. 9 JU1422, limiting the analysis to the orthologs of the sequences used in the computation of the SNP density. Interspecies divergence was measured with a Jukes–Cantor distance in MEGA 5, excluding indels (Kumar et al. 2008).
Results
C. briggsae diversity
C. briggsae is distributed all over the world, and previous analyses of nucleotide diversity showed that most strains cluster in one of two main phylogeographic groups, with a few strains from several locations having divergent haplotypes with little genetic affinity to either clade (Cutter et al. 2006, 2010; Dolgin et al. 2008; Howe and Denver 2008; Raboin et al. 2010). We first sought to test whether 17 newly collected strains from 12 locations conform to this established pattern of phylogeography, by comparing them to a worldwide dataset of 117 C. briggsae isolates (Table 1 and Figure S1).
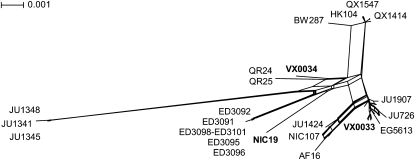
Among 17 new strains, we identified 14 distinct haplotypes, of which 9 were not described previously (Figure S1). Most of the newly collected strains fall into either the temperate or the tropical clades, with 2 notable exceptions (Figure 1). Nearly all strains from the tropical latitudes form a phylogeographic group that also includes a newly identified strain (VX0033) from a more northerly location in mainland China (30.61°N), suggesting that the geographic range of this clade extends farther than previously thought, beyond the formal definition of tropics (23°N or S latitude). Remarkably, none of the new isolates from tropical locations fall in the temperate clade, supporting the idea that most C. briggsae strains are separated by latitude. We also identified 2 genetically distinct strains from Taiwan (NIC19) and Guangshui, China (VX0034) that fall outside of the latitudinal dichotomy and that show little evidence of recombination with the remaining isolates (Figure 1), consistent with additional sampling continuing to discover rare, novel, geographically restricted genetic backgrounds in C. briggsae (Cutter et al. 2010). These results are consistent with most C. briggsae strains being genetically differentiated on the basis of their latitudinal geographical origin, but that pockets of divergent genotypes occur both in high and low latitude parts of the world that evolve essentially independently from the bulk of C. briggsae genotypes.
Figure 1 .
Neighbor network for the worldwide distribution of C. briggsae isolates based on concatenated sequences from six nuclear loci. Only a few strains are labeled for reference. Reticulation denotes potential recombination among strains. Note the divergence of the newly collected strains VX0034 and NIC19 and the grouping of strain VX0033 with the tropical strains, suggesting a greater range distribution for this clade.
Patterns of miRNA sequence evolution in C. briggsae
With our understanding of the phylogeographic relationships among C. briggsae strains in mind, we then investigated nucleotide variation at miRNA loci for a subset of them from across the world (Table S1). We sequenced 25–48 strains for each of 18 miRNAs, representing 12% of the miRNA complement in the C. briggsae genome (de Wit et al. 2009). These miRNAs are located on five chromosomes (none on chromosome V; Table 2) and 12 of them occur in three clusters (on chromosomes II, III, and X).
