Abstract
The yeast transcriptional activator Gal4 localizes to UASGAL sites even in the absence of galactose but cannot activate transcription due to an association with the Gal80 protein. By 4 min after galactose addition, Gal4-activated gene transcription ensues. It is well established that this rapid induction arises through a galactose-triggered association between the Gal80 and Gal3 proteins that decreases the association of Gal80 and Gal4. How this happens mechanistically remains unclear. Strikingly different hypotheses prevail concerning the possible roles of nucleocytoplasmic distribution and trafficking of Gal3 and Gal80 and where in the cell the initial Gal3–Gal80 association occurs. Here we tested two conflicting hypotheses by evaluating the subcellular distribution and dynamics of Gal3 and Gal80 with reference to induction kinetics. We determined that the rates of nucleocytoplasmic trafficking for both Gal80 and Gal3 are slow relative to the rate of induction. We find that depletion of the nuclear pool of Gal3 slows the induction kinetics. Thus, nuclear Gal3 is critical for rapid induction. Fluorescence-recovery-after-photobleaching experiments provided data suggesting that the Gal80–Gal4 complex exhibits kinetic stability in the absence of galactose. Finally, we detect Gal3 at the UASGAL only if Gal80 is covalently linked to the DNA-binding domain. Taken altogether, these new findings lead us to propose that a transient interaction of Gal3 with Gal4-associated Gal80 could explain the rapid response of this system. This notion could also explain earlier observations.
GENETIC regulation of transcriptional initiation occurs through environmentally responsive molecular switching systems that facilitate cellular differentiation (Reece 2000; Forsberg and Ljungdahl 2001). One such system in eukaryotes is the Saccharomyces cerevisiae GAL gene switch that regulates galactose-responsive transcription through the interplay of the Gal4, Gal80, and Gal3 proteins (Carlson 1987; Johnston 1987; Lohr et al. 1995; Traven et al. 2006). Gal4 is a DNA-binding transcription activator that has a single DNA sequence-specific binding domain (Pan and Coleman 1989; Reece and Ptashne 1993; Schjerling and Holmberg 1996) and a strong transcription activation domain (ADII; aa 768–881) (Giniger and Ptashne 1987; Johnston et al. 1987; Melcher and Johnston 1995; Ansari et al. 1998). Gal4 is found associated with the UASGAL DNA site(s) of the GAL genes in both absence and presence of galactose, but in the absence of galactose it is unable to activate transcription due to masking of its ADII by Gal80 (Torchia et al. 1984; Lue et al. 1987; Ma and Ptashne 1987; Chasman and Kornberg 1990; Leuther and Johnston 1992; Mizutani and Tanaka 2003). Galactose facilitates association of the Gal3 and the Gal80 proteins, an event that brings about dissociation of Gal80 from Gal4 (Peng and Hopper 2002; Jiang et al. 2009) and subsequent Gal4-mediated recruitment of RNA polymerase to UASGAL-associated promoters (Torchia and Danzinger 1980; Johnston et al. 1987; Lue et al. 1987; Chasman and Kornberg 1990; Yun et al. 1991; Leuther and Johnston 1992; Parthun and Jaehning 1992; Melcher and Johnston 1995; Suzuki-Fujimoto et al. 1996; Wu et al. 1996; Blank et al. 1997; Yano and Fukasawa 1997; Koh et al. 1998; Platt and Reece 1998; Vollenbroich et al. 1999; Bhaumik and Green 2001; Jeong et al. 2001; Larschan and Winston 2001; Melcher and Xu 2001; Carrozza et al. 2002; Timson et al. 2002; Bryant and Ptashne 2003; Menezes et al. 2003).
A hallmark of the GAL gene switch is its rapid induction response. A de novo increase in GAL1 and GAL10 mRNAs is observed within 4–6 min of galactose addition to cells cultured at 30° (Yarger et al. 1984). This rapid induction of transcription stems in part from the fact that a round of protein synthesis subsequent to galactose addition is not required for target gene activation (Perlman and Hopper 1979). The rapid response correlates well with the documented rapid recruitment of RNA Pol II to the GAL1 promoter that begins ∼2–4 min after galactose addition (Bryant and Ptashne 2003). Because Gal4-mediated recruitment of SAGA and Mediator complexes precedes recruitment of Pol II (Bryant and Ptashne 2003), it is apparent that galactose-activated Gal3 begins to relieve Gal80 inhibition of Gal4AD by 1–3 min following galactose addition. Accordingly, the mechanisms by which the galactose-activated Gal3 brings about Gal80 dissociation from Gal4 must be fast acting.
The inherent mechanisms and molecular species involved in the process by which Gal3–Gal80 interaction overcomes Gal80 inhibition of Gal4 remain unclear and subject to conflicting hypotheses. In this work, we addressed two unresolved questions: Where in the cell does Gal3 initially bind to Gal80 to trigger induction—the cytoplasm, the nucleus, or both? And, does nucleocytoplasmic trafficking of Gal3 and/or Gal80 play a role in the switch? There are two published hypotheses concerning the first question. One suggests that Gal3 interacts with Gal80 exclusively in the cytoplasm and sequesters it away from nuclear Gal4 (Peng and Hopper 2002). A strikingly different hypothesis specifies that cytoplasmic Gal3 binds galactose and then moves into the nucleus to bind to Gal80 (Wightman et al. 2008). These two hypotheses specify not only different subcellular compartments in which the initial interaction of Gal3 and Gal80 occurs, but also different nucleocytoplasmic trafficking dynamics for these proteins.
Deciphering the mechanisms that overcome Gal80’s inhibition of Gal4 in live cells requires imaging the Gal3, Gal80, and Gal4 molecules, their associations, and their subcellular distributions and dynamics prior to and during the first few minutes of galactose addition. However, imaging these molecules expressed from their native promoters under such conditions has been hampered, as their levels in uninduced cells are very low (Wightman et al. 2008). Here we have employed spinning disc confocal microscopy (SDCM) to alleviate this problem and facilitate critical tests of competing hypotheses specifying GAL gene switch mechanisms. SDCM requires less excitation light to achieve an equivalent quality of signal compared to conventional confocal microscopy and thus provides a high quality of spatial and temporal resolution with relatively low photobleaching. These features make SDCM optimally suited for fluorescence recovery after photobleaching (FRAP) analyses and for tracking of small foci in live cells (Graf et al. 2005).
Using SDCM-based FRAP, we tested nucleocytoplasmic exchange rates of Gal80 and Gal3 molecules and dynamics of protein–protein interactions at the uninduced GAL1 promoter. Our FRAP experiments showed that neither Gal80 nor Gal3 rapidly shuttle between nucleus and cytoplasm. These results challenge central mechanistic elements of two current hypotheses concerning the GAL gene switch, one of which stems from this lab. Moreover, we discovered that a marked reduction of Gal3 levels in the nucleus diminishes the rate of induction from GAL1 promoters. These results strongly suggest that the nuclear concentration of Gal3 is a critical feature of the GAL gene switch regardless of whether Gal3 interacts with Gal4-associated Gal80 or free Gal80. We also show that, in the absence of the galactose-induced Gal3–Gal80 interaction, the Gal80–Gal4 complex at the promoter-associated UASGAL is kinetically stable relative to the induction rate. The results are discussed in light of the older models of the GAL gene switch and lead us to propose a new model that highlights the importance of nuclear Gal3.
