Abstract
Here we review recent advances in understanding the regulation of mRNA synthesis in Saccharomyces cerevisiae. Many fundamental gene regulatory mechanisms have been conserved in all eukaryotes, and budding yeast has been at the forefront in the discovery and dissection of these conserved mechanisms. Topics covered include upstream activation sequence and promoter structure, transcription factor classification, and examples of regulated transcription factor activity. We also examine advances in understanding the RNA polymerase II transcription machinery, conserved coactivator complexes, transcription activation domains, and the cooperation of these factors in gene regulatory mechanisms.
IN concluding his 1995 review of yeast transcriptional regulation, Kevin Struhl (1995) proposed three major questions to direct future research: (1) How do activators and repressors affect the basic transcriptional machinery of the cell? (2) How are the activities of regulators themselves regulated? and (3) How are the various regulatory pathways integrated to coordinate cell growth and response to external signals? Studies in yeast have made seminal contributions in each of these areas in the subsequent 15 years. Enormous strides have been made to answer the first question. The structures of RNA polymerase (Pol) II and several general transcription factors have been determined, and the yeast system has been at the forefront in discoveries of fundamental transcription mechanisms. Transcription activators and repressors act, in part, by recruitment of the transcription machinery or repression complexes to gene regulatory regions. How the regulators are regulated (question 2) surprisingly seems to have almost as many answers as there are regulators in the cell. Question 3 is the subject of current “omics” research. Genome-wide methods are being used to analyze global expression and DNA binding by many transcription factors assayed under varied growth conditions and in multiple yeast species. Below, we provide an outline of recent advances in understanding the mechanisms of Saccharomyces cerevisiae transcription factors, the general transcription machinery, and the cooperation of these factors in transcriptional regulation.
Upstream Activation Sequence Elements
Transcriptional regulation begins with sequence-specific recognition of unique DNA elements by transcription factors (TFs), either transcription activators (TAs) binding to upstream activation sequences (UASs) or repressors (TRs) binding to upstream repression sequences (URSs). UASs and their associated TFs are probably required for expression of all protein-coding genes transcribed by Pol II, which contain one or more UASs. Although there is a low level of non-activator-dependent “basal” transcription in vitro, basal transcription is repressed when a chromatin template, rather than free DNA, is used, suggesting that nucleosomes repress non-TF-dependent transcription initiation in vivo (Juan et al. 1993). Thus, the “ground-state” of a yeast promoter is inactive and transcription must be promoted by one or more TFs (Struhl 1999). Poly(dA:dT) elements (Iyer and Struhl 1995b) and altered chromatin (Han and Grunstein 1988; Han et al. 1988) may stimulate activator-independent transcription.
Identification of UAS elements
The classical method of UAS (or URS) identification utilizes fusions of regulatory region DNA or deletion derivatives to a core promoter and reporter gene such as Escherichia coli lacZ followed by expression analysis (Sentenac and Hall 1982; Struhl 1989, 1995). A technique for generalizing this experimental approach has recently been described (Doyon and Liu 2007). However, sequencing of multiple yeast genomes has allowed a bioinformatics approach to largely supplant the classical approach for discovering and characterizing regulatory motifs. The development of algorithms that can find conserved or known motifs in promoter sequences has been instrumental, particularly when combined with global TF-binding data (Hu et al. 2010; Reid et al. 2010). Phylogenetic footprinting (Cliften et al. 2003; Kellis et al. 2003; Borneman et al. 2007; Tuch et al. 2008; Lavoie et al. 2010) and nucleosome location data (Narlikar et al. 2007; Gordan et al. 2009) can enhance the probability that conserved motifs represent functional UASs (Wingender et al. 1996; Monteiro et al. 2008; Park et al. 2008). Genome-wide mapping of 5′ mRNA ends (Miura et al. 2006; Nagalakshmi et al. 2008) and newly discovered or revised binding site preferences for >100 yeast transcription factors have also improved the ability to associate conserved motifs with known or suspected TFs. Proof of UAS/URS function still requires chromatin immunoprecipitation (ChIP) experiments to demonstrate TF occupancy and mutation of the binding site to demonstrate function.
UAS conservation and evolution
Evolution can be studied in yeast because yeast has a relatively short doubling time and can utilize a variety of nutrients, and its genome can be analyzed rapidly and completely (Dunham et al. 2002). The current paradigm in molecular evolution is that phenotypic diversity, and perhaps speciation, has primarily occurred by altering gene regulation rather than by altering the function of individual proteins. The results in yeast demonstrate that regulatory circuits are evolving rapidly using conserved TFs to regulate different sets of genes and are being driven by changes in both binding motifs and TFs (Tsong et al. 2003, 2006; Borneman et al. 2007; Chang et al. 2008; Tuch et al. 2008; Zheng et al. 2010).
UAS function and combinatorial control
UAS/URS function is generally orientation independent but dependent on being 5′ of the promoter with a few exceptions (Errede et al. 1984; Mellor et al. 1987; Company and Errede 1988; Fantino et al. 1992; Gray and Fassler 1996). Their number and position may influence the level of expression (Swamy et al. 2009). It is unknown why most yeast enhancers (UASs) cannot function 3′ of the promoter, but enhancers in higher eukaryotes can function both 5′ and 3′ of the promoter. The TF Gal4 can operate from both up- and downstream of +1 in higher eukaryotes (Webster et al. 1988) and can also function from a site >1 kb downstream of the start site of transcription when it binds near a telomere (de Bruin et al. 2001). Thus the position-dependent activity of UASs in yeast does not reside in the TF and may be due to unknown differences in either chromatin or the core promoter.
Although the exact distance of the UAS from the transcription start site may not be important, most functional UASs appear to be located in the nucleosome-depleted region of yeast promoters directly upstream of the transcription start site or are exposed on the surface of nucleosomes (Xue et al. 2004; Albert et al. 2007; Lee et al. 2007; reviewed in Li et al. 2007). One interpretation of this observation is that TF accessibility is adversely affected by the presence of a nucleosome, and thus the depleted region presents unimpeded access to its binding site.
When different types of UAS are present in the same promoter, they allow combinatorial control of transcription. The complex dynamics of gene expression during the cell cycle and after subjecting cells to stress have been assayed by ChIP analysis of the regulatory TFs and coactivators (McBride et al. 1997; Bhoite et al. 2001; Simon et al. 2001; Horak et al. 2002; Tan et al. 2008; Ni et al. 2009). Similarly, during sporulation (Kahana et al. 2010) and glucose starvation (Young et al. 2003; Tachibana et al. 2005; Ratnakumar and Young 2010) multiple TFs bind upstream of the promoter of coregulated genes. In the sulfur metabolic network, the non-DNA-binding Met4 activator interacts with multiple cofactors that stabilize DNA binding (Lee et al. 2010). Mcm1 heterodimerizes with different TFs to regulate cell-type and other processes (Tan and Richmond 1998; Hollenhorst et al. 2001; Tuch et al. 2008).
Transcription Factors
At present there are 169 genes designated as TFs in the Yeastract database (http://www.yeastract.com; Teixeira et al. 2006), making TFs one of the most abundant classes of proteins in the yeast genome. Analysis of other Saccharomyces genomes has allowed a comprehensive comparison of TFs between species (http://www.cssm.info/priloha/fm2008_drobna_tab2.pdf; Drobna et al. 2008). Many yeast TFs were discovered by genetic means because they influence expression of downstream target genes. Additional TFs were identified by homology. Whether all yeast TFs can be identified by these approaches is an open question. Recently, a metabolic enzyme involved in ornithine biosynthesis, Arg5,6 was identified in a screen for DNA-binding proteins (Hall et al. 2004). Arg5,6 represents just one of several examples in yeast of so-called “moonlighting” proteins, in this case a DNA-binding protein that also serves as an enzyme of intermediary metabolism (Gancedo and Flores 2008).
TF classification
TFs are grouped into three general classes on the basis of the type of DNA-binding domain (DBD) that they contain: zinc (Zn)+2 stabilized, helix-turn-helix, and zipper type; the classes were updated in 2000 using analysis of structural information in the Brookhaven Protein Database (http://www.biochem.ucl.ac.uk/bsm/prot_dna/prot_dna.html; Luscombe et al. 2000). Figure 1, A–E, illustrates five yeast TF DBDs.
Figure 1.
DNA-binding domains of five yeast transcription factors. Blue spheres, Zn; gray spheres, DNA. Protein data bank (http://www.biochem.ucl.ac.uk/bsm/prot_dna/prot_dna.html) accession codes are in parentheses. (A) C2H2 zinc fingers of Adr1 (2ADR); (B) C6 (zinc knuckle) of Gal4 (1D66); (C) bZIP structure of Gcn4 (1YSA); (D) bHLH of Pho4 (1AOA); (E) helix-turn-helix of Matα2 and winged helix of Mcm1 (1MNM).