Table 2 . Summary statistics of nucleotide variation in Caenorhabditis briggsae miRNA loci.
| Pre-miRNA sequence | Mature sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | LG | N | L | S | π (10−3) | SNP density (10−3) | K | L | S | SNP density (10−3) | K |
| Cbr-mir-1817 | I | 48 | 108 | 4 | 8.39 | 9.53 | 0.047 | 23 | 1 | 11.29 | 0 |
| Cbr-mir-40 | II | 31 | 116 | 5 | 6.79 | 12.67 | 0.063 | 22 | 1 | 13.36 | 0.047 |
| Cbr-mir-41 | II | 31 | 110 | 2 | 1.64 | 5.30 | 0.047 | 21 | 0 | 0 | 0 |
| Cbr-mir-41b | II | 31 | 100 | 1 | 0.65 | 2.94 | 0.010 | 20 | 0 | 0 | 0 |
| Cbr-mir-2222-1 | II | 43 | 115 | 0 | 0 | 0 | 0.063 | 23 | 0 | 0 | 0 |
| Cbr-mir-2225 | II | 30 | 100 | 0 | 0 | 0 | 0 | 21 | 0 | 0 | 0 |
| Cbr-mir-2227 | II | 35 | 109 | 1 | 1.91 | 2.60 | 0.047 | 21 | 0 | 0 | 0 |
| sblock26 | III | 38 | 95 | 1 | 0.55 | 2.91 | 0.035 | 25 | 1 | 7.48 | 0.085 |
| Cbr-mir-64 | III | 38 | 109 | 4 | 4.76 | 10.07 | 0.057 | 22 | 0 | 0 | 0 |
| Cbr-mir-64b | III | 38 | 117 | 7 | 9.36 | 16.57 | 0.094 | 20 | 1 | 13.85 | 0.107 |
| Cbr-mir-2226 | III | 32 | 99 | 0 | 0 | 0 | 0.031 | 24 | 0 | 0 | 0 |
| Cbr-mir-2223 | IVa | 25 | 114 | 3 | 1.4 | 7.67 | 0.027 | 21 | 0 | 0 | 0 |
| Cbr-mir-54a | X | 29 | 118 | 2 | 3.26 | 5.09 | 0.109 | 22 | 0 | 0 | 0 |
| Cbr-mir-54b | X | 29 | 112 | 5 | 5.37 | 13.40 | 0.028 | 23 | 0 | 0 | 0 |
| Cbr-mir-54c | X | 30 | 96 | 2 | 0.44 | 6.19 | 0.043 | 20 | 0 | 0 | 0 |
| Cbr-mir-54d | X | 48 | 114 | 2 | 1.42 | 4.48 | 0.064 | 22 | 0 | 0 | 0 |
| Cbr-mir-55 | X | 29 | 112 | 1 | 2.2 | 2.68 | 0.085 | 23 | 0 | 0 | 0.045 |
| Cbr-mir-2224 | X | 47 | 120 | 5 | 4.1 | 10.88 | 0.258 | 21 | 1 | 12.44 | 0.286 |
| Average | 35 | 109 | 2.5 | 2.9 | 6.28 | 0.062 | 22 | 0.28 | 3.25 | 0.032 | |
LG, linkage group; N, number of strains sequenced; L, length (bp) of the AF16 allele (de Wit et al. 2009); S, number of mutations, including indels; π, nucleotide diversity; K, nucleotide divergence between C. briggsae AF16 and C. sp. 9 JU1422.
The exact location on chromosome IV is unknown.
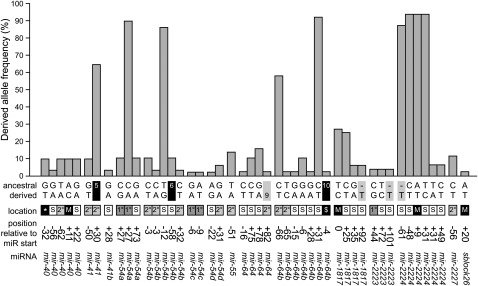
Overall, we discovered a total of 45 polymorphisms in miRNA genes, including 38 single base-pair mutations and seven insertion/deletions of ≥1 bp, the majority of which are located in the stem region of the hairpin structure (Figure 2). A fifth of the polymorphisms (9/45) are derived alleles that segregate at high frequency in C. briggsae, with one derived high-frequency allele located in the mature sequence of Cbr-mir-2224 (Figure 3).
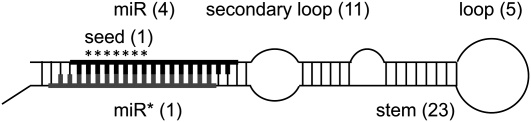
Figure 2 .
Diagram of a stereotypical miRNA hairpin with a stem-loop structure. The hairpin is processed to produce a heteroduplex comprising the mature miRNA (miR) and its complementary sequence (miR*). After processing, the mature sequence integrates the RNA-induced silencing complex (RISC) and binds to its target mRNA through exact base pairing in the 7-bp-long seed sequence (denoted by asterisks). The number of polymorphisms found in the different regions of the 18 miRNAs analyzed in this study is shown in parentheses.
Figure 3 .