Materials and Methods
Strains
Genotypes of all the strains used in this study are presented in Table 1. MATa and MATα strains used in this study were derived from S. cerevisiae ScTEB652 (MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1) and FY630 (MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63), respectively (Arndt et al. 1995; Blank et al. 1997). Details of strain construction will be provided upon request. The array strains were constructed by consecutive insertion of pFJ58×64 and pFJ57×8 (Jiang et al. 2009) into Sc652 (MATa strains) and FY650 (MATα strains). The GAL3, GAL80, and GAL4 genes were tagged in the genome with the green fluorescent protein (GFP) ORF using insertion cassettes obtained from pFJ120N, pFJ109N, and pFJ110N, respectively.
Table 1 . Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| RLY2800 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Fus3-GFP | Slaughter et al. (2007) |
| MS739 | MATα ura3-52 leu2-3,112 ade2-101 kar1-1 | Vallen et al. (1992) |
| Sc723 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 PGAL1-HIS3 | Blank et al. (1997) MS730 |
| Sc724 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 PGAL1-HIS3 gal3Δ | Blank et al. (1997) |
| Sc859 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 GAL3-2GFP::NAT | Jiang et al. (2009) |
| Sc862 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 GAL80-2GFP::NAT | This study |
| Sc920 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 LacOx64::LEU2 (PGAL1-GST)x8::Kanr GAL80-2GFP::NAT | This study |
| Sc930 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 LacOx64::LEU2 (PGAL1-GST)x8::Kanr GAL4-2GFP::CaURA3 | This study |
| Sc932 | MATaade1 ile leu2-3,112 ura3-52 trp1-HIII his3-1 LacOx64::LEU2 (PGAL1-GST)x8::Kanr GAL80-GFP::NAT gal3Δ::CaURA3 Gal80-2GFP::NAT | This study |
| Sc963 | MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 LacOx64::LEU2 (PGAL1-GST)x8::Kanr Gal80-2GFP::NAT | This study |
| Sc963 | MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 LacOx64::LEU2 (PGAL1-GST)x8::Kanr Gal80-2GFP::NAT | This study |
| Sc965 | MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 LacOx64::LEU2 (PGAL1-GST)x8::Kanr Gal4-2GFP::NAT | This study |
| Ds300 (Sc920 × Sc963) | MATa/α ADE1/ade1 ile/ILE his3-Δ1/HIS3 HIS4/his4-917δ LYS2/lys2-173R2 leu2-3,112/leu2Δ1, ura3-52/ura3-52 trp1-HIII/trp1Δ63 LacOx64::LEU2 (PGAL1-GST)x8::Kanr/LacOx64::LEU2 (PGAL1-GST)x8::Kanr Gal80-2GFP::NAT/Gal80-2GFP::NAT | This study |
| Ds301 (Sc930 × Sc965) | MATa/α ADE1/ade1 ile/ILE his3-Δ1/HIS3 HIS4/his4-917δ LYS2/lys2-173R2 leu2-3,112/leu2Δ1, ura3-52/ura3-52 trp1-HIII/trp1Δ63 LacOx64::LEU2 (PGAL1-GST)x8::Kanr/LacOx64::LEU2 (PGAL1-GST)x8::Kanr Gal4-2GFP::NAT/Gal4-2GFP::NAT | This study |
Plasmids
The plasmids used in this study are listed in Table 2. The NAT selection marker in plasmids that were used as a source of genomic insertion cassettes was isolated from Toda10 (a gift from Jian-Qiu Wu). pFJ04 was constructed by insertion of SmaI/XhoI fragments of pGP15 (Peng and Hopper 2000) and pSA80N1 (S. Alam and J. E. Hopper, unpublished results) into pRS416 (New England Biolabs). pFJ109N was constructed by simultaneous insertion of the NAT marker and a 2GFP cassette obtained from pKT209 (Sheff and Thron 2004) into pFJ04 by PCR-based and other molecular methods. pFJ110N was constructed by insertion of the NAT marker and the 2GFP cassette into the GAL4-containing plasmid, pLM34 (L. Mylin and J. E. Hopper, unpublished results). pFJ120N was constructed by insertion of the NAT cassette into pGP17 (Peng and Hopper 2000). Plasmids pOE180 [nuclear export sequence (NES)-Gal3] and pOE182 [mutant NES (mNES)-Gal3] were constructed as follows. To introduce Rna1 NES and mNES (L326, I328) into Gal3, the region of RNA1-encoding residues (AA 316–357) was isolated from pGEM-T Rna1(AA 316–357) and pGEM-T Rna1(AA 316–357) (Feng et al. 1999) by PCR with primers OE167 (5′-CTCGTCGACGTCGAAAAGGGAAATTTACCTGA-3′) and OE168 (5′-CTCGTCGACGTCCTCTTCAAAATCGTCAACCT-3′). The resulting PCR products were inserted into the AatII site of pAKS15 to yield pOE180 (NES-Ga3) and pOE182. A single GFP ORF was inserted at the 3′ end of Gal3 in pOE180 and pOE182 to make pOE183 and pOE185, respectively. The PGAL1-2GFP reporter plasmid, pOE33, was constructed by PCR-based and other molecular methods in pRS416 (New England Biolabs). Construction of pFJ35 was described previously (Jiang et al. 2009). All PCRs for plasmid construction purposes were carried out with high-fidelity Pfu DNA polymerase (Stratagene). The oligonucleotides were purchased from Integrated DNA Technologies.
Table 2 . Plasmids used in this study.
| Plasmid | Genotype | Source |
|---|---|---|
| pAKS15 | CEN ARS1 TRP1 PGAL3-GAL3 | Sil et al. (1999) |
| pOE33 | CEN ARS1 URA3 PGAL1-2GFP | This study |
| pOE180 | CEN ARS1 TRP1 PGAL3-NES-GAL3 | This study |
| pOE182 | CEN ARS1 TRP1 PGAL3-mNES-GAL3 | This study |
| pOE183 | CEN ARS1 TRP1 PGAL3-NES-GAL3-GFP | This study |
| pOE185 | CEN ARS1 TRP1 PGAL3-mNES-GAL3-GFP | This study |
| pGP15Δ | CEN ARS1 URA3 PGAL80-GAL80 | Peng and Hopper (2000) |
| pGP17 | CEN ARS1 TRP1 PGAL3-GAL3-GFP | Peng and Hopper (2000) |
| pGP56GFP | CEN ARS1 TRP1 PGAL3-myr-GAL3-GFP | Peng and Hopper (2002) |
| PGP57GFP | CEN ARS1 TRP1 PGAL3-mom-GAL3-GFP | Peng and Hopper (2002) |
| pFJ35 | CEN ARS1 TRP1 PADH2-Htb2-mCherry | Jiang et al. (2009) |
| pFJ109N | CEN ARS1 TRP1 PGAL80-GAL80-2GFP::NAT | This study |
| pFJ110N | CEN ARS1 TRP1 PGAL4-GAL4-2GFP::NAT | This study |
| pFJ120N | CEN ARS1 TRP1 PGAL3-GAL3-GFP::NAT | This study |
| pME9 | CEN ARS1 HIS3 PADH2-LacI-mCherry | This study |
| pPS1739 | CEN ARS1 URA3 PHOG1-Hog1-GFP | Ferrigno et al. (1998) |
Microscopy
All microscopy experiments were performed with a Nikon TE-2000U spinning disc confocal microscope that was equipped with a ×100/1.4 N.A. objective lens (Nikon, Melville, NY); 488-, 514-, and 568-nm argon ion lasers; and a charge-coupled device camera (ORCA-AG, Hamamatsu, Bridgewater, NJ). Live yeast cells were concentrated to appropriate cell density by centrifugation and immobilized on Superfrost slides (Fisher Scientific) coated with 2% gelatin mix. The experiments in which we monitored the effect of different media on a single cell were carried out using Y04C Cellasik Microfluidics plates (Cellasik) mounted onto the microscope stage.