The Zn+2-stabilized DBD is the most abundant in all organisms and can be subdivided (Krishna et al. 2003) into C2H2 zinc fingers (Bohm et al. 1997), C6 (zinc knuckle or Zn2Cys6 binuclear zinc cluster) (MacPherson et al. 2006), and C4 or GATA fingers (Scazzocchio 2000). Yeast also has at least one TF whose DNA-binding domain is stabilized by Cu+2 (e.g., Ace1/Cup2). C2H2 zinc-finger proteins are ubiquitous as are C4 (GATA) factors. The C6 or zinc-knuckle type is unique to fungi.
C2H2 zinc fingers (53 members—e.g., Adr1, Mig1, Zap1) have a modular structure that is stabilized by tetrahedral coordination of a zinc ion by Cys and His ligands and by a hydrophobic core of conserved Phe and Leu residues (Bohm et al. 1997) (Figure 1A). The C terminus is an α-helix whose N-terminal amino acids confer DNA-binding specificity. The C2H2 TFs generally bind DNA as monomers with each finger recognizing consecutive triplets of bases; specificity and high affinity are achieved by multiple fingers.
The C6 proteins (55 members—e.g. Gal4, Mal63, Hap1, Leu3) have a DBD containing two zinc ions liganded to six Cys residues (MacPherson et al. 2006) (Figure 1B). The DBD is N-terminal in most C6 TFs with the DNA-binding residues C-terminal to the C6 region. The C6 TFs bind DNA sites containing CGG triplets flanking a region containing a variable number of residues. The orientation of the CGG triplets and their spacing are the primary determinants of DNA-binding specificity (Reece and Ptashne 1993). The C6 proteins bind as dimers to symmetric sites, utilizing a dimerization domain C-terminal to the DBD. Many C6 proteins bind DNA exclusively as homodimers, such as Gal4 and Leu3, whereas others, such as Oaf1 and Pip2 and Pdr1 and Pdr3, can form heterodimers as well. A few, such as Rgt1, are thought to bind DNA as monomers.
The C4 or GATA class of zinc-stabilized TFs consists of proteins that are primarily involved in nitrogen metabolism in S. cerevisiae (Cooper 2002) (5 members—Gln3, Gat1, Nil1, Dal80, Ash1). Ash1 represses HOmothallic switching endonuclease (HO) transcription specifically in daughter cells through cytoskeletal-regulated partitioning of its mRNA in the nucleus (Cosma 2004), where it activates the filamentation pathway. In other fungi, the GATA factors are involved in multiple pathways that include mating-type switching (Scazzocchio 2000).
The second most abundant class of TF is the zipper type (22 members). This class is characterized by a DBD consisting of a dimerization motif and a basic region. The first subclass, bZIP proteins (14 members—e.g., Gcn4, Yap1-8, Sko1), also called leucine zippers, have a dimerization domain consisting of multiple Leu residues (Fernandes et al. 1997; Moye-Rowley 2003) (Figure 1C). The second subclass, bHLH proteins (9 members—e.g., Ino2, Ino4, Pho2, Pho4), has paired amphipathic α-helices separated by a loop of variable length followed by a basic region (Robinson and Lopes 2000) (Figure 1D). These proteins generally bind DNA as heterodimers, giving rise to a multitude of different complexes (Fernandes et al. 1997; Robinson and Lopes 2000; Moye-Rowley 2003; Chen and Lopes 2007, 2010; Tan et al. 2008).
The third most abundant class, helix-turn-helix (HTH) TFs (Matα1, Matα2, Mata1; eight members), are most closely related to homeodomain-containing proteins in higher eukaryotes and to prokaryotic repressors and activators. They form both homo- and heterodimers. The classical HTH protein in yeast is Matα2, which, together with Mcm1, represses a-specific genes in Matα haploids (Figure 1E) and forms a heterodimer with Mata1 to repress haploid-specific genes in MATa/Matα diploid cells.
The forkhead (Fkh) or MADS-box transcription factors Mcm1, Fkh1, Fkh2, and Hcm1 and the heat-shock factor Hsf1 are related to the HTH proteins (Tan and Richmond 1998; Kaestner et al. 2000). Three helices and two large loops or “wings,” which led to the name “winged helix,” form the DBD. Mcm1 associates with several proteins and acts as both an activator and a repressor to control cell-specific gene expression (Elble and Tye 1991). Fkh proteins are involved in numerous processes, including cell cycle regulation, where they appear to endow other transcription factors with promoter specificity (Hollenhorst et al. 2001; Voth et al. 2007).
Modularity
TFs have a modular structure that consists of multiple, independently functioning domains. One consequence of this modularity is that, unlike globular proteins, they have been resistant to structural analysis as a single entity. Some proteins classified as TFs, such as Met4 (Lee et al. 2010) and Swi6 (Sidorova and Breeden 1993), lack a DBD motif and interact with DNA through a binding partner. Other TFs, such as Gcr1 and Dal81, have a DBD that is dispensable, apparently because it forms a heterodimer and the other subunit is sufficient for DNA binding (Bricmont et al. 1991; Scott et al. 2000; Tornow et al. 1993). The effector domains of a TF may include an activation/repression domain (AD/RD), a nuclear localization sequence (NLS), and a regulatory domain. The first evidence suggesting the modular nature of eukaryotic TFs accompanied the discovery of the Gcn4 and Gal4 ADs (Hope and Struhl 1986; Ma and Ptashne 1987b). The ability of Gal4 to function universally attests to the conserved nature of the activation process in fungi, plants, insects, and metazoans (Fischer et al. 1988; Kakidani and Ptashne 1988; Ma et al. 1988; Webster et al. 1988).
ADs, sometimes more than one, have been documented in numerous TAs (Table 1, “Activation domains”). Although most yeast TAs possess both a DBD and an AD within the same polypeptide, in some heterodimeric TFs only one subunit has an AD. The ReTroGrade (RTG) response, a mechanism by which the mitochondria and nuclear compartments communicate (Butow and Avadhani 2004), is orchestrated by two TFs, Rtg1 and Rtg3. These bHLH proteins form a heterodimer in which only Rtg3 contains an AD that responds to the RTG signal (Rothermel et al. 1997). RDs have been found in many TFs that have a negative role in gene expression (Table 1, “Repression domains”). Some of these proteins, including Ash1, Rgt1, and Rap1, have the ability to both activate and repress transcription, depending on the promoter, chromatin context, and growth conditions.
Table 1. Yeast transcription factors with demonstrated activation or repression domains.
NLSs have been identified by a similar approach by making deletion mutations in the native protein and/or by creating chimeras, usually by fusion to E. coli β-galactosidase, and assessing its intracellular location (Silver et al. 1984, 1986). This approach has identified NLSs on numerous yeast TFs (Hahn et al. 2008). The receptors for classical NLSs are nuclear importins functioning as α/β heterodimers where the α-subunit recognizes the NLS and the β-subunit recognizes the nuclear pore complex (Silver et al. 1989; Brodsky and Silver 1999).
Functions of TF-DNA binding
The DNA-binding specificity of TFs largely determines where it acts in the genome. Recent high-throughput biochemical screens identified new and revised binding sequences for a large number of TFs (Badis et al. 2008; Zhu et al. 2009). Gene expression analysis in strains deleted for or overexpressing specific TFs have identified both direct and indirect TF targets. Short-term overexpression appears to identify direct targets more reliably than analysis of deletion strains (Chua et al. 2006; Sopko et al. 2006). In some cases, direct and indirect effects can be distinguished by integrating binding-site information with data on nucleosome positions and gene expression (Beyer et al. 2006; Gordan et al. 2009).
ChIP-chip and ChIP-Seq (chromatin IP followed by hybridization to microarrays or used in high-throughput DNA sequencing, respectively) are the gold standard for determining TF-binding sites in vivo. ChIP-Seq is particularly valuable because it can provide nucleotide-level resolution of TF-binding sites (Guo et al. 2010). Condition-dependent binding of a TF is one potential complication of ChIP analysis as is the possibility that a bound TF may be inactive (Gao et al. 2004). Genome-wide localization analysis has been reported for TFs involved in numerous pathways [Saccharomyces Genome Database, (http://www.yeastgenome.org/); Yeastract (http://www.yeastract.com)]. Determining how TFs achieve their remarkable promoter-binding specificity is an important goal (Georges et al. 2010).
Functions of TF-activating and -repressing transcription
As described more fully below, much data support the recruitment model of transcription activation proposed by Ptashne (Ptashne 1988; Ptashne and Gann 1997). It has also been proposed that gene localization within the nucleus is sometimes regulated in response to regulatory signals (Menon et al. 2005; Sarma et al. 2007). According to this model, genes and their associated activators move to the nuclear periphery where they encounter the transcriptional machinery at nuclear pore complexes. It has been suggested that retention at the nuclear periphery could explain the phenomenon of transcriptional memory, the ability of an induced but subsequently repressed gene to be rapidly reactivated (Brickner 2009).