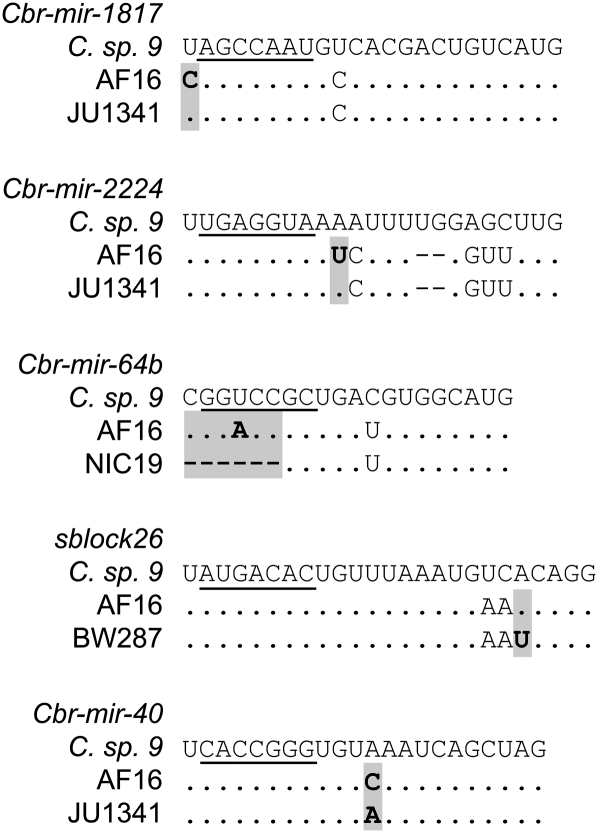
Polymorphisms identified in C. briggsae miRNAs. Most derived alleles, inferred with C. sp. 9 as outgroup, are found either at high or low frequency. For each polymorphism, the ancestral and derived states are shown below the allelic frequencies. Black boxes represent deletions, with numbers indicating the size of the deletion. Gray boxes indicate insertions with numbers showing the length of the insertion. The structural context and the position of each polymorphism relative to the start of the mature sequence are shown in the intermediate panel. M, mature sequence; $, seed sequence; S, stem; *, complementary mature sequence; 1°, primary loop; 2°, secondary loop (see Figure 2).
The distribution of substitutions between orthologs of C. briggsae and C. sp. 9 shows the following pattern of conservation across the miRNA regions (number of substitutions shown in parentheses): miR* (5) < miR (14) < loop (29) < secondary loop (39) < stem (60). This pattern of constraint among regions of miRNAs is fully consistent with the polymorphism data as well (Figure 2). Surprisingly, the complementary mature sequences miR* have less substitutions than the functional mature sequences. Nevertheless, the complementary sequence miR* is degraded soon after the mature miR sequence loads in the RISC and is often present in the cell at levels too low for detection (Bartel 2004). For instance, miR* was cloned for only a third of the miRNAs analyzed here (de Wit et al. 2009). Comparison of the mean divergence between the miR and miR* sequences suggests that long-term selection pressure is higher on the mature miR sequence (K = 0.032 ± 0.017) than on the miR* sequence (K = 0.042 ± 0.025), similar to the SNP pattern observed across miRNA loci in Arabidopsis and humans (Saunders et al. 2007; Ehrenreich and Purugganan 2008). Clearly, the high conservation of the seed sequence reflects strong purifying selection associated with its requirement for target recognition.
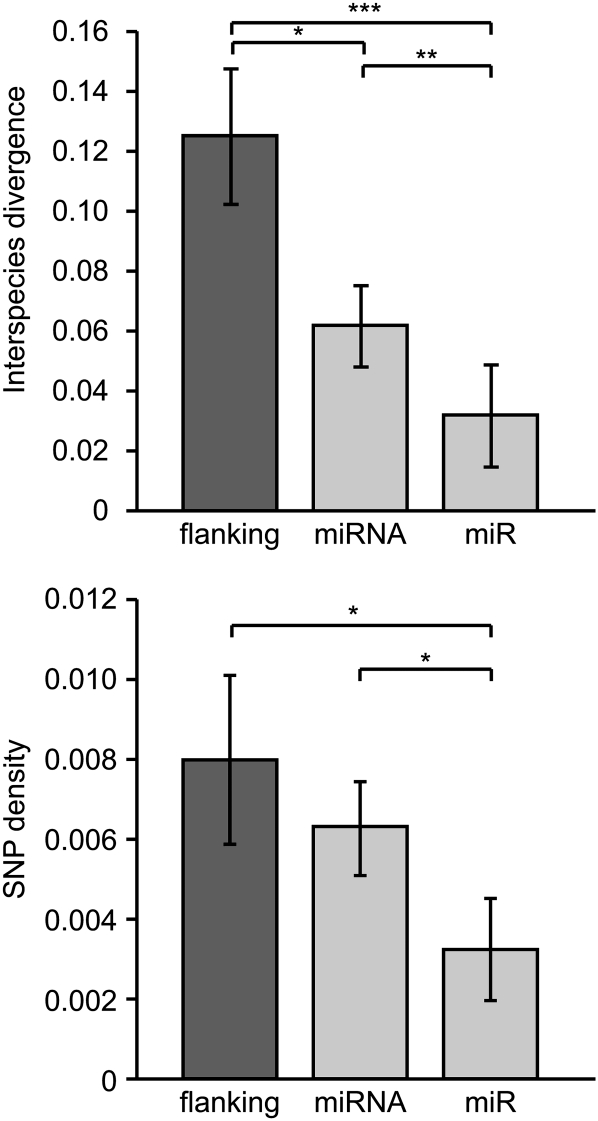
The level of nucleotide diversity at pre-miRNA loci ranges from 0 to 9.36 × 10−3 per site (Table 2). Three miRNAs, Cbr-mir-2222-1, Cbr-mir-2225, and Cbr-mir-2226, contained no variants even with ≥30 strains sampled, suggesting that purifying selection may be strong across the entire miRNA sequence. Lending further support to this idea, nucleotide diversity at miRNA loci (n = 18, mean π = 2.902 × 10−3) is significantly lower than the background nucleotide diversity in C. briggsae (n = 29, mean π = 5.531 × 10−3) (Wilcoxon two-sample P = 0.019). However, nucleotide diversity was sampled in a different set of strains for the miRNAs and non-miRNAs (this work and Cutter and Choi 2010). To further investigate selective constraints acting on C. briggsae miRNA loci, we compared interspecific and intraspecific nucleotide variation in miRNAs and in mature miRs with their flanking sequence. Overall, there is two and four times, respectively, more interspecies nucleotide divergence in flanking sequence than in the pre-miRNA and mature sequences, consistent with purifying selection generally eliminating variants in miRNA loci, with strongest selective constraints on the mature sequence involved in post-transcriptional regulation (Figure 4).