The FRAP experiments testing the nucleocytoplasmic mobility of Gal3-GFP and Gal80-2GFP molecules were carried out with Sc859 and Sc862 cells, respectively. The cells were grown to mid-log phase in glycerol–lactic acid media. To produce sufficient levels of Gal80-2GFP signal, the Sc862 cells were induced with 2% galactose for 4 hr and then shifted back to glycerol–lactic acid media for 4 hr to establish nonexpressing conditions for GAL genes. It was previously demonstrated that Gal4–Gal80 interaction is reconstituted 2 hr after the cells were shifted from galactose to non-inducing conditions (Jiang et al. 2009). The nuclear or cytoplasmic regions of these cells were then bleached for 100 ms at 20% laser power to ablate the GFP signal in the target area. The cells were then monitored for signal recovery for indicated time periods. All fluorescence quantifications of the acquired images were carried out with the software ImageJ (Wayne Rasband, National Institutes of Health).
The photobleaching experiments that tested the stability of Gal80–Gal4 and Gal4–DNA interactions were carried out with diploid cells (Ds300 and Ds301, Table 1). A small subnuclear region overlapping one of the GFP spots and excluding the other spot was briefly bleached at 30% laser power for 100 ms.
GFP reporter assay
Sc724 cells expressing 2GFP from the native GAL1 promoter (PGAL1) on pOE33 and wild-type Gal3 or NES-Gal3 or mNES-Gal3 from pAKS15, pOE180, or pOE182, respectively, were grown in 25 ml of glycerol–lactic acid media. Galactose was added (2% final) to induce synthesis of reporter GFP molecules. At indicated time points after induction, 3 ml of cells at OD600= 0.5 was taken from the culture, and cell pellets were collected. Protein extracts were prepared using the sodium hydroxide method, as described elsewhere (Kushnirov 2000). Proteins were separated with sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis, and GFP was detected by Western blotting using anti-GFP (Invitrogen).
Results
Two previous studies failed to detect Gal3 in the nucleus of uninduced cells (Peng and Hopper 2000; Wightman et al. 2008). The apparent lack of Gal3 in the nucleus of uninduced cells in conjunction with other data led to two strikingly different hypotheses concerning how Gal3 works. One hypothesis posits that Gal3 works primarily, if not exclusively, in the cytoplasm to bind to Gal80, which at the time was thought to shuttle between cytoplasm and nucleus (Peng and Hopper 2002). Accordingly, it was proposed that cytoplasmic Gal3–Gal80 interaction would sequester Gal80 in the cytoplasm to free Gal4 from Gal80’s inhibitory effect. In another study, Gal3 could not be detected anywhere in the uninduced cells (Wightman et al. 2008). Those authors hypothesized that galactose-bound Gal3 protein moves from the cytoplasm into the nucleus (Wightman et al. 2008). These hypotheses are called into question by recent observations. First, making Gal3 predominantly nuclear by fusing it to a strong nuclear localization sequence (NLS) does not alter the kinetics of induction (Jiang et al. 2009). Second, Gal3 expressed from its native gene is detectable within both the nucleus and the cytoplasm of live yeast cells grown in the absence of galactose (uninduced cells) under similar conditions as in the previous works mentioned above (Jiang et al. 2009). Considering these new observations, we used SDCM to evaluate the subcellular dynamics of Gal3 and Gal80 and to determine whether nuclear Gal3 plays a role in induction.
Cytoplasmic:nuclear ratio of Gal3 does not change in response to galactose
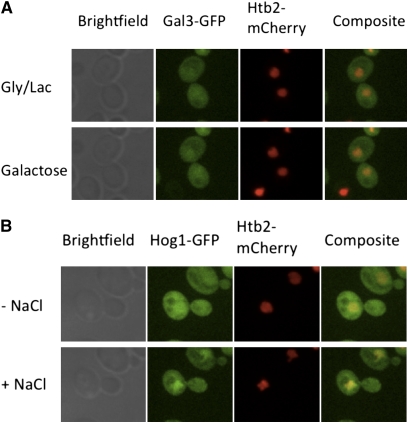
In a recent study, Gal3 was not detectable in uninduced cells but was readily detected in both cytoplasm and nucleus 45 min following galactose addition (Wightman et al. 2008). Accordingly, those authors proposed that, upon binding galactose, Gal3 translocates from the cytoplasm to the nucleus where it binds to Gal80. Here we used SDCM and a highly sensitive camera capable of detecting uninduced levels of wild-type Gal3-GFP in living cells to evaluate whether Gal3 moves from cytoplasm to nucleus in the absence or presence of galactose. In one series of experiments, we imaged Gal3-GFP nucleocytoplasmic distribution in cycloheximide-treated Sc859 cells before and after galactose induction. Htb2-mCherry expressed from pFJ35 served as a nuclear marker. In glycerol–lactic acid media, the Gal3-GFP was uniformly distributed between the two compartments, and this did not notably change throughout the 45 min following the addition of galactose (Figure 1A). In contrast, our imaging setup readily detected rapid subcellular redistribution of Hog1-GFP, which rapidly concentrated in the nucleus within 10 min after NaCl was added to the media (final: 500 mm) (Figure 1B) as was previously demonstrated (Ferrigno et al. 1998).
Figure 1 .
Gal3 distribution does not change in the course of early induction. (A) Sc859 (Gal3-2GFP) cells expressing Htb2-mCherry from pFJ35 were grown to mid-log phase in glycerol–lactic acid media and were placed into the Cellasik Onyx microfluidics system. Prior to imaging, cells were treated with cycloheximide for 15 min to prevent galactose-induced increases in Gal3-GFP. Images were acquired before and 45 min after galactose induction. (B) Sc723 cells expressing Hog1-GFP from pPS1739 and Htb2-mCherry from pFJ35 were grown to mid-log phase and were placed into the Cellasik Onyx microfluidics system. Images were acquired before and 10 min after osmotic stress with 500 mM NaCl.
In another series of experiments, we utilized FRAP to evaluate whether Gal3 rapidly enters the nucleus. The nucleus of Sc859 cells as marked by Htb2-mCherry was photobleached briefly to eliminate the nuclear Gal3-GFP signal. Limited recovery of the GFP fluorescence in the nucleus was detectable beginning only after 450 sec, and the compartments did not fully re-equilibrate by 10 min (Figure 2A). This result was obtained for both cells in glycerol–lactic acid and galactose media. As a positive control, we performed FRAP with cells expressing Fus3-GFP, which is a known nucleocytoplasmic shuttling protein (Blackwell et al. 2003). The GFP signal in the bleached nuclei of these cells exhibited apparently full re-equilibration within 15 sec (Figure 2C). These results provide direct evidence that Gal3 does not exhibit rapid nuclear import and challenge the hypothesis that Gal3 moves from the cytoplasm into the nucleus in response to galactose.