The recruitment model has also been invoked to explain the function of RDs. The TRs Matα2, Mig1, Rgt1, Rox1, Rfx1, and Sko1 repress transcription by recruiting the Tup1–Ssn6/Cyc8 complex to promoters (Smith and Johnson 2000; Courey and Jia 2001). Repression occurs by a histone-dependent pathway (Wu et al. 2001; Davie et al. 2002) and through interactions with RNA Pol II and the coactivator termed Mediator (Gromoller and Lehming 2000; M. Lee et al. 2000; Papamichos-Chronakis et al. 2000). The extent of the repressed chromatin domain can be quite large, as at the silent copies of the MAT locus and at telomeres. Repression at these loci is due in part to the Rap1-dependent recruitment of the Sir2/3/4 complex (Wyrick et al. 1999). The extent of the repressed chromatin domain, characterized by hypoacetylated histones, is controversial (Ducker and Simpson 2000; Courey and Jia 2001; Wu et al. 2001).
Ume6, Opi1, and Ash1 modify chromatin and repress transcription by recruiting the Sin3–Rpd3 histone deacetylase complex to create a localized domain of hypoacetylated histones H3 and H4 (Kadosh and Struhl 1998; Carrozza et al. 2005). Ume6 represses meiotic genes during vegetative growth but activates them during sporulation (Rubin-Bejerano et al. 1996; Kassir et al. 2003). Ash1 similarly has both repressive and activating functions (Chandarlapaty and Errede 1998; Maxon and Herskowitz 2001). The Rme1 repressor is unusual in that it has overlapping repression and activation domains and no apparent requirement for specific DNA binding (Blumental-Perry et al. 2002). All known TRs recruit complexes that maintain chromatin in a generally repressive state that may inhibit the binding of an activator and the recruitment of coactivators and prevent chromatin remodeling or block a subsequent step in transcription.
Transcriptional Regulation
Some pathways in yeast are transcriptionally controlled by altering the expression level rather than the activity of the TFs involved. The prototypical yeast activator Gcn4 is one such factor whose level is controlled by translational readthrough of short upstream open reading frames and by ubiquitination-dependent turnover (Hinnebusch 2005). Cell-type determination is another example of complex regulation that occurs at multiple levels (Galgoczy et al. 2004).
The nuclear localization of numerous TFs is altered by regulated interaction with nuclear importins and karyopherins (Komeili and O’Shea 2000). In contrast, the TFs described below represent TAs whose activation function is regulated by inter- or intramolecular interactions triggered by ligand binding, phosphoryation, or stress.
Adr1 Phosphorylation-dependent inhibition of Adr1 by Bmh
Adr1 activates catabolic pathways that are essential for growth in the absence of a fermentable sugar (derepression) (Young et al. 2003). The AMP-activated, protein kinase homolog Snf1 is essential for promoter binding of Adr1, Adr1-dependent chromatin remodeling, and transcription when glucose is exhausted (Di Mauro et al. 2002; Young et al. 2002; Agricola et al. 2004, 2006; Biddick et al. 2008a,b). Glucose repression inhibits Adr1 activity by multiple mechanisms, including ADR1 expression (Blumberg et al. 1988), DNA binding (Sloan et al. 1999; Kacherovsky et al. 2008), and transcription activation (Cherry et al. 1989; Cook et al. 1994; Tachibana et al. 2007; Ratnakumar et al. 2009). PKA phosphorylates S230 in the regulatory domain in vitro (Denis et al. 1992), but its activity is not required for phosphorylation (Ratnakumar et al. 2009) or glucose inhibition of Adr1 activity in vivo (Denis et al. 1992; Dombek et al. 1993).
Promoter binding of Adr1 is also inhibited by hypoacetylation of the histone tails (Verdone et al. 2002). Loss of histone deacetylase activity and consequent hyperacetylation of the histone tails overcomes glucose inhibition of promoter binding, chromatin remodeling, and preinitiation complex recruitment, but not transcription (Verdone et al. 2002; Tachibana et al. 2007). Activated Adr1 and activation of Snf1 stimulate the poised, inactive preinitiation complex and together allow complete escape from glucose repression (Tachibana et al. 2007).
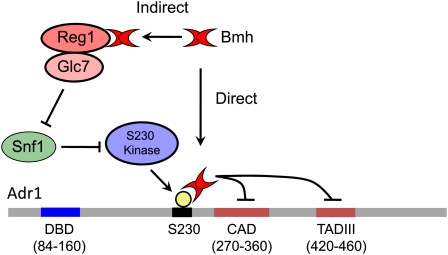
Recent evidence suggests that yeast 14-3-3 (Bmh) proteins inhibit Adr1 activation function by binding to the S230-phosphorylated regulatory domain (Parua et al. 2010). 14-3-3 proteins are components of multiple signaling pathways and affect protein localization or activity by binding to phosphorylated domains (Yaffe 2002; Van Heusden and Steensma 2006). ADR1c alleles, such as ADR1-S230A, disrupt this inhibition as would be expected if phosphorylation were important for Bmh binding (Parua et al. 2010). As illustrated in Figure 2, Bmh has direct and indirect roles in Adr1 regulation, and both roles involve Snf1. Identifying the S230 kinase and the mechanism whereby Snf1 promotes its dephosphorylation (Ratnakumar et al. 2009) might link Adr1 activity to other nutrient-signaling pathways.
Figure 2.
Model for Bmh regulation of Adr1 activity that could explain the direct and indirect roles of Bmh in the repression of Adr1-dependent gene expression. The direct role of Bmh involves binding to the S230-phosphorylated regulatory domain and inhibition of Adr1 ADs. Snf1 is implicated in reversing this direct inhibition because it is required to promote the dephosphorylation of S230 and thus inhibit Bmh binding. Snf1 is also involved in the indirect role of Bmh because Snf1 is inactivated by the Reg1–Glc7 complex in which Bmh plays an unknown role.
Gal4 regulation by intermolecular AD masking
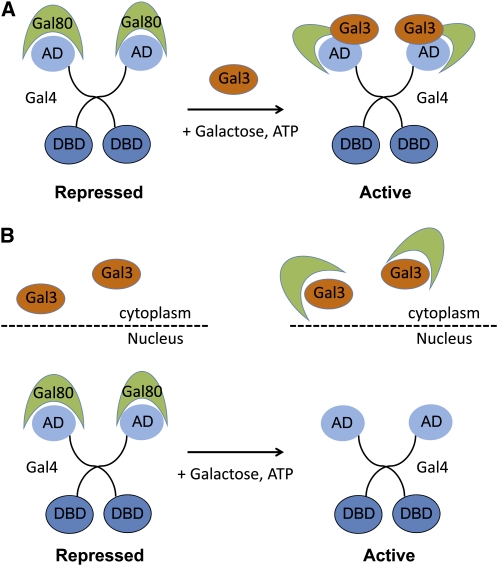
The regulation of GAL gene expression and the description of the regulators involved constitute an important paradigm for eukaryotic gene regulation (Johnston and Carlson 1992). One focus of current research is the nature of promoter-bound but transcriptionally inactive Gal4 (Wightman et al. 2008; Jiang et al. 2009). In the absence of galactose, the C-terminal AD cannot recruit coactivators because it is occluded by Gal80 (Johnston et al. 1987; Ma and Ptashne 1987a). The sensor is Gal3, which binds and inactivates Gal80 when galactose is present. Genetic and structural analyses indicate that Gal4 and Gal3 bind to Gal80 at distinct but overlapping sites (Pilauri et al. 2005; Thoden et al. 2007). Thus, Gal3 holds the key to understanding how galactose is sensed by Gal80 and ultimately by Gal4.
Two opposing models have been proposed to explain Gal3 inhibition of Gal80 (Figure 3). A nondissociation model suggests that Gal3 binds Gal80 but does not remove it from Gal4 at the promoter (Johnston et al. 1987; Ma and Ptashne 1988; Chasman and Kornberg 1990; Parthun and Jaehning 1992; Platt and Reece 1998). Although this tripartite complex has been observed in vitro in the presence of galactose and ATP (Platt and Reece 1998), it has eluded detection in vivo. Recent fluorescence resonance energy transfer (FRET) experiments showed that a Gal80–Gal3 complex is present in both the cytoplasm and the nucleus after galactose induction; however, association of Gal80-Gal3 with Gal4 at a promoter has not yet been observed (Wightman et al. 2008). Other conflicting evidence supports a dissociation model. Cytoplasmic Gal3 can sequester Gal80 away from the nuclear compartment in galactose-inducing conditions, suggesting that Gal80 is not bound to Gal4 at the promoter (Peng and Hopper 2000; Peng and Hopper 2002; Pilauri et al. 2005). Using live-cell imaging and a novel array of Gal4-binding sites, Gal80 was shown to rapidly dissociate from Gal4 in a Gal3, galactose-dependent manner (Jiang et al. 2009), suggesting that there is not a stable Gal3–Gal80–Gal4 complex at the promoter.