Figure 4 .
MicroRNA loci are subject to purifying selection, as evident from comparisons of mean polymorphism and divergence between the mature miR sequence, the full pre-miRNA, and their flanking sequence. Error bars indicate ± 1 SEM. Wilcoxon two-sample tests (n = 18 loci): *P = 0.05, **P = 0.01, ***P = 0.001.
Despite strong purifying selection operating on mature miRNAs, we discovered that five mature miR sequences nevertheless harbor polymorphisms. Three of the variants are transversion SNPs from an adenine and one is a thymine-to-cytosine transition (Figure 3). Interestingly, one of the polymorphisms is a 10-bp-long deletion in Cbr-mir-64b that overlaps five nucleotides of the seed sequence in the divergent strain NIC19 from Taiwan (Figure 5). Exact matching of the seed sequence with the target messenger RNA is required for proper miRNA regulation (Bartel 2004), although some classes of miRNA binding sites also require pairing in the 3′ end or in the center of the mature sequence (Brennecke et al. 2005; Shin et al. 2010). Consequently, the deletion of ∼70% of the seed sequence strongly suggests that the NIC19 allele of Cbr-mir-64b is not functional. Additionally, Cbr-mir-64b harbors the most segregating polymorphisms within C. briggsae of any of the miRNA loci we examined, implying that it might be subject to less stringent purifying selection than other C. briggsae miRNAs. Among the 18 miRNAs analyzed here, Cbr-mir-64b also is the only miRNA with a substitution in the seed sequence between C. briggsae and C. sp. 9 orthologs (Figure 5), suggesting divergent mRNA target sets between the two species (Table 2).
Figure 5 .
Polymorphisms in C. briggsae miRNA mature sequences. The C. sp. 9 orthologous outgroup miR is shown in the top row of each miRNA, with the reference C. briggsae strain AF16 in the middle row. The seed sequence is underlined and dots represent identical nucleotides relative to the top sequence. Shaded boxes highlight polymorphic sites between C. briggsae alleles. Dashes indicate indels. Note the deletion of the seed sequence in Cbr-mir-64b in the divergent strain NIC19 from Taiwan.
The strains that have been isolated from a few locations also contribute to the higher level of polymorphism observed at Cbr-mir-64b. Among the seven polymorphisms found in Cbr-mir-64b, four are specific to the divergent strains from Kerala, Taipei, and Nairobi. This is a general pattern across miRNA loci: 47 and 60% of the polymorphisms found in miRNAs and miRs, respectively, are specific to the divergent strains from Kerala, Nairobi, Montreal, Taipei, and Guangshui, indicating relatively high divergence of miRNA sequences for these strains that have distinctive genetic backgrounds.