Figure 2 .
Gal3 does not move into the nucleus rapidly. (A and B) Yeast strain Sc859 nuclei, defined by the nuclear marker Htb2-mCherry from pFJ35, were bleached briefly. The recovery of GFP signal from Gal3-GFP was monitored up to 10 min, at which point only 19% re-equilibration was observed. (C and D) RLY2800 (Fus3-GFP) cells were grown to mid-log phase in glycerol–lactic media. Distribution of Fus3-GFP was imaged, and the indicated nucleus was bleached. The nuclear:cytoplasmic ratio of the molecules completely re-equilibrated <20 sec after bleaching.
Diminishing the nuclear pool of Gal3 impairs the kinetics of induction
In earlier work from this lab, we were unable to detect Gal3 in the nucleus (Peng and Hopper 2000). Subsequently, it was found that fusing Gal3 to peptides for targeting to outer mitochondrial membranes (mom-Gal3) or vesicular membranes (myr-Gal3) outside the nucleus did not diminish maximal levels of induction (Peng and Hopper 2002). Taken together, those results led to the hypothesis that Gal3 acts in the cytoplasm and need not enter the nucleus to promote Gal4 activation (Peng and Hopper 2002). That hypothesis was called into question by the recent detection of Gal3 in the nucleus of uninduced cells (Jiang et al. 2009). However, the possible relevance of the nuclear Gal3 to the regulatory mechanism has not been addressed to date.
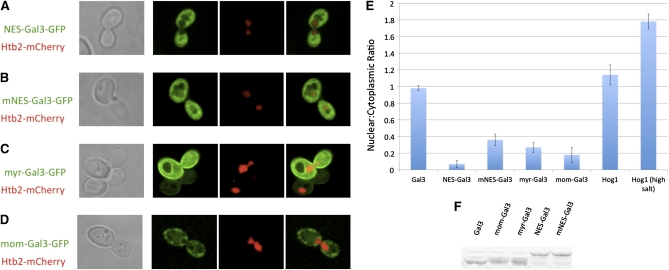
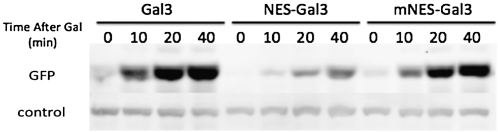
Here, we directly addressed whether depletion of the nuclear Gal3 affects induction. For this test, we fused Gal3-GFP to the strong NES consisting of residues 316–357 of Rna1 (Feng et al. 1999). The NES-Gal3-GFP was observed only in the cytoplasmic region (Figure 3A). This striking exclusion from the nucleus was, as expected, attenuated in control experiments with the L326A, L328A double-mutant version of the NES (mNES) (Figure 3B) previously shown to impair NES activity (Feng et al. 1999). We then assessed the galactose-induction activity of NES-Gal3 compared to wild-type Gal3 and mNES-Gal3 using a galactose-inducible PGAL1-2GFP reporter. Cells with NES-Gal3 showed impaired induction kinetics when compared to cells with wild-type Gal3. Moreover, the effect of the NES tag was alleviated when the leucine residues were replaced by alanines (Figure 4). These results indicate that a critical level of nuclear Gal3 is required for the normally very rapid GAL gene switch response to galactose.
Figure 3 .
Effect of different tags on Gal3 distribution. Sc724 (gal3Δ) cells expressing (A) NES-Gal3-GFP from pOE183, (B) mNES-Gal3-GFP from pOE185, (C) myr-Gal3-GFP from pGP56GFP, or (D) mom-Gal3-GFP from pGP57GFP. All cells were grown to mid-log phase in glycerol–lactic media. (E) The ratio of the nuclear:cytoplasmic signal exhibited by the indicated molecule tagged with GFP was determined by measuring signal density. Ten cells were evaluated for each molecular species, and the results were averaged. The cells were induced for 4 hr in 2% galactose to enhance the GFP signal and were shifted back to gly/lac for 4 hr, at which time image acquisition began. The nuclear regions were marked by Htb2-mCherry that was expressed from pFJ35. (F) Total cellular levels of the various Gal3 molecules in the whole-cell extracts are demonstrated by the Western blot.
Figure 4 .
Effect of depleting nuclear Gal3 on induction kinetics. Sc724 (gal3Δ) cells were transformed with pOE33 (PGAL1-2GFP) and pAKS15 (Gal3WT), pOE180 (NES-Gal3), or pOE182 (mNES-Gal3). Cultures were grown to mid-log phase in glycerol–lactic acid media and induced for indicated times. The GFP produced at each time point was determined by a Western blot. A nonspecific band from the same blot served as a loading control.
The finding that some critical level of Gal3 within the nucleus is required for rapid induction challenges the previous hypothesis that Gal3 need not enter the nucleus to support induction (Peng and Hopper 2002). That hypothesis stemmed in part from results of studies using myr- and mom-tagged versions of Gal3 that highly localized Gal3 to vesicular membranes and mitochondrial outer membrane, respectively (Peng and Hopper 2002). Such tagged versions of Gal3 were shown to be similar to wild-type Gal3 in supporting maximal levels of induction as determined at 6 hr following galactose addition.
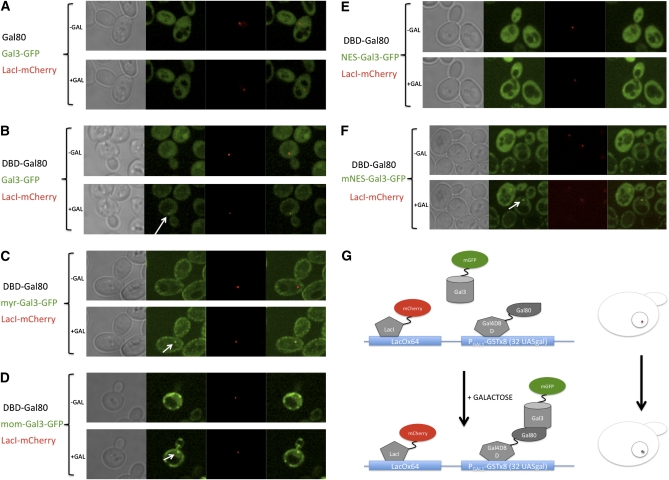
Although the myr-Gal3-GFP and mom-Gal3-GFP signals were seen in those previous experiments to be highly localized to the appropriate membranes outside the nucleus (Peng and Hopper 2002), the imaging sensitivity at the time did not permit detection of residual nuclear pools of these molecules. With our current greatly improved imaging capabilities afforded by SDCM and a more sensitive camera, we sought to determine the effectiveness of the myr, mom, NES, and mNES tags in depleting the nuclear pool of Gal3. We found that all of these tagged forms of Gal3-GFP, when compared to Gal3-GFP, show reductions of the nuclear Gal3-GFP pool (Figure 3). To better assess the relative effectiveness of the various tags in reducing nuclear levels of Gal3, we developed a novel sensitive assay for nuclear Gal3-GFP. This assay utilizes yeast containing a previously developed chromosomal UASGAL array (Jiang et al. 2009) consisting of 32 Gal4 UASGAL-binding sites associated with eight tandem GAL1 promoter-GST genes. The Gal3-GFP expressed in such cells formed a fluorescent spot at the site of the chromosomal UASGAL array within the nucleus only in galactose-induced cells that also expressed Gal80 fused to the Gal4 DNA-binding domain (DBD-Gal80) (Figure 5, A and B). When mNES-Gal3-GFP, myr-Gal3-GFP, and mom-Gal3-GFP were expressed, each of these formed spots at the array (Figure 5, C, D, and F). In contrast, NES-Gal3-GFP did not (Figure 5E). These results indicate that the myr, mom, and mNES tags are not as effective as NES in depleting the nuclear pool of Gal3 and thus provide a plausible explanation for the erroneous earlier proposal (Peng and Hopper 2002) that normal induction does not require Gal3 in the nucleus.