Figure 3.
Two models for regulation of Gal4 by Gal80 and Gal3. (A) In the nondissociation model (Wightman et al. 2008), Gal80 may remain bound to DNA-bound Gal4 in the nucleus when it interacts with Gal3 and a structural change allows the Gal4 AD to interact with coactivators. (B) The dissociation model (Peng and Hopper 2000, 2002) suggests that Gal3 bound to galactose and ATP in the cytoplasm sequesters Gal80 from the nucleus and thus frees Gal4 AD for interaction with coactivators.
The GAL system has provided extraordinary insights into eukaryotic gene regulation and the evolution of a regulatory system. The galactose sensor Gal3 is a homolog of the Gal1 galactokinase (Bajwa et al. 1988; Suzuki-Fujimoto et al. 1996), and Gal80 is structurally related to a glucose–fructose oxidoreductase (Thoden et al. 2007). Thus, these two regulatory genes appear to have evolved from genes involved in intermediary metabolism. The GAL system has also demonstrated how yeast genetics and molecular biology can be combined with biochemistry and structural biology to reveal fundamental mechanisms of eukaryotic transcription.
Hap1 and Mal63 regulation by chaperones
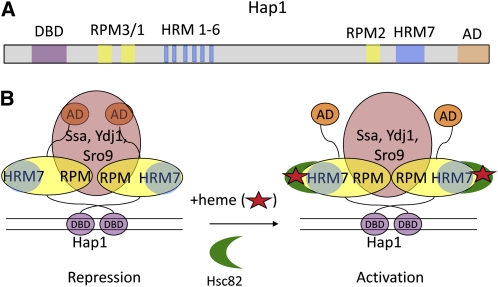
Hap1 and Mal63 are regulated by molecular chaperones as are steroid hormone receptors in mammalian cells (Pearl and Prodromou 2006; Wandinger et al. 2008). Hap1, the major oxygen sensor in the yeast cell, is regulated by the Hsp70/Hsp90 chaperone machine as illustrated in Figure 4. In the absence of heme, repressive regions in the central part of Hap1 keep it in an inactive state (Hach et al. 1999). In response to heme, a biosynthetic precursor of active cytochromes, repression is relieved and genes in the Hap1 regulon are activated (Lee and Zhang 2009). Heme binds in the central regulatory region of Hap1 (Zhang et al. 1998; Hon et al. 2000), allowing transcription activation by the C-terminal AD.
Figure 4.
Model for the regulation of Hap1 by heme and molecular chaperones. (A) The diagram of the domain structure of Hap1 shows the DNA-binding domain (DBD, purple), activation domain (AD, tan), repression modules (RPM3/1, yellow), and heme-responsive motifs (HRM1-6, blue) that bind heme and recruit Hsp90 (Hsc82, green crescent in B) to activate transcription. (B) Model for the repression and activation of Hap1. In the absence of heme (red star), Hap1 is held in an inactive conformation by Ssa proteins (Hsp70) and co-chaperones Ydj1 and Sro9 (orange) that bind to repression modules (RPM). Activation occurs when heme binds to heme-responsive motifs (particularly HRM7), causing association with Hsc82 (Hsp90, green crescent), apparently without dissociation of the repressive chaperones (Lee and Zhang 2009).
Hap1 forms subcomplexes with Hsp70 and the co-chaperones Ydj1 and Sro9 that facilitate inhibition of Hap1 (Lan et al. 2004). One possibility is that heme binds Hap1 in the inhibited complex, alters its conformation such that it can bind Hsp90, which converts Hap1 to an activation-competent state (Hon et al. 2001). The nature of the functional activator, whether free or bound to Hsp90 and the other chaperones, is unresolved, as is the location of the inhibited complex when expressed at the endogenous level.
Mal63, the activator of the MAL genes, is also a client protein of the Hsp70/Hsp90 chaperone machine (Bali et al. 2003), but two different co-chaperones, Sti1 and Aha1, facilitate inhibition (Ran et al. 2010). In the presence of maltose, Hsp90 apparently displaces the inhibitors and converts Mal63 to an active form (Ran et al. 2008). The step(s) in transcription activation that are inhibited by the chaperone complex is unclear.
Hsf1 regulation through conformational change
Yeast Hsf1 is constitutively bound to some target genes but is inactive in the absence of stress (Jakobsen and Pelham 1988). Stress, such as increased temperature or oxidative damage, induces binding to new targets (Hahn et al. 2004) and activation of Hsf1 through two differentially acting ADs, one N- and one C-terminal to the central DBD (Nieto-Sotelo et al. 1990; Sorger 1990). The essential C-terminal AD modulates the response to high temperature (Nieto-Sotelo et al. 1990; Sorger 1990) and is indispensable for response to heat stress for genes containing a single heat shock element (Hashikawa et al. 2006). The N- and C-terminal ADs activate different sets of genes in the Hsf1 regulon during heat shock (Eastmond and Nelson 2006). Stress-induced activation is accompanied by Hsf1 hyperphosphorylation, but whether it is important for activation or for return to an inactive state is unclear (Sorger 1990; Jakobsen and Pelham 1991; Hoj and Jakobsen 1994; Hashikawa et al. 2006).
The sensor for both transient and sustained response appears to be the DNA-binding and oligomerization domains of Hsf1 on the basis of heterologous fusion proteins and other studies (Bonner et al. 1994, 2000a,b). Temperature-resistant mutations near the DBD enable Hsf1 lacking its C-terminal AD to activate transcription (Hashikawa et al. 2006). These suppressors implicate the DBD in mediating the response, as did single point mutations that constitutively enhanced transcription (Bulman et al. 2001). A conformational change accompanying activation can be detected by electrophoretic mobility shift in vitro and requires two trimers of Hsf1 bound to DNA (Bonner et al. 2000a; S. Lee et al. 2000), but determining whether a similar alteration occurs in vivo is challenging. Genetic evidence suggests that repression of DNA-bound Hsf1 may be mediated by chaperones (Duina et al. 1998; Bonner et al. 2000a). Thus, mutations in Hsf1 that cause constitutive activity could alter its interactions with inhibitory chaperones.
Leu3 binding to a metabolite activates transcription
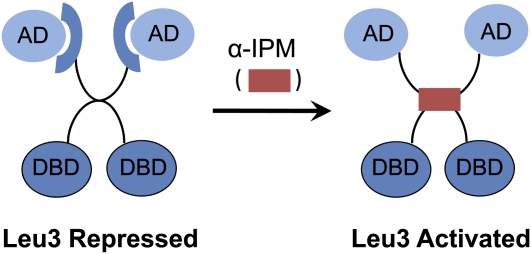
Leu3 is a zinc-knuckle TF that acts as both repressor and activator of genes encoding enzymes involved in branched-chain amino acid biosynthesis (Kohlhaw 2003) The immediate signal for up-regulation of transcription is an intermediate in the pathway, α-isopropylmalate (α-IPM), that accumulates during leucine starvation. Leu3 is apparently DNA bound in the absence of α-IPM because it represses the low level of constitutive gene expression that occurs in the presence of leucine by an unknown mechanism. ChIP experiments are needed to confirm its direct action.
As illustrated in Figure 5, internal masking of the Leu3 AD, as opposed to recruitment of a corepressor, is favored by the Kohlhaw group primarily because Leu3 is regulated by α-IPM in both mammalian cells and mammalian cell-free extracts (Wang et al. 1997, 1999; Kohlhaw 2003). This interpretation assumes that corepressors able to bind Leu3 are absent in mammalian cells. Genetic studies of mutant Leu3 activators are consistent with intramolecular as opposed to intermolecular interactions regulating the Leu3 AD (Friden et al. 1989; Wang et al. 1997, 1999). Other TFs that activate specific metabolic pathways (Arg81, Bas1, Lys14, Ppr1, Put3, War1) are also regulated by metabolites (Sellick and Reece 2005). In the case of Put3, direct binding of proline has been demonstrated (Sellick and Reece 2003).
Figure 5.
Leu3 regulation by intramolecular masking of the activation domain. In the absence of α-isopropylmalate (α-IPM), the inducing metabolite produced during leucine biosynthesis, Leu3 acts as a repressor. When α-IPM is present, Leu3 becomes an activator. Mutations in different parts of the central region either can make Leu3 a constitutive activator or inhibit Leu3 activity independently of α-−IPM. These mutations and the ability of Leu3 to be regulated by α-IPM in mammalian cell-free extracts suggest that its activation does not require another protein (Kohlhaw 2003).