Effects of temperature and polymorphisms on miRNA folding structures
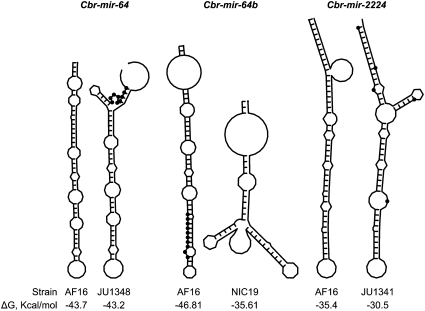
The patterns of nucleotide polymorphism and divergence we documented for miRNAs clearly implicate purifying selection to maintain the integrity of the hairpin structure (Figure 4). We investigated further whether the polymorphisms found within C. briggsae might nevertheless identify obvious candidates for functional differentiation as a consequence of how the polymorphisms affect RNA folding. Most polymorphisms have little effect on the hairpin structure (Table 3). However, several alleles from the geographically restricted C. briggsae strains carry mutations that alter the secondary structure and reduce its themodynamic stability (Figure 6). The 10-bp deletion in NIC19’s allele of Cbr-mir-64b not only removes most of the seed sequence but also results in a clover leaf-like secondary structure that is very different from the typical miRNA hairpin structure. Similarly, a 9-bp insertion in JU1348's allele of Cbr-mir-64 modifies the shape of the hairpin, resulting in a complex secondary structure with a bifurcating stem. Several point mutations in the JU1341 allele of Cbr-mir-2224 also lead to the formation of an additional stem (Figure 6). These results indicate that the strains showing divergent haplotypes based on neutral markers (Figure 1) also harbor genetic differences in miRNAs that are indicative of functional differentiation from the other C. briggsae isolates.
Table 3 . Effect of temperature and polymorphisms on RNA folding.
| SNPs affecting secondary structure | ||||
|---|---|---|---|---|
| miRNA domain | 14° | 20° | 30° | TS |
| Stem | 14 | 14 | 13 | 3 |
| Secondary loop | 4 | 4 | 6 | 1 |
| Loop | 2 | 2 | 1 | 0 |
| miR | 4 | 4 | 4 | 0 |
| miR* | 1 | 1 | 1 | 0 |
| Total | 25 | 25 | 25 | 4 |
| Frequency (%) | 55.6 | 55.6 | 55.6 | 8.9 |
The MFE structure of each allele is compared to the AF16 MFE structure for three temperatures. Temperature-sensitive (TS) variants are polymorphisms that have temperature-dependent effects on the MFE structure.
Figure 6 .
Effect of sequence polymorphism on miRNA folding in the divergent strains from Kerala and Taiwan. The typical hairpin structure of the miRNAs Cbr-mir-64, Cbr-mir-64b, and Cbr-mir-2224 in C. briggsae reference strain AF16 is shown for comparison. The minimum free energy (ΔG in Kcal/mol) is given for each allele. Solid dots indicate the position of the polymorphic sites. A 9-bp insertion in JU1348 and a 10-bp deletion comprising most of the seed sequence in NIC19 result in dramatic changes of the miRNA hairpin structure. A divergent secondary structure also results from multiple mutations in strain JU1341.
Although we detected allelic variants both in the temperate and the tropical strains, no polymorphism was found exclusively in all strains within one of these two phylogeographic clades. Most of the alleles segregate either at high or at low frequency (Figure 3), and the two intermediate-frequency variants do not differentiate the tropical and temperate groups of strains. Thus, the hypothesis that selection could have favored distinct alleles in the temperate and tropical clades does not hold for this collection of 18 miRNAs.
We next investigated how temperature might affect RNA folding by comparing the MFE structure of each allele to the AF16 reference strain allele’s MFE structure at three different temperatures. First, we noted that temperatures from 14° to 30° do not change the predicted hairpin structure, or alter it only slightly, for the majority of the miRNAs (Table 3). However, there was a strong effect on the predicted AF16 fold for three miRNAs (not shown): temperatures below 30° result in complex secondary structures for the AF16 allele of Cbr-mir-64b, Cbr-mir-54d, and Cbr-mir-41b, whereas the stereotypical hairpin structure is restored at 30°. Second, for each temperature that we explored, half of the variants (45%) do not influence the miRNA secondary structure (Table 3). Nearly all of the variants that do alter the miRNA secondary structure have little impact on overall hairpin formation, and either create bulges or extend the length of the secondary and primary loops. Two polymorphisms do strongly change the hairpin structure at 14°, however: a G→A transition at position +78 in Cbr-mir-64 for strain EG5612 and a 6-bp deletion at position −54 in Cbr-mir-54d for strain JU1341, but these effects disappear when temperature is increased to 20° and 30°, respectively. Two other mutations in strain JU1341 have temperature-dependent effects (Table 3). There is a T→C transition that increases the length of a secondary loop in Cbr-mir-64b at 30° and the effect of a C→T transition disappears in Cbr-mir-2224 at 30°, although other mutations in this miRNA do alter the hairpin structure (Figure 6). These results suggest that C. briggsae miRNAs generally are tolerant to variation in temperature even in the face of sequence polymorphism, which may explain the observed lack of correlation between miRNA genotype and geographical origin.