Figure 5 .
Effect of mis-localization tags on the nuclear Gal3 pool. In Sc932 (gal3Δ, [PGAL1-GSTx8]::[LacOx64]::LEU2) cells, (A) Gal80 or (B–F) DBD-Gal80 was expressed from pOE142 or pOE165, respectively. The indicated Gal3 species were expressed from (A and B) pGP17, (C) pGP56GFP, (D) pGP65, (E) pOE183, or (F) pOE185. (G) Schematic presentation of the array system. The cells were grown in glycerol–lactic to mid-log phase and placed into chambers of Cellasik Microfluidics plates. Synthetic drop-out media with 3% glycerol and 2% lactic acid as carbon source were fed through the chambers for 20 min. Images of cells were obtained before and at 1 hr after galactose addition. The same cells were monitored for Gal3-GFP spots, which colocalized with LacI-mCherry expressed from pME9.
Exchange between nuclear and cytoplasmic pools of Gal80 is slow
Earlier studies showed that either one of two distinct regions of Gal80, amino acids 1–321 and amino acids 341–423, were able to localize β-galactosidase to the yeast nucleus in long-term grown cultures (Nogi and Fukasawa 1989). Those results suggested that Gal80 has two independent NLSs. More recent studies showed movement of NLSSV40-Gal80-GFP molecules from one nucleus to the other within the binucleate heterokaryon zygote formed in a kar1-1 cross, suggesting the presence of an NES (Peng and Hopper 2000). These previous data led us to hypothesize that Gal80 shuttles between nucleus and cytoplasm. Thus, we surmised that galactose-activated Gal3, which was not detected in the nucleus at the time, sequestered Gal80 in the cytoplasm (Peng and Hopper 2002). The rapid shuttling of Gal80 molecules is one central feature of such a model; however, no evaluation of the kinetics of Gal80 nucleocytoplasmic trafficking has been reported.
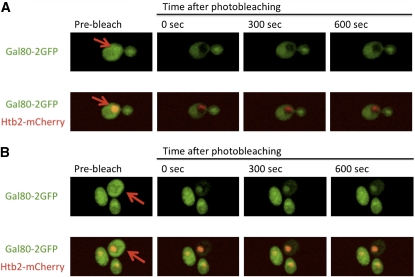
Here we used FRAP to directly assess the subcellular dynamics of Gal80 in Sc862 cells. We bleached Gal80-2GFP in the cytoplasm and observed only ∼15% recovery 10 min after bleaching (Figure 6B). When we bleached Gal80-2GFP in the nucleus, we observed only ∼8% recovery 10 min after bleaching (Figure 6A). These results were in stark contrast to our FRAP results with Fus3-GFP, a known shuttling protein, that showed apparently full recovery 15 sec after nuclear bleaching (Figure 2C). We also used FRAP to test the kinetics of NLS-Gal80-GFP exchange between the two nuclei in a heterokaryon resulting from a kar1 cross performed identically to that in the work of Peng and Hopper (2000). In the present work, and as previously reported (Peng and Hopper 2000), the NLS-Gal80-GFP appeared within both nuclei of the heterokaryons, even though it was expressed in only one of the two mating haploids and its expression was terminated immediately prior to the kar1-1 cross (Figure 7). However, the FRAP analyses clearly showed that the NLS-Gal80-GFP in the unbleached nucleus did not re-equilibrate with the molecules within the bleached nucleus for up to 300 sec post bleach (Figure 7). Taken together, our FRAP results with the NLS-Gal80-GFP in heterokaryon zygotes and Gal80-2GFP in vegetatively grown haploid cells reveal that Gal80 does not shuttle rapidly between the nucleus and cytoplasm.
Figure 6 .
Subcellular distribution and dynamics of Gal80. Sc862 (Gal80-2GFP) cells expressing Htb2-mCherry from pFJ35 were grown to mid-log phase in glycerol–lactic acid media. The cells were induced with galactose for 4–6 hr to enhance the GFP signal and then shifted back to non-inducing media for 4 hr. The (A) nuclear or (B) cytoplasmic region was briefly bleached.
Figure 7 .
NLS-Gal80-GFP does not shuttle rapidly in heterokaryons. NLS-Gal80-GFP was expressed in wild-type MATa cells from a GAL1/CYC1 promoter. The expression was repressed with 2% glucose for 2 hr, after which the cells were mated with MATα kar1-1 cells. Binucleate heterokaryons appeared within 2 hr. The GFP signal was found in both nuclei of the heterokaryons. One of the two nuclei was bleached briefly, and the heterokaryon nuclei were monitored for 5 min.
Assessment of the dynamics of the Gal80–Gal4–UASGAL and Gal4–UASGAL complexes
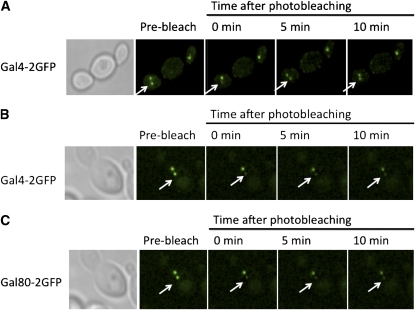
Because Gal3 is in the nucleus prior to galactose addition (Figure 1) (Jiang et al. 2009) and the nuclear pool of Gal3 is important for rapid induction kinetics (Figure 4), we wondered whether Gal3 might work by contacting Gal4-associated Gal80, as posited earlier in a nondissociation model (Platt and Reece 1998). Although it is now clear that Gal80 does dissociate from Gal4, a galactose-triggered interaction of nuclear Gal3 with Gal4-associated Gal80 could be the event that triggers such dissociation. In such a case, rapid induction would be expected to be independent of the kinetic stability of the Gal80–Gal4 complex. If, on the other hand, Gal3 binds exclusively to free Gal80, we would expect that high kinetic stability (slow off-rate) for the Gal80–Gal4 complex at the UASGAL site would be rate limiting for induction. Accordingly, we evaluated the kinetic stability of Gal80–Gal4 complexes at UASGAL sites using a novel genetic assay consisting of diploid cells that have two of the above-mentioned UASGAL chromosomal arrays (Jiang et al. 2009). Expression of Gal80-2GFP or Gal4-2GFP in these cells allowed us to visualize Gal4–Gal80 complexes as two separated nuclear spots, one for each array (Figure 8). We bleached one of the spots and monitored the fluorescence recovery in the bleached array as well as loss of fluorescence in the unbleached array. The signal in the targeted spots of Gal4–Gal80-2GFP did not recover >10% within 10 min after bleaching. The lack of re-equilibration between the two spots within the same nucleus signifies very little dissociation of either the Gal4–Gal80-2GFP complex from the DNA or the Gal80-2GFP from the Gal4–DNA complex within 10 min (Figure 8C). In contrast, our experiments with Gal4-2GFP:Gal80 showed that the Gal4-2GFP molecules re-equilibrated between the two arrays within 5 min after photobleaching when the cells were in galactose media (Figure 8A), but not when they were in glycerol–lactic acid media (Figure 8B). These results indicate that the Gal4–Gal80 complex is kinetically stable.