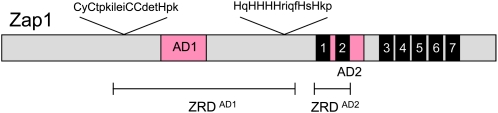
Zap1: AD regulation by Zn+2 binding
Zap1 is the TF for a regulon that responds to limiting amounts of the essential but potentially toxic metal ion Zn+2 (Eide 2009). As shown in Figure 6, Zap1 has two ADs embedded in zinc regulatory regions that function independently of one another in response to the level of Zn+2 (Bird et al. 2000). Both ADs have been highly conserved in the Hemiascomycetes, indicating that both are important for Zap1 function. Mutation of potential Zn+2 ligands in AD1 blocks the inhibition of Zap1 activity that normally occurs in response to high Zn+2 levels (Herbig et al. 2005). Purified AD1 binds multiple Zn+2 atoms, consistent with a direct role for metal binding. AD2 consists of finger 2 of the seven canonical C2H2 zinc fingers, the last five of which constitute the DBD. When Cys and His residues in zinc finger 1 and zinc finger 2 are mutated to abolish Zn+2 binding, AD2 is constitutively active, suggesting that Zn+2 binding stabilizes the interaction of zinc fingers 1 and 2 and that this interaction is essential to inhibit Zap1 AD function (Wang et al. 2006). Genetic and biochemical studies suggest a model for the structure of this pair of fingers that implicates hydrophobic residues in Zap1 regulation (Eide 2009). The coactivator targets of the Zap1 ADs and the Zap1 residues involved in the interaction have not been determined.
Figure 6.
Zap1 regulation is mediated by Zn+2. The DNA-binding domain consists of C2H2 zinc fingers 3–7 (black boxes). AD1 is within the zinc-responsive domain (ZRD) that is Cys and His rich (Herbig et al. 2005). AD2 is within finger 2 but both fingers 1 and 2 are required for zinc responsiveness (Bird et al. 2000).
In most of the examples described above, transcriptional activity is inhibited or activated by direct ligand binding. In some cases, the ligand is a protein (Gal4, Adr1, Hap1); in other cases, it is a small molecule, either a metabolite (Leu3, Put3) or a metal ion (Zap1). In the case of Hsf1, a conformational change may be ligand independent although this has yet to be directly demonstrated. It is assumed that the step in activation that is inhibited is recruitment of coactivators although this remains to be directly demonstrated. Structural studies showing how ligand binding influences the interaction of coactivators with ADs should lead to new insights into the mechanism of transcription activation.
Core Promoter Architecture
Regulatory signals from UAS and URS elements converge at the core promoter, the site where RNA Pol II and the general transcription factors assemble in the transcription preinitiation complex (PIC) before transcription initiation begins. The core promoter was first identified in mammalian gene regulatory regions and is defined as “the minimal DNA element required for basal transcription” (Smale and Kadonaga 2003). At least 60 bp of promoter DNA is occupied in the PIC, where nearly every base pair is in contact with Pol II and/or the general transcription factors (Douziech et al. 2000; Kim et al. 2000; Miller and Hahn 2006). Work with yeast has been especially important in defining different types of core promoters, the pathways of activator-stimulated PIC assembly, the structure of the PIC, and the role of chromatin structure in different classes of promoters.
Functional sequence elements in core promoters include the TATA element, initiator (INR), downstream promoter element (DPE), motif 10 element (MTE), and TFIIB recognition element (BRE) (Smale and Kadonaga 2003; Juven-Gershon and Kadonaga 2010). TATA is the recognition site for the general transcription factor TATA-binding protein (TBP), while INR, DPE, and MTE are recognition sites for the TBP-associated factors (Taf) subunits of the coactivator TFIID and BRE is a recognition site for the general factor TFIIB. All of these core promoter elements are short, degenerate, low-specificity elements. The combination of these elements varies among promoters and can determine activator and enhancer specificity.
Of these metazoan motifs, TATA is the only one clearly conserved in yeast (Basehoar et al. 2004; Sugihara et al. 2011). Since ∼90% of yeast genes are TFIID dependent (Shen et al. 2003; Huisinga and Pugh 2004), it is likely that yeast-specific TFIID recognition elements exist, but have not yet been recognized—perhaps because they are degenerate or have significantly diverged in sequence from their metazoan counterparts. Additional conserved yeast core promoter elements may be identified in future work. For example, a recent study found functionally redundant A- and T-rich sequences within the TATA-less RPS5 gene that may be recognition sites for a component of the transcription machinery (Sugihara et al. 2011).
Two promoter assembly pathways
There are two pathways for assembly of the PIC that use the coactivators TFIID or SAGA (Kuras et al. 2000; Li et al. 2000; Bryant and Ptashne 2003; Qiu et al. 2004). These coactivators contact activators at UAS elements and are responsible for recruitment of TBP to promoters. Genome-wide analysis showed that TATA-containing promoters are primarily SAGA dependent, highly regulated, and generally stress responsive (Huisinga and Pugh 2004). Only ∼19% of yeast promoters contain TATA elements, and many of these (∼10% of all yeast promoters) are dependent on SAGA coactivator function (Basehoar et al. 2004). In contrast, ∼90% of yeast promoters are primarily TFIID dependent, are usually more constitutively active, and generally lack TATAs. Efficient transcription requires coupling of compatible UAS and core promoters that recruit the appropriate coactivator. The first example of this was two elements, TC and TR, described at HIS3 (Iyer and Struhl 1995a). TR has a consensus TATA and responds to activation by Gcn4 while TC does not contain the TATA sequence and may correspond to a TFIID-binding site. TC is responsible for basal HIS3 transcription and does not respond to activation by Gcn4. Nearly all studies to define core promoter function have been done with TATA-containing promoters, but it will be important in future work to examine the mechanism of initiation at TATA-less, TFIID-dependent promoters.
Transcription start site determinants
One important difference between yeast and metazoan promoters is the site of transcription initiation with respect to TATA. In metazoan and yeast TATA-containing promoters, the PIC is assembled around the TBP–TATA complex (Douziech et al. 2000; Kim et al. 2000; Miller and Hahn 2006). From this location, the Pol II active site is positioned ∼30 bp downstream of TATA, which coincides with the metazoan transcription initiation site. In contrast, S. cerevisiae Pol II initiates transcription at preferred sequences [consensus A(Arich)5NYA(A/T)NN(Arich)6] within a window of ∼50–120 bp downstream of TATA (Hampsey 1998; Zhang and Dietrich 2005). Regulation of at least one yeast gene, IMD2, occurs by modulation of the transcription start site from a single promoter, which is regulated by intracellular guanine levels (Jenks et al. 2008; Kuehner and Brow 2008).
Nucleosome-depleted regions and noncoding transcripts
An important aspect of promoter function is the regulation of nucleosome occupancy and positioning. It was first recognized in S. cerevisiae that promoter regions were generally nucleosome depleted (Bernstein et al. 2004; Lee et al. 2004; Sekinger et al. 2005; Yuan et al. 2005), and genome-wide studies found a conserved chromatin organization at most yeast promoters (Cairns 2009; Jiang and Pugh 2009). This involves a nucleosome-depleted region within the promoter of ∼140 bp bounded by well-positioned nucleosomes termed −1 and +1. At many promoters, the presence of the nucleosome-depleted region does not correlate with transcription status and is found at active and inactive promoters. However, recent results showed that at least one of these presumed nucleosome-depleted regions at the GAL1,10 UAS contains a modified nucleosome bound by the chromatin remodeler RSC (Floer et al. 2010). This nucleosome, important in transcriptional regulation of the GAL1,10 genes, was missed in earlier studies because it occupies less than the expected 150 bp of DNA. Future studies will need to examine whether nucleosome-depleted regions are really nucleosome free or contain alternative forms of nucleosomes.
The −1 and +1 nucleosomes often contain the alternative histone Htz1 (H2A.Z) and are frequently highly acetylated (Cairns 2009; Jiang and Pugh 2009). Yeast gene regulatory regions often contain poly(A-T) sequences that disfavor nucleosome binding (Russell et al. 1983; Struhl 1985), and these are components of the nucleosome-depleted regions at many genes. Positioning of the +1 nucleosome is important because it results in ordering of neighboring nucleosomes over the open reading frame, where positioning is strongest at +1 and then becomes progressively less ordered toward the 3′ end of the gene. The +1 nucleosome may contribute to transcription start site usage. However, since initiation occurs in vitro in the absence of chromatin at many of the same sites as on chromatin templates, DNA sequences at the transcription start site seem to be the primary determinant of initiation (Ranish et al. 1999; Herbig et al. 2010). A consequence of nucleosome-depleted regions, often found at the 5′ and 3′ end of genes, is noncoding transcripts that include both divergent and antisense RNAs (Seila et al. 2009). Several recent genome-wide studies have found that nearly 75% of the yeast genome is transcribed and that most noncoding transcripts initiate from nucleosome-depleted regions associated with the 5′ or 3′ end of genes (Nagalakshmi et al. 2008; Xu et al. 2009). Recent work has shown that several of these noncoding RNAs play important regulatory roles, and more examples of this are certain to emerge in future studies (Berretta and Morillon 2009).