Discussion
Mutations generated in the laboratory during forward genetic screens or using reverse genetics are powerful means to investigate gene function and have proven to be particularly effective in understanding developmental mechanisms and their evolution (Brenner 1974; Nüsslein-Volhard and Wieschaus 1980; Nüsslein-Volhard et al. 1984; Jansen et al. 1997). However it is unclear how such mutations, often with strong deleterious effects, may contribute to the long-term evolution of the trait they affect. Investigation of allelic variation in natural populations is a valuable complement to understanding the evolution of particular gene function (Kammenga et al. 2008; Jovelin 2009). We applied this philosophy to the nematode C. briggsae by examining nucleotide polymorphism and divergence at miRNA loci, a class of regulatory molecules with important roles in development. The patterns of polymorphism and divergence clearly demonstrate that purifying selection is the dominant mode of natural selection acting on miRNA genes (Figure 2 and Figure 4), similar to the pattern observed in humans and Arabidopsis (Chen and Rajewsky 2006; Saunders et al. 2007; Ehrenreich and Purugganan 2008; Quach et al. 2009). In particular, the mature sequence of miRNAs is subject to strong purifying selection, consistent with its important role in gene regulation. Despite strong constraint overall, we identified notable examples of functionally relevant polymorphisms in C. briggsae miRNAs.
SNPs altering miRNA structure
Although most polymorphisms have little effect on miRNA folding (Table 3), we identified some mutations that likely cause complex and atypical secondary structures (Figure 6), suggesting that these polymorphisms may have functional effects. At least one such allele, in Cbr-mir-64b, is clearly not functional due to a 10-bp deletion that removes most of the seed sequence that is required for interaction with target mRNAs (Figure 5 and Figure 6). Interestingly, the common allele of this miRNA also harbors a substitution in the seed sequence between C. briggsae and C. sp. 9 (Figure 5), suggesting that Cbr-mir-64b orthologs in these two species regulate distinct sets of target genes. Because Cbr-mir-64b is the only miRNA with seed sequence GGACCGC in the C. briggsae genome (de Wit et al. 2009) its function cannot be rescued by same-family miRNAs (Alvarez-Saavedra and Horvitz 2010).
One possible explanation for the pattern of nucleotide variation at Cbr-mir-64b is its recent origin in the lineage leading to C. briggsae and C. sp. 9. Orthologs of Cbr-mir-64b were not found in other nematode species (de Wit et al. 2009) and there is evidence that young miRNAs evolve faster than more phylogenetically conserved miRNAs (Lu et al. 2008a; Fahlgren et al. 2010; Nozawa et al. 2010). It is conceivable that young miRNAs have limited functional scope and contribute little to organismal fitness and, consequently, are more subject to genetic drift until they either become inactive or are coopted into regulatory networks. Lending support for this view, most newly arisen miRNAs become inactive and disappear during evolution (Lu et al. 2008b; Nozawa et al. 2010). Alternatively, gene loss may be an important mechanism of phenotypic diversification (Aboobaker and Blaxter 2003). In contrast to Cbr-mir-64b, the miRNA Cbr-mir-64 is not a young miRNA and orthologs are present in other nematode species (de Wit et al. 2009). However, its seed sequence is conserved in two other miRNAs found together in the same cluster on chromosome III (de Wit et al. 2009), suggesting that if the 9-bp insertion found in strain JU1348 is deleterious (Figure 6), then the function of Cbr-mir-64 could potentially be rescued by other members of the mir-64 family (Alvarez-Saavedra and Horvitz 2010).
Phylogeography of miRNA polymorphism in C. briggsae
The background level of nucleotide diversity at silent sites in C. briggsae is similar to that observed in A. thaliana (Nordborg et al. 2005; Cutter et al. 2010). And yet, we found that 28% (5/18) of the C. briggsae miRNAs surveyed contained a polymorphism in the mature sequence compared to only 3% (2/66) of miRNAs in a global sample of A. thaliana accessions (Ehrenreich and Purugganan 2008). Notably, plant miRNAs typically require perfect or near-perfect complementarity with their target sites, whereas animal miRNAs preferentially bind to their targets through complementarity of the seed sequence (Axtell et al. 2011). The difference between A. thaliana and C. briggsae in polymorphism levels within mature miRNA sequences could be due in part to differences in pairing requirements with their target sites, with stronger purifying selection on the overall mature sequence in plant miRNAs.