Figure 8 .
Dynamics of Gal4–DNA and Gal80–Gal4 interactions. (A and B) Sc930 (MATa Gal4-2GFP [PGAL1-GSTx8]::[LacOx64]::LEU2) and (C) Sc920 (MATa Gal80-2GFP [PGAL1-GSTx8]::[LacOx64]::LEU2) strains were mated with Sc965 (MATα Gal4-2GFP [PGAL1-GSTx8]::[LacOx64]::LEU2) and Sc963 (MATα Gal80-2GFP [PGAL1-GSTx8]::[LacOx64]::LEU2), respectively. The resulting diploid cells containing two arrays were grown to mid-log phase in (A) galactose or (B and C) glycerol–lactic acid media. One of the GFP spots in the cells was briefly bleached, and the cells were monitored for 10 min. (A) The Gal4-2GFP signal in the bleached array of the cells grown with galactose recovered completely within 5 min, while no detectable re-equilibration was observed for (B) Gal4-2GFP and (C) Gal80-2GFP spots in cells grown in glycerol–lactic acid media.
Discussion
It is well established and widely agreed that the GAL gene switch functions through a galactose-triggered binding of Gal3 to Gal80 that relieves Gal80 inhibition of Gal4. How this occurs mechanistically is controversial (Platt and Reece 1998; Peng and Hopper 2002; Wightman et al. 2008; Jiang et al. 2009). Lack of agreement centers primarily on three issues: whether the molecular events that initiate induction involve the movement of Gal80 or Gal3 molecules between cytoplasm and nucleus; where in the cell the initial, induction-triggering interaction between Gal3 and Gal80 occurs; and whether Gal3 binds to Gal4-associated Gal80, to exclusively free Gal80, or to both forms of Gal80. To address these issues, we evaluated the kinetics of Gal80 and Gal3 movements between cytoplasm and nucleus and the dynamics of the Gal80–Gal4 complex.
Gal80-2GFP movement between subcellular compartments is relatively slow
Our FRAP analyses involving Gal80-GFP in haploid cells and in heterokaryons revealed very slow kinetics for nucleocytoplasmic transport of Gal80-GFP molecules. This was in striking contrast to our results with Fus3-GFP, a protein known to exhibit rapid nucleocytoplasmic shuttling (Blackwell et al. 2003). These results, considered with respect to the rapid response of the GAL gene switch, challenge the hypothesis that nucleocytoplasmic shuttling of Gal80 is a central feature of the GAL gene switch mechanism (Peng and Hopper 2002). That hypothesis stemmed in large part from two studies. One previous study identified two different fragments of Gal80, designated NLS I and NLS II, which independently localized the Escherichia coli β-galactosidase protein to the yeast nucleus (Nogi and Fukasawa 1989). However, in that study, the subcellular localization was determined following several generations of growth. Consequently, those results could be explained by slow entry of the Gal80–β-galactosidase fusion protein into the nucleus. The other previous study determined that NLSSV40-Gal80-GFP moved from one nucleus (donor) to the other nucleus in a yeast heterokaryon, leading to the conclusion that Gal80 has an NES (Peng and Hopper 2000). In that work and in identical experiments carried out here, the time lag between the formation of the heterokaryon and image acquisition of the NLSSV40-Gal80-GFP was 1–2 hr. Consequently, the results of the heterokaryon experiments can be explained by slow movement of NLSSV40-Gal80-GFP from the one nucleus to the cytoplasm, followed by rapid entry into the other nucleus, due to the strong SV40 NLS tag on Gal80. Therefore, we conclude that Gal80 does not exhibit a rate of nucleocytoplasmic exchange typical of shuttling proteins.
Gal3-GFP does not move rapidly into the nucleus
If Gal3 enters the nucleus upon binding galactose, we expected to see evidence of such redistribution by a decrease in cytoplasmic fluorescence and a corresponding increase in nuclear fluorescence in cells expressing Gal3-GFP. Our results show that the subcellular distribution observed for preexisting Gal3 (in non-induced cells) does not change rapidly in response to galactose. This conclusion is corroborated by the results of our FRAP experiments in which nuclear Gal3-GFP was bleached, and only a small fractional recovery was detected 10 min after bleaching. Thus, it is very unlikely that galactose causes entry of Gal3 into the nucleus to account for the rapid Gal4-mediated gene activation (induction).
Nuclear-localized Gal3 is required for rapid induction
The fact that Gal3 is in the nucleus prior to galactose addition and does not show rapid movement from cytoplasm to the nucleus in response to galactose prompted us to test the significance of nuclear Gal3. We find that strongly reducing the nuclear level of Gal3 by fusing it to an exogenous NES impairs the kinetics of induction. Thus, some level of Gal3 is required in the nucleus for the normal rapid induction kinetics. This conclusion is at odds with the conclusion from previous work that Gal3 entry to the nucleus is not required for maximal levels of induction (Peng and Hopper 2002). Peng and Hopper’s work showed that the levels of MEL1 gene-encoded α-galactosidase enzyme activity produced by cells at 6 hr after galactose addition was similar for cells expressing wild-type Gal3, myr-, or mom-tagged Gal3 variants (Peng and Hopper 2002). To explain the contradictory conclusions, we reasoned that the myr and mom tags might not be as effective as the NES tag in excluding Gal3 from the nucleus and that the magnitude of induction might not be as sensitive as the kinetics of induction to reduced nuclear levels of Gal3. The effectiveness of misplacing Gal3 with myr and mom tags was determined using conventional wide-field imaging (Peng and Hopper 2002). In addition, in this study when we used confocal imaging of non-array cells to compare the myr, mom, and NES tags, we found that they similarly depleted nuclear levels of Gal3-GFP. However, when we used a more sensitive photon-concentrating assay employing cells with the 32 Gal4-binding site (UASGAL) array and expressing DBD-Gal80, we detected a difference in the effects of the tags. The myr- and mom-Gal3-GFP but not NES-Gal3-GFP formed spots at the array, indicating that the myr and mom tags are not as effective as the NES tag in reducing the nuclear concentration of Gal3. Thus, we surmise that, in Peng and Hopper (2002), small pools of myr- or mom-tagged Gal3 in the nucleus were not detectable but were sufficient to support the maximal levels of induction observed at 6 hr, leading to the view at the time that Gal3 need not act in the nucleus.
That even a small fraction of myr- or mom-tagged Gal3 in the nucleus is likely sufficient to support high levels of GAL gene expression is suggested also by our comparison of NES and mNES versions of Gal3-GFP. We found that mNES, like NES, diminished nuclear Gal3 in non-array cells (Figure 3B), but mNES-Gal3 supported induction kinetics very similarly to wild-type Gal3 (Figure 4). As expected, our sensitive array system showed mNES-Gal3-GFP spots at the Gal4-binding sites occupied by DBD-Gal80 (Figure 5F). Taken together, these results indicate that some minimal nuclear level of Gal3 is required to support the rapid induction kinetics that is characteristic of the GAL gene switch.