An exception to the uniformly nucleosome-depleted regions is found at some regulated promoters where modulation of chromatin structure is part of the gene regulatory mechanism. For example, a positioned nucleosome at PHO5 must be removed prior to gene activation because it covers a binding site for the Pho4 activator (Almer et al. 1986; Fascher et al. 1990). Expression of genes containing nucleosomes positioned within the promoter are highly dependent on recruitment of chromatin-remodeling factors by transcription activators (Cairns 2009).
Pol II Transcription Machinery and the Mechanism of Initiation
Specific transcription initiation by eukaryotic and archaeal RNA polymerases requires the general transcription factors (Hahn 2004; Thomas and Chiang 2006). These factors recognize core promoters, recruit RNA Pol into an active transcription initiation complex, and interact with coactivators and repressors to modulate transcription. Each of the three nuclear Pols (Pol I, II, and III) has its own set of general factors, sharing only TBP, which is required for transcription by all three enzymes. Many key advances in understanding the mechanism and regulation of Pol II and the general factors have been made in the yeast system. These include structure determination of Pol II in many different forms, the first isolation of genes encoding general transcription factors (TBP and TFIIA), models for assembly of the PIC, and using a combination of genetics and biochemistry to determine conserved mechanisms of gene regulation.
RNA Pol II
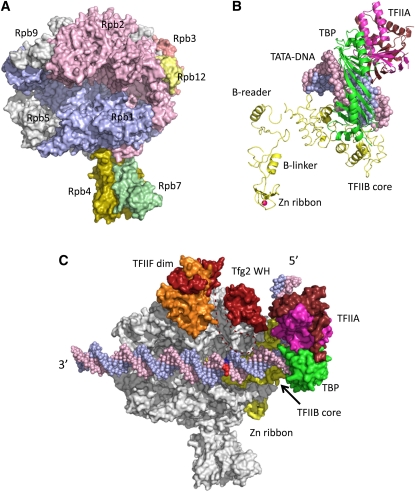
All multi-subunit RNA Pols are related in sequence and structure (Lane and Darst 2010a,b). Pol II is composed of 12 subunits, termed Rpb1–12, and numbered from largest to smallest (Cramer et al. 2008). All of these subunits except Rpb4 and Rpb9 are essential. Much of our detailed understanding about the mechanism of eukaryotic Pol II is derived from the groundbreaking structural work on yeast Pol II from the Kornberg and Cramer laboratories (Cramer et al. 2001, 2008; Gnatt et al. 2001). The structures of Pol II, either free or bound to other transcription factors or in various elongation complexes, form an invaluable framework for understanding mechanisms of gene regulation by all nuclear Pols (Figure 7A). The two largest Pol II subunits, Rpb1 and Rpb2, correspond to the β′- and β-subunits of bacterial Pol; together, these two subunits form the active site, the pore for entering nucleotide triphosphates, and the binding sites for DNA and the DNA–RNA hybrid in the transcription elongation complex. Rpb3 and Rpb11 correspond to the dimer of bacterial α-subunits, and Rpb6 corresponds to the bacterial ω-subunit, important for assembly and stability of bacterial Pol. The remainder of the Pol II subunits have no homology to bacterial subunits and are distributed around the surface of the enzyme where they perform roles in interaction with general factors, nucleic acids, and/or coactivators (Werner and Grohmann 2011). Eukaryotic Pols share 5 subunits (Rpb5, -6, -8, -10, -12) with 7 other Pol II subunits having sequence similarity with their Pol I and III counterparts. Pol I and III also have subunits that are similar to two of the Pol II general factors (Carter and Drouin 2010; Geiger et al. 2010), representing general factors that were stably incorporated into an ancestral form of Pol I and III (Werner and Grohmann 2011).
Figure 7.
(A) RNA Pol II structure. Selected subunits are labeled. (B) Model for arrangement of TBP-TFIIB-TFIIA and TATA-DNA in the PIC. Red sphere, Zn. (C) Model for structure of the yeast Pol II PIC. The DNA template and nontemplate strands are blue and pink, and the base pair where DNA melting initiates is in dark blue and red; the TFIIF large and small subunits (Tfg1 and Tfg2) are orange and red; the TBP conserved domain (TBP) is green; TFIIB is yellow; and the large and small TFIIA subunits (Toa1 and Toa2) are brown and magenta. TFIIF dim, TFIIF dimerization domain; Tfg2 WH, Tfg2 winged helix domain; Zn ribbon, TFIIB N-terminal Zn ribbon domain. Dashed line represents a Tfg2 loop connecting the dimerization and winged helix domains. The 5′ and 3′ ends of the noncoding strand of promoter DNA are indicated.
Pol II is unique among the multi-subunit Pols in containing a repeated seven-residue motif (YSPTSTS) at the C terminus of Rpb1, termed the C-terminal domain (CTD) (Buratowski 2009). The unstructured CTD plays a role in assembly of the PIC, functionally interacting with the coactivator Mediator, and it is a target of several kinases that phosphorylate Ser at positions 2, 5, and 7 of the repeat. Pol II with a nonphosphorylated CTD is preferentially assembled in the PIC and then phosphorylated at Ser5 and -7 during initiation, principally by Kin28/Cdk7, a subunit of the general factor TFIIH (Feaver et al. 1994; Akhtar et al. 2009), although Srb10/Cdk9 can functionally substitute upon inhibition of Kin28 (Liu et al. 2004; Kanin et al. 2007). Upon transition of Pol II to the elongating mode, Ctk1 and Bur1, both related to the mammalian kinase Cdk9, phosphorylate the CTD at Ser2 (Buratowski 2009; Liu et al. 2009; Qiu et al. 2009; Zhou et al. 2009). The levels of CTD phosphorylation are precisely modulated during initiation, elongation, and termination, and these modifications function in regulating the association of many important factors with elongating Pol, including mRNA capping factors, chromatin modifiers, mRNA export, and transcription termination factors.
General transcription factors and PIC assembly
The human Pol II general factors were discovered as factors essential for specific transcription initiation from a TATA-containing core promoter (Matsui et al. 1980). A subset of the general factors recognize promoter DNA and form an asymmetric platform for the binding of Pol II and the incorporation of the remaining general factors (Buratowski et al. 1989). These factors include TBP, TFIIA, and TFIIB, all of which form a complex with promoter DNA (Figure 7B). As described below, TFIIB directly contacts Pol II and, along with TFIIF, is critical for Pol II recruitment, initiation activity, and start site recognition. The remaining factors, TFIIE and TFIIH, play key roles in separation and stabilization of promoter DNA strands during transition of the transcription machinery into the active open complex state. Several major advances have been made in understanding the function of the general factors since recent extensive reviews (Hahn 2004; Thomas and Chiang 2006), and these are summarized below.
TFIIB is a key component in assembly of the PIC, contacting TBP, DNA, and Pol II. TFIIB consists of an N-terminal Zn ribbon domain that contacts the Pol II dock domain, an unstructured segment termed the B-reader and linker regions, and two cyclin folds that form the TFIIB core domain, binding TBP and DNA on either side of the TATA (Hahn 2004; Thomas and Chiang 2006). Positioning of TFIIB on Pol II has been visualized by site-directed crosslinking and targeted hydroxyl radical cleavage assays and by several X-ray structures of the Pol II–TFIIB complex (Chen and Hahn 2003, 2004; Bushnell et al. 2004; Kostrewa et al. 2009; Liu et al. 2010). Two recent structures of Pol II–TFIIB show the B-reader and linker regions near two critical regions in the Pol II active site. In the complex, a helical portion of the B-reader is in position to interact with single-stranded DNA in the open complex state and is proposed to “read” the DNA sequence, contributing to start site recognition (Kostrewa et al. 2009). A helical portion of the B-linker region in the complex is positioned near the presumed site of DNA strand unwinding and proposed to contribute to DNA melting (Kostrewa et al. 2009; Liu et al. 2010). As first shown by site-specific biochemical probes (Chen and Hahn 2004; Chen et al. 2007), the N-terminal cyclin fold (core) of TFIIB binds the Pol II wall domain where it functions to position the TBP–DNA complex over the Pol II active site cleft (Figure 7C). This positioning is the key to setting the architecture of the PIC and to positioning promoter DNA directly over the Pol II cleft (Miller and Hahn 2006).
Yeast TFIIF is composed of two conserved subunits, Tfg1 and Tfg2, as well as a third nonconserved subunit that is a component of several other complexes involved in gene regulation (Henry et al. 1994). TFIIF is involved in stabilizing Pol II in the PIC, setting the transcription start site, stabilizing the RNA–DNA hybrid in early elongation complexes, and stimulating transcription elongation in vitro (Hahn 2004; Thomas and Chiang 2006). Two structured domains of TFIIF, the dimerization domain and the Tfg2 winged helix domain, are located on two separate sites on the Rpb2 surface above the active site cleft (Figure 7C) (Eichner et al. 2010; A. Chen et al. 2010). The winged helix domain is in position to contact DNA upstream of the TATA and bend it over the top of Pol II, possibly contributing to stabilization of the PIC. These two TFIIF domains are connected by an essential nonstructured linker that may play a direct role in initiation (Eichner et al. 2010).