Strikingly, pre-miRNAs in C. briggsae harbor twice as much nucleotide diversity as pre-miRNAs in A. thaliana (Ehrenreich and Purugganan 2008), despite the fact that background levels of polymorphism at neutral sites are very similar between the two species. This surprisingly higher incidence of polymorphisms among the miRNAs of C. briggsae suggests that miRNA-mediated gene expression variation could be particularly important in C. briggsae, perhaps facilitated by the high differentiation of the geographically restricted isolates. Remarkably, all mutations having strong effects on miRNA folding (Figure 6), as well as half of the total polymorphisms, are found in the few divergent C. briggsae strains that have been isolated from only a few locations. Although mutations in the seed sequence are a clear indication of functional divergence, nucleotide variation at other positions in the mature sequence and in the miRNA precursor also can have consequences on miRNA function (Duan et al. 2007; de Meaux et al. 2008; Jazdzewski et al. 2008, 2009), and some miRNA binding sites require base pairing beyond the seed sequence (Brennecke et al. 2005; Shin et al. 2010). Thus, the polymorphisms in the geographically restricted strains suggest that distinct miRNA-mediated gene regulation might be operating and raises the possibility that these strains differ in some aspects of their biology relative to the vast majority of C. briggsae strains. Close examination of phenotypic differences in the newly identified divergent genotypes might provide insights into the microevolution of developmental processes and/or life-history traits within this species (Figure 1 and Cutter et al. 2010).
Most strains of C. briggsae from around the world show a distinct haplotype structure based on geographical origin that correlates with temperature-dependent effects on fitness (Figure 1, Figure S1, and Cutter et al. 2006, 2010; Dolgin et al. 2008; Howe and Denver 2008; Prasad et al. 2010; Raboin et al. 2010). Because RNA folding also is temperature dependent, we hypothesized that miRNAs with fixed nucleotide differences between the temperate and tropical phylogeographic groups might confer distinct miRNA secondary structures (de Meaux et al. 2008). Surprisingly, none of the polymorphisms for the 18 miRNA loci that we identified were specific to either of these two phylogeographic groups, as most alleles segregate at high or low frequency (Figure 3). Thus, the pattern of nucleotide variation in miRNA loci contrasts sharply with the pattern of differentiation at putatively neutral loci that have been investigated to date. One reason for this disparity may be that the strong selective pressure on miRNA sequence keeps most new mutations at low frequency, preventing fixation in any one clade (Figure 4). Nevertheless, we found that most variants did not change the secondary structure when we computationally manipulated temperature (Table 3), suggesting that there may be little or no selective pressure to maintain alternative variants in each phylogeographic group. This contrasts with the situation of temperature-sensitive balancing selection for MIR824 in A. thaliana (de Meaux et al. 2008). Although we did not detect a correlation between miRNA genotype and strain latitudinal origin, future investigation of the remaining miRNAs and other small RNAs in the C. briggsae genome merits additional exploration of the potential for local adaptation mediated by small RNA divergence.
In conclusion, we document strong overall purifying selection on miRNAs within contemporary populations of C. briggsae. Nevertheless, our analysis of polymorphism in 12% of C. briggsae’s miRNAs uncovered substantial nucleotide variation in mature sequences. Our results motivate further investigation of miRNA sequence variation in C. briggsae and identify strong candidates for experimental work to dissect the functional consequences of allelic variation on gene expression regulation among divergent strains of C. briggsae.
Acknowledgments
We thank Erik Andersen, Christian Braendle, Marie-Anne Felix, Leonid Kruglyak, Matt Rockman, and Guo-Xiu Wang for kindly providing C. briggsae strains. Some of the C. sp. 9 sequence data were produced by The Genome Center at Washington University School of Medicine in St. Louis. R.J. is supported by a postdoctoral fellowship from the Ontario Ministry of Research and Innovation and A.D.C. is supported by the Natural Sciences and Engineering Research Council of Canada and a Canada Research Chair in Evolutionary Genomics.