Stable Gal3–Gal80–Gal4 complex is not detectable in galactose-induced cells
Our finding that a reduction in the nuclear pool of Gal3 slows the induction kinetics together with our evidence against rapid movements of Gal80 and Gal3 between nucleus and cytoplasm implies that the nuclear pool of galactose-activated Gal3 might be critical for initiating rapid relief of Gal80 inhibition. Accordingly, nuclear Gal3 might act through binding to only free Gal80 or to both free Gal80 and Gal80 that is in complex with Gal4. Good evidence exists for Gal3 binding to free Gal80 in vivo, as Gal3-YFP–Gal80-CFP complexes have been detected in both cytoplasm and nucleus 25 min after galactose addition (Wightman et al. 2008). Also, Gal3 is able to bind to Gal80 in vitro in the absence of Gal4 (Yano and Fukasawa 1997). On the other hand, direct evidence for in vivo association of Gal3 with Gal4-associated Gal80 is lacking. In the course of these investigations, we attempted to detect such a complex. We first confirmed that we could detect Gal3-GFP as a fluorescent spot at the array in galactose-exposed cells expressing DBD-Gal80 (Figure 5B). However, when such cells expressed Gal80 instead of DBD-Gal80, a Gal3-GFP spot did not appear at the array in response to galactose (Figure 5A). This result is inconsistent with a previously proposed model specifying nondissociation of Gal80 from Gal4 in response to galactose (Leuther and Johnston 1992; Platt and Reece 1998), but is consistent with the observed galactose- and Gal3-dependent dissociation of Gal80 from Gal4 in live cells (Jiang et al. 2009). While our results are negative, they are not inconsistent with a mechanism wherein Gal3 interacts with Gal4-associated Gal80 to cause immediate dissociation of Gal80 (presumably as a Gal3–Gal80 complex) from Gal4.
Gal80 complexed with Gal4 on DNA does not exchange rapidly with free Gal80
We lack any direct evidence that would distinguish between transient Gal3 binding and Gal4-associated Gal80 and Gal3 binding to only free Gal80. However, if the GAL gene switch works by Gal3 binding to only free Gal80, we would expect to observe rapid exchange between Gal4-associated Gal80 and free Gal80. Otherwise, a kinetically stable Gal80–Gal4 complex would be rate limiting for induction. To our knowledge, the rate of exchange, or dynamics, between Gal4-associated Gal80 and free Gal80 has not been determined. To evaluate the kinetic stability of the Gal80–Gal4 complex on the DNA, we carried out FRAP experiments with diploid array cells. We monitored re-equilibration between bleached and unbleached Gal80-2GFP spots. Re-equilibration is possible only if either the Gal4–Gal80-2GFP complex dissociates from a UASGAL or the Gal80-2GFP molecules dissociate from the Gal4–UASGAL complex at the array. Our results show that a bleached Gal80-2GFP spot did not re-equilibrate substantially with the unbleached spot over 10 min (Figure 8C). This lack of evident re-equilibration challenges a mechanism wherein Gal3 binds to only free Gal80.
On the other hand, Gal4 molecules exchanged between the two arrays within 5 min, when the cells were in galactose, but not in glycerol–lactic acid media. According to a previously proposed Gal4 cycle model, it was suggested that Gal4 molecules became more dynamically associated with the DNA-binding sites after galactose induction (Muratani et al. 2005). However, other more recent work counters this idea by showing that Gal4 became more stable on the GAL promoters of induced cells (Ferdous et al. 2007). Our results from the FRAP experiments using diploid array strains are consistent with Gal4 becoming less stable on the DNA upon induction.
Transient association of Gal3 with Gal4-associated Gal80: a plausible mechanistic feature of the GAL gene switch
In light of the very rapid induction in this system, the relatively slow exchange of Gal80 between its Gal4-bound and free states observed here and the importance of nuclear Gal3 suggests a new plausible model. Transient interaction of nuclear Gal3 with Gal4-associated Gal80 might be what initiates the rapid galactose-mediated dissociation of Gal80 from Gal4. This notion is seemingly at odds with the previously reported detection of a Gal3–Gal80:Gal4 ternary complex by DNA-electrophoretic mobility shift analyses (EMSA) in polyacrylamide gels (Platt and Reece 1998). However, the form of Gal3 used in those EMSA experiments was not the wild-type Gal3 protein but rather a misense mutant Gal3 protein identified as not requiring galactose to bind to Gal80 (Blank et al. 1997). Moreover, it has been shown that exchange rates for macromolecular complexes can be 100-fold slower in gels than in free solution (Fried and Crothers 1981; Fried and Bromberg 1997; Vossen and Fried 1997). Thus, off-rates can be substantially underestimated by EMSA (Yang et al. 2002). Accordingly, we suggest the possibility that the association of Gal3 with Gal80–Gal4:DNA complexes observed previously might have been artificially stabilized by the gel environment and possibly also by the use of the Gal3 mutant protein.
The possibility of transient interaction of Gal3 with Gal4-associated Gal80 as a mechanistic feature of the GAL gene switch has two important testable implications. First, Gal3 interaction with Gal80 must occur on a surface separate from that to which Gal4 binds. Although the binding surface of Gal80 for Gal4 has been determined partially (Kumar et al. 2008), its surface for binding to Gal3 has not. Second, Gal3 transient binding to Gal80 must alter some property of Gal80 that is essential to its high-affinity binding with Gal4. One property of Gal80 that has been implicated in its association with Gal4 is its capacity to dimerize (Pilauri et al. 2005; Kumar et al. 2008), but no direct evidence exists for whether Gal3 interaction with Gal80 affects Gal80 self-association. These issues highlight the fact that much remains to be learned concerning the mechanisms involved in this seemingly simple gene switch.
Acknowledgments
We thank Jian-Qiu Wu for his help in the use of the spinning disc confocal microscope, Rong Li and Brian Slaughter for providing the yeast strain RLY2800, and Sudip Goswami for critical reading of the manuscript. This work was supported by grant GM027925 from the National Institutes of Health (to J.E.H.).
Literature Cited
- Ansari A. Z., Reece R. J., Ptashne M., 1998. A transcriptional activating region with two contrasting modes of protein interaction. Proc. Natl. Acad. Sci. USA 95: 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. M., Ricuperohovasse S., Winston F., 1995. Tbp mutants defective in activated transcription in-vivo. EMBO J. 14: 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E., Halatek I. M., Kim H. J. N., Ellicott A. T., Obukhov A. A., et al. , 2003. Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol. Cell. Biol. 23: 1135–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank T. E., Woods M. P., Lebo C. M., Xin P., Hopper J. E., 1997. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant G. O., Ptashne M., 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11: 1301–1309 [DOI] [PubMed] [Google Scholar]
- Carlson M., 1987. Regulation of sugar utilization in Saccharomyces species. J. Bacteriol. 169: 4873–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., John S., Sil A. K., Hopper J. E., Workman J. L., 2002. Gal80 confers specificity on HAT complex interactions with activators. J. Biol. Chem. 277: 24648–24652 [DOI] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D., 1990. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol. Cell. Biol. 10: 2916–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W. Q., Benko A. L., Lee J. H., Stanford D. R., Hopper A. K., 1999. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 112: 339–347 [DOI] [PubMed] [Google Scholar]
- Ferdous A., Sikder D., Gillette T., Nalley K., Kodadek T., et al. , 2007. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding promoters. Genes Dev. 21: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A., 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17: 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H., Ljungdahl P. O., 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40: 91–109 [DOI] [PubMed] [Google Scholar]
- Fried M. G., Bromberg J. L., 1997. Factors that affect the stability of protein-DNA complexes during gel electrophoresis. Electrophoresis 18: 6–11 [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M., 1981. Equilibria and kinetics of Lac repressor-operator interactions by polyacrylamide-gel electrophoresis. Nucleic Acids Res. 9: 6505–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Ptashne M., 1987. Transcription in yeast activated by a putative amphipathic alpha-helix linked to a DNA-binding unit. Nature 330: 670–672 [DOI] [PubMed] [Google Scholar]
- Gräf R., Rietdorf J., Zimmermann T., 2005. Live cell spinning disk microscopy. Adv. Biochem. Eng. Biotechnol. 95: 57–75 [DOI] [PubMed] [Google Scholar]
- Jeong C. J., Yang S. H., Xie Y., Zhang L., Johnston S. A., et al. , 2001. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry 40: 9421–9427 [DOI] [PubMed] [Google Scholar]
- Jiang F. L., Frey B. R., Evans M. L., Friel J. C., Hopper J. E., 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol. Cell. Biol. 29: 5604–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51: 458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Dincher S. S., 1987. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell 50: 143–146 [DOI] [PubMed] [Google Scholar]
- Koh S. S., Ansari A. Z., Ptashne M., Young R. A., 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1: 895–904 [DOI] [PubMed] [Google Scholar]
- Kumar P. R., Yu Y., Sternglanz R., Johnston S. A., Joshua-Tor L., 2008. NADP regulates the yeast GAL induction system. Science 319: 1090–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Larschan E., Winston F., 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuther K. K., Johnston S. A., 1992. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science 256: 1333–1335 [DOI] [PubMed] [Google Scholar]
- Lohr D., Venkov P., Zlatanova J., 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9: 777–787 [DOI] [PubMed] [Google Scholar]
- Lue N. F., Chasman D. I., Buchman A. R., Kornberg R. D., 1987. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol. Cell. Biol. 7: 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M., 1987. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 50: 137–142 [DOI] [PubMed] [Google Scholar]
- Melcher K., Johnston S. A., 1995. Gal4 interacts with tata-binding protein and coactivators. Mol. Cell. Biol. 15: 2839–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K., Xu H. E., 2001. Gal80-Gal80 interaction on adjacent Gal4p binding sites is required for complete GAL gene repression. EMBO J. 20: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R. A., Amuel C., Engels R., Gengenbacher U., Labahn J., 2003. Sites for interaction between Gal80p and Gal1p in Kluyveromyces lactis: structural model of galactokinase based on homology to the GHMP protein family. J. Mol. Biol. 333: 479–492 [DOI] [PubMed] [Google Scholar]
- Mizutani A., Tanaka M., 2003. Regions of GAL4 critical for binding to a promoter in vivo revealed by a visual DNA-binding analysis. EMBO J. 22: 2178–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Kung C., Shokat K. M., Tansey W. P., 2005. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120: 887–899 [DOI] [PubMed] [Google Scholar]
- Nogi Y., Fukasawa T., 1989. Functional domains of a negative regulatory protein, GAL80, of Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 3009–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Coleman J. E., 1989. Structure and function of the Zn(Ii) binding-site within the DNA-binding domain of the Gal4 transcription factor. Proc. Natl. Acad. Sci. USA 86: 3145–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M. R., Jaehning J. A., 1992. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol. Cell. Biol. 12: 4981–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Hopper J. E., 2000. Evidence for Gal3p’s cytoplasmic location and Gal80p’s dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 5140–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Hopper J. E., 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. USA 99: 8548–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Hopper J. E., 1979. Constitutive synthesis of the Gal4 protein, a galactose pathway regulator in Saccharomyces cerevisiae. Cell 16: 89–95 [DOI] [PubMed] [Google Scholar]
- Pilauri V., Bewley M., Diep C., Hopper J., 2005. Gal80 dimeirization and the yeast GAL gene switch. Genetics 169: 1903–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A., Reece R. J., 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 17: 4086–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., 2000. Molecular basis of nutrient-controlled gene expression in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 57: 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M., 1993. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science 261: 909–911 [DOI] [PubMed] [Google Scholar]
- Schjerling P., Holmberg S., 1996. Comparative amino acid sequence analysis of the C-6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24: 4599–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M. A., Thron K. S., 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670 [DOI] [PubMed] [Google Scholar]
- Slaughter B. D., Schwartz J., Li R., 2007. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc. Natl. Acad. Sci. USA 104(51): 20320–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Fujimoto T., Fukuma M., Yano K. I., Sakurai H., Vonika A., et al. , 1996. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol. Cell. Biol. 16: 2504–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson D. J., Ross H. C., Reece R. J., 2002. Gal3p and Gal1p interact with the transcriptional repressor Gal80p to form a complex of 1:1 stoichiometry. Biochem. J. 363: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia M. G., Danzinger R. G., 1980. Reversed-phase high-performance liquid-chromatographic assay for cefoxitin in proteinaceous biological samples. J. Chromatogr. A 181: 120–122 [DOI] [PubMed] [Google Scholar]
- Torchia T. E., Hamilton R. W., Cano C. L., Hopper J. E., 1984. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol. Cell. Biol. 4: 1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Jelicic B., Sopta M., 2006. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 7: 496–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E. A., Hiller M. A., Scherson T. Y., Rose M. D., 1992. Separate domains of Kar1 mediate distinct Functions in mitosis and nuclear fusion. J. Cell Biol. 117: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenbroich V., Meyer J., Engels R., Cardinali G., Menezes R. A., et al. , 1999. Galactose induction in yeast involves association of Gal80p with Gal1p or Gal3p. Mol. Gen. Genet. 261: 495–507 [DOI] [PubMed] [Google Scholar]
- Vossen K. M., Fried M. G., 1997. Sequestration stabilizes lac repressor-DNA complexes during gel electrophoresis. Anal. Biochem. 245: 85–92 [DOI] [PubMed] [Google Scholar]
- Wightman R., Bell R., Reece R. J., 2008. Localization and interaction of the proteins constituting the GAL genetic switch in Saccharomyces cerevisiae. Eukaryot. Cell 7: 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Reece R. J., Ptashne M., 1996. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 15: 3951–3963 [PMC free article] [PubMed] [Google Scholar]
- Yang E., Henriksen M. A., Schaefer O., Zakharova N., Darnell J. E., 2002. Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J. Biol. Chem. 277: 13455–13462 [DOI] [PubMed] [Google Scholar]
- Yano K., Fukasawa T., 1997. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarger J. G., Halvorson H. O., Hopper J. E., 1984. Regulation of galactokinase (GAL1) enzyme accumulation in Saccharomyces cerevisiae. Mol. Cell. Biochem. 61: 173–182 [DOI] [PubMed] [Google Scholar]
- Yun S. J., Hiraoka Y., Nishizawa M., Takio K., Titani K., et al. , 1991. Purification and characterization of the yeast negative regulatory protein GAL80. J. Biol. Chem. 266: 693–697 [PubMed] [Google Scholar]