Open complex formation and transcription initiation
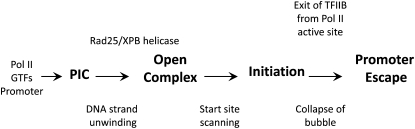
Transition of the PIC into the open complex involves a dramatic conformational change (Murakami and Darst 2003) requiring insertion of double-stranded promoter DNA into the jaw and downstream cleft of Pol II, the TFIIH helicase-dependent separation of DNA strands surrounding the transcription start site from ∼ −9 to +1 (with respect to the transcription start) (Wang et al. 1992; Holstege et al. 1997; Revyakin et al. 2004), and insertion of the single-stranded DNA template strand into the active site of Pol II. This step can be highly regulated in bacteria and the mechanism of open complex formation is one of the major unanswered questions for all multi-subunit RNA polymerases (Figure 8).
Figure 8.
Steps in the pathway of yeast Pol II transcription initiation.
Recently, the yeast PIC structure model was merged with the structure of yeast-elongating Pol II to generate the first structural model of the open complex (Kostrewa et al. 2009; Liu et al. 2010). In this model, there are 14 unwound bases with the TFIIB B-reader segment interacting directly with single-stranded DNA upstream of the transcription start site (Kostrewa et al. 2009). Future advances in understanding the open complex will emerge from structural and biochemical studies examining other general factors situated within the enzyme active site and determining how they interact with each other and DNA to promote start site selection, initiation, and the initial steps of elongation.
Transcription start site scanning
Yeast Pol II scans downstream sequences for a suitable transcription start (Giardina and Lis 1993; Kuehner and Brow 2006; Steinmetz et al. 2006). Consistent with this, DNA in vivo at the GAL1 and GAL10 promoters is unwound from ∼20 bp downstream of TATA (the approximate site of initial strand unwinding for metazoan Pol II) through the transcription start ∼90 bp distant from the TATA (Giardina and Lis 1993). This scanning mechanism does not involve transcription of the DNA between the TATA and initiation site (Khaperskyy et al. 2008). Since the cost of disrupting a DNA base pair is ∼2 kcal/mol, there is a significant energetic cost of unwinding 10–70 bp. A reasonable model to explain these findings (Miller and Hahn 2006) is that the yeast-scanning mechanism involves strand unwinding and DNA translocation promoted by the TFIIH Rad25/XPB helicase using the energy of ATP hydrolysis (Figure 8). There is no evidence yet to determine whether the DNA between the initial melting site and the transcription start site is unwound all together or whether an ∼10-bp bubble translocates downstream. Answering this question will likely require single molecule studies of the initiation reaction.
Pioneering genetic experiments by Hampsey, as well as later work, suggested that TFIIB, TFIIF, and Pol II are all involved in start site selection since mutations in any of these factors can alter the normal start site distribution (Hampsey 1998; Faitar et al. 2001; Ghazy et al. 2004; Chen et al. 2007). Mutations in both the TFIIB B-reader and the switch 2 segment of Rpb1 cause transcription to start farther away from TATA but still at sequences matching the initiator consensus (Faitar et al. 2001; Kostrewa et al. 2009). These mutations act as though they decrease the efficiency of initiator recognition. In contrast, mutations in the TFIIF dimerization domain or in two subunits of Pol II that interact with TFIIF, Rpb2, and Rpb9, start transcription closer to TATA than in wild-type cells and behave as though they have relaxed specificity for initiator recognition (Khaperskyy et al. 2008). The study of this mechanistic step in S. cerevisiae will undoubtedly reveal important details about start site selection in other eukaryotes.
Transcription Coactivators
Transcription activation is one of the most important mechanisms for control of gene regulation and is a common endpoint for many signaling pathways, including those controlling cell growth and the response to environmental or metabolic stress. The principal targets of activators are coactivators, large protein complexes that enhance activated transcription by direct contact with the basal transcription machinery and/or by chromatin modification. These two activities cooperate to stimulate PIC formation, leading to increased transcription. Although much progress has been made in defining coactivators and their mechanism of action, there is much to be learned about the architecture of coactivator complexes and how they interact with the transcription machinery and integrate signals to modulate transcription. The coactivators discussed below are conserved in eukaryotes, and several of the coactivators (Mediator, SAGA, NuA4) were first discovered in yeast. The yeast system has often led the way in understanding the nature and mechanism of coactivators and how these complexes function in other eukaryotes.
Mediator
Mediator is a 25-subunit complex that functions as an intermediate between transcription regulators and the general transcription factors (Biddick and Young 2005; Bjorklund and Gustafsson 2005). All yeast Mediator subunits have homologs in insects and mammals, and a common nomenclature has been developed for Mediator subunits (Bourbon et al. 2004; Bourbon 2008). A set of 17 mediator subunits is conserved in all eukaryotes and forms a core for assembly of other organism-specific subunits (Bourbon 2008). Human Mediator is more complex than its yeast counterpart, containing additional subunits, and exists in multiple forms with variable subunit composition (Conaway et al. 2005).
Mediator binds transcription activation domains and Pol II, allowing activator-dependent Pol II recruitment (Bjorklund and Gustafsson 2005; Malik and Roeder 2005), but its role in gene regulation is much more complex than simply linking activators and polymerase. Mediator also stimulates basal transcription, at least in part, by stabilization of the PIC and by stimulation of TFIIH-dependent Pol II CTD phosphorylation. Mediator can positively or negatively affect transcription and seems to cooperatively interact with other coactivators (Bryant and Ptashne 2003; X. Liu et al. 2008). Finally, it has been proposed that Mediator may be a direct target of signaling pathways, although this remains to be firmly established (Malik and Roeder 2005; Taatjes 2010). Because of these diverse roles, Mediator is thought to be a major target of transcriptional regulatory signals that are integrated and transmitted to the transcription machinery in a promoter and gene-specific manner.
Mediator was discovered in yeast in a classic example of biochemistry and genetics converging to identify the same factor, with the sum of functional insights greater than could be revealed by either approach alone. In a biochemical tour de force, Roger Kornberg’s laboratory developed a yeast basal transcription system using purified general factors and Pol II that did not respond to activators. A factor that they termed Mediator was purified that stimulated basal transcription and allowed stimulation by the activators Gal4-VP16 and Gcn4 (Flanagan et al. 1991). At the same time, in an elegant series of genetic experiments, Rick Young’s lab isolated suppressors of cold-sensitive yeast mutants containing a shortened Pol II CTD. Young’s group found that these Srb proteins (Suppressor of RNA polymerase B) copurified with Pol II and were identical to some of the Mediator subunits (Thompson et al. 1993). Further comparison of the Mediator subunits with other regulatory genes showed that many of the Mediator subunits had been identified in genetic screens for defects in specific regulatory pathways [Table 1 in Biddick and Young 2005; (Bjorklund and Gustafsson 2005)].
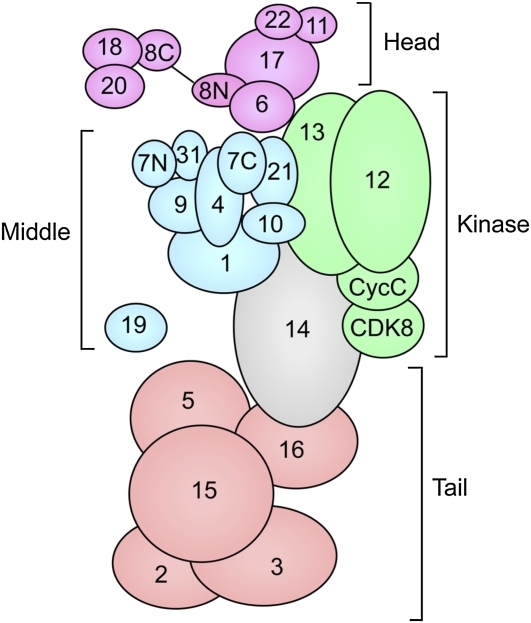
Structural organization of Mediator
Mediator can be subdivided into four distinct modules: head, middle, tail, and kinase (Figure 9) (Davis et al. 2002; Cai et al. 2009). Extensive biochemical, genetic, and two-hybrid studies have mapped protein–protein interactions and the four modules within Mediator (Kang et al. 2001; Guglielmi et al. 2004). Electron microscopy revealed that the modules form separate domains that are highly flexible with respect to each other (Davis et al. 2002; Cai et al. 2009). Well-diffracting crystals from Mediator modules have not yet been obtained because of the flexibility of Mediator (Toth-Petroczy et al. 2008). Sequence analysis predicts that many Mediator subunits are composed of protein–protein interaction motifs connected by large inherently unstructured segments. To date, X-ray and NMR structures have been determined for five relatively small and well-ordered Mediator domains (Baumli et al. 2005; Hoeppner et al. 2005; Lariviere et al. 2006; Koschubs et al. 2009; Thakur et al. 2009).
Figure 9.
Organization of yeast Mediator and Mediator modules. Numbers indicate the Mediator subunit name. Head module, purple; middle module, blue; kinase module, green; tail module, orange. Med14 (Rgr1), in gray, is at the interface of the middle, kinase, and tail modules. Adapted from Koschubs, T., K. Lorenzen, S. Baumli, S. Sandstrom, A. J. Heck et al., 2010, Preparation and topology of the Mediator middle module. Nucleic Acids Res. 38: 3186–3195; by permission of Oxford University Press.
The head module can be reconstituted upon coexpression of its seven individual subunits, Med6, -8, -11, -17, -18, -20, and -22 (Takagi et al. 2006; Lariviere et al. 2008; Cai et al. 2010). The N terminus of Med8 is reported to bind TBP (Lariviere et al. 2006), but it has not yet been shown if this interaction occurs in functional transcription complexes. From electron microscopy (EM) studies, the head module seems to closely interact with the back surface of Pol II, closely approaching the subunits Rpb3/11 and the protruding Rpb4/7 subunits that modulate closing of the Pol II cleft (Davis et al. 2002; Cai et al. 2010). Recent studies suggest that the head module binds weakly to a minimal PIC consisting of Pol II, TFIIF, TFIIB, TBP, and promoter DNA (Takagi et al. 2006). Mediator does not appear to bind directly to the Pol II CTD, so the molecular basis by which Srb mutations were originally isolated is still unclear; most of the Srb subunits are located within the head module.
The middle module is composed of eight to nine subunits with Med14/Rgr1 connecting the middle and tail modules. A seven-subunit recombinant complex lacking Med19 and Med14 was analyzed using protein biochemistry (Koschubs et al. 2010), revealing that the middle module is elongated and highly flexible. Part of this is likely due to the Med7/21 interface composed of a four-helix bundle and a flexible protrusion connected by a flexible linker (Baumli et al. 2005). This structure is very elongated, nearly one-third the length of Mediator, and the linker may contribute to Mediator conformational changes upon binding Pol II.
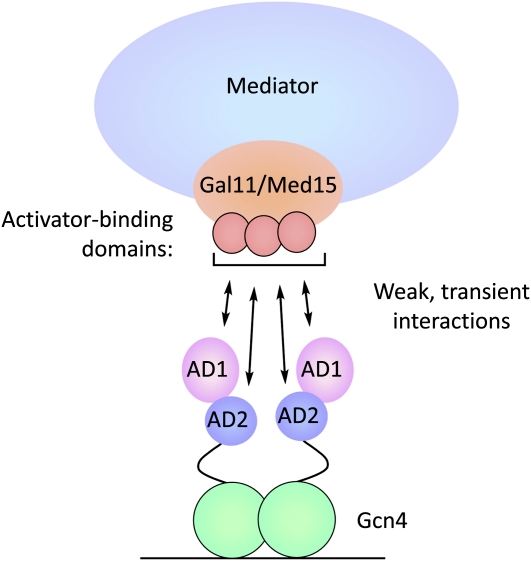
The tail module is composed of four to five subunits and is a target of at least several transcriptional activators (Kang et al. 2001). The subunits Med2, -3, and -15 (Gal11) form a submodule that can be recruited in vivo to a DNA-bound activator in a strain containing a MED16/SIN4 deletion (Zhang et al. 2004). Mutation of any one of these subunits disrupts this submodule. The best-characterized tail subunit is Med15/Gal11, containing four N-terminal activator-binding domains separated by glutamine or glutamine–asparagine-rich flexible linkers (Herbig et al. 2010; Jedidi et al. 2010).
The kinase module is composed of four subunits including Cdk8 and cyclinC (Liao et al. 1995). This module has both positive and negative effects on expression (Bjorklund and Gustafsson 2005; van de Peppel et al. 2005; Taatjes 2010). Early work showed that Cdk8 levels are reduced as cells reach stationary phase, leading to expression of genes induced by nutrient deprivation (Holstege et al. 1998). Studies with the human Mediator suggest that the kinase module inhibits a Mediator conformational change that opens up a pocket containing the Pol II-binding surface (Taatjes 2010). Conversely, it was demonstrated that Cdk8 can act positively to promote initiation and CTD phosphorylation in the absence of Cdk7 kinase activity (Liu et al. 2004), and studies in human cells showed that Cdk8 activity stimulates transcription of genes in the serum response network (Donner et al. 2010).
Mediator targets
Mediator gene targets are still somewhat controversial. Pioneering genome-wide studies using a temperature-sensitive MED17 allele showed that transcription of nearly all Pol II-transcribed genes was rapidly shut down upon heat shock (Holstege et al. 1998), a powerful argument for direct action of Mediator at all Pol II genes. Genome-wide ChIP studies later reported that Mediator is located in control regions of nearly all Pol II-transcribed genes (Andrau et al. 2006). This finding was challenged by results suggesting that Mediator strongly crosslinked to only a small subset of genes in cells grown in rich media and that the original ChIP analysis was flawed due to low signal-to-noise ratios (Fan et al. 2006; Fan and Struhl 2009). Also puzzling are the findings that Mediator nearly always crosslinks to the UAS element rather than to the promoter, where it is expected to interact with Pol II. These results may be explained in part by inefficient crosslinking of Mediator to promoter DNA, a result expected if Mediator is located several interactions away from proteins that directly bind promoter DNA.
In contrast to the model that Mediator is a simple link between activators and Pol II, mutations in the nonessential Mediator subunits result in both positive and negative effects on mRNA expression. This was first observed when a mutation in MED16/RGR1 was found to increase expression of HO in the absence of its activator Swi5 (Stillman et al. 1994). These effects were examined systematically using microarray analysis to study genome-wide changes in mRNA levels upon deletion of the nonessential Mediator subunits (Holstege et al. 1998; van de Peppel et al. 2005). Although mutation of each of these subunits has both positive and negative effects, elimination of some subunits predominantly increases gene expression while elimination of others generally decreases expression. For example, deletion of MED19 or kinase module subunits results in increased expression from a large set of genes. Conversely, mutation of tail or head subunits results in predominantly decreased gene expression. Mutation of middle module subunits falls somewhere in between. Although not yet understood, these complex phenotypes may be due to changes in Mediator that preclude or enhance interaction with specific transcription factors or the general transcription machinery in a promoter-specific fashion. Understanding these mechanisms in more detail will greatly help in understanding how Mediator integrates inputs from various signaling pathways and why Mediator is so complex.
TFIID
TFIID is composed of TBP and 14 Tafs (Matangkasombut et al. 2004; Cler et al. 2009). Thirteen of the yeast TAFs are conserved in eukaryotes, with one yeast-specific Taf (Taf14) a subunit of other multi-subunit complexes involved in transcriptional regulation such as TFIIF, INO80, and SWI/SNF (Hahn 2004). The strong sequence conservation of Tafs has allowed a unified Taf nomenclature for all eukaryotes (Tafs 1–14) (Tora 2002). Although TBP is sufficient to promote basal transcription from a TATA-containing promoter when combined with purified Pol II and the other general transcription factors, transcription from TATA-less promoters and, in many cases, response to activators requires the TFIID complex.
The subunit composition of TFIID was first revealed in human and Drosophila cells, where TBP is tightly associated with Tafs (Dynlacht et al. 1991; Kokubo et al. 1993). Yeast TBP is not as stably bound to the Tafs compared to other eukaryotes, explaining why yeast TBP was originally purified as a single polypeptide (Buratowski et al. 1988). Yeast Tafs were isolated later using streamlined and gentler purification methods such as TBP affinity columns or immune purification of TBP (Reese et al. 1994; Poon et al. 1995). Depletion of yeast Tafs from whole-cell extracts abolished activation by the strong heterologous activator Gal4-VP16, and purified yeast TFIID allowed modest transcription stimulation by activators using purified general factors and Pol II (Reese et al. 1994; Poon et al. 1995). Yeast Tafs are encoded by single-copy genes, while several human and Drosophila Tafs are encoded by multiple genes. In these more complex systems, one Taf allele is typically expressed only in specific cell types, contributing to tissue and developmental-specific gene expression (D’Alessio et al. 2009).
TFIID structure and TBP–DNA binding
Nine of the 13 conserved Taf subunits contain histone fold domains (HFDs), and biochemical and structural analysis showed that these domains are used for dimerization of specific Taf pairs: Tafs 6–9, 11–13, 8–10, 3–10, and 4–12 (Cler et al. 2009). These Tafs are all thought to be present in at least two copies per TFIID complex. The coactivator SAGA contains a related subset of these HFD Tafs except that Taf4–12 has been replaced by Ada1-Taf12; Taf13–11 has been replaced by Spt3, which contains two HFDs; and Taf3–10 has been replaced by Spt7-Taf10. Whether these HFD Tafs