Literature Cited
- Aboobaker A. A., Blaxter M. L., 2003. Hox gene loss during dynamic evolution of the nematode cluster. Curr. Biol. 13: 37–40 [DOI] [PubMed] [Google Scholar]
- Alonso C. R., Wilkins A. S., 2005. The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 6: 709–715 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Anderson J. L., Albergotti L., Proulx S., Peden C., Huey R. B., et al. , 2007. Thermal preference of Caenorhabditis elegans: a null model and empirical tests. J. Exp. Biol. 210: 3107–3116 [DOI] [PubMed] [Google Scholar]
- Axtell M. J., Westholm J. O., Lai E. C., 2011. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel D. P., Chen C. Z., 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396–400 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M., 2005. Principles of microRNA-target recognition. PLoS Biol. 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N., 2006. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 38: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Choi J. Y., 2010. Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae. Genome Res. 20: 1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Felix M. A., Barriere A., Charlesworth D., 2006. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Yan W., Tsvetkov N., Sunil S., Felix M. A., 2010. Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenorhabditis briggsae. Mol. Ecol. 19: 798–809 [DOI] [PubMed] [Google Scholar]
- de Meaux J., Hu J. Y., Tartler U., Goebel U., 2008. Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8994–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Linsen S. E., Cuppen E., Berezikov E., 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. S., Felix M. A., Cutter A. D., 2008. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity 100: 304–315 [DOI] [PubMed] [Google Scholar]
- Duan R., Pak C., Jin P., 2007. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 16: 1124–1131 [DOI] [PubMed] [Google Scholar]
- Ehrenreich I. M., Purugganan M. D., 2008. Sequence variation of MicroRNAs and their binding sites in Arabidopsis. Plant Physiol. 146: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K. D., Sullivan C. M., Chapman E. J., et al. , 2010. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix M. A., Braendle C., 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20: R965–R969 [DOI] [PubMed] [Google Scholar]
- Graustein A., Gaspar J. M., Walters J. R., Palopoli M. F., 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Lorenz R., Bernhart S. H., Neubock R., Hofacker I. L., 2008. The Vienna RNA websuite. Nucleic Acids Res. 36: W70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W., Artieri C., Khezri N., Singh R. S., Gupta B. P., 2008. Comparative analysis of function and interaction of transcription factors in nematodes: extensive conservation of orthology coupled to rapid sequence evolution. BMC Genomics 9: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- Herranz H., Cohen S. M., 2010. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24: 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D. K., Denver D. R., 2008. Muller's Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 8: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D., 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Scornavacca C., 2011. A survey of combinatorial methods for phylogenetic networks. Genome Biol. Evol. 3: 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Hazendonk E., Thijssen K. L., Plasterk R. H., 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17: 119–121 [DOI] [PubMed] [Google Scholar]
- Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R., et al. , 2008. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 105: 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K., Liyanarachchi S., Swierniak M., Pachucki J., Ringel M. D., et al. , 2009. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl. Acad. Sci. USA 106: 1502–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., 2009. Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis. Mol. Biol. Evol. 26: 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., Phillips P. C., 2011. Expression level drives the pattern of selective constraints along the insulin/Tor signal transduction pathway in Caenorhabditis. Genome Biol. Evol. 3: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., Dunham J. P., Sung F. S., Phillips P. C., 2009. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in Caenorhabditis. Genetics 181: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga J. E., Phillips P. C., De Bono M., Doroszuk A., 2008. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends Genet. 24: 178–185 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K., 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V., 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lu J., Fu Y., Kumar S., Shen Y., Zeng K., et al. , 2008a. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol. Biol. Evol. 25: 929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Shen Y., Wu Q., Kumar S., He B., et al. , 2008b. The birth and death of microRNA genes in Drosophila. Nat. Genet. 40: 351–355 [DOI] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York [Google Scholar]
- Nokes E. B., Van Der Linden A. M., Winslow C., Mukhopadhyay S., Ma K., et al. , 2009. Cis-regulatory mechanisms of gene expression in an olfactory neuron type in Caenorhabditis elegans. Dev. Dyn. 238: 3080–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Miura S., Nei M., 2010. Origins and evolution of microRNA genes in Drosophila species. Genome Biol. Evol. 2: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., Kluding H., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 193: 267–282 [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., 2006. Micro RNAs in animal development. Cell 124: 877–881 [DOI] [PubMed] [Google Scholar]
- Prasad A., Croydon-Sugarman M. J., Murray R. L., Cutter A. D., 2010. Temperature-dependent fecundity associates with latitude in Caenorhabditis briggsae. Evolution 65: 52–63 [DOI] [PubMed] [Google Scholar]
- Quach H., Barreiro L. B., Laval G., Zidane N., Patin E., et al. , 2009. Signatures of purifying and local positive selection in human miRNAs. Am. J. Hum. Genet. 84: 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboin M. J., Timko A. F., Howe D. K., Felix M. A., Denver D. R., 2010. Evolution of Caenorhabditis mitochondrial genome pseudogenes and Caenorhabditis briggsae natural isolates. Mol. Biol. Evol. 27: 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M. A., Liang H., Li W. H., 2007. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 104: 3300–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C., Nam J. W., Farh K. K., Chiang H. R., Shkumatava A., et al. , 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L., Bao Z., Blasiar D., Blumenthal T., Brent M., et al. , 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: 166–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chamberlin H. M., 2002. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev. 16: 2345–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Felix M. A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Wu C. I., Shen Y., Tang T., 2009. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 19: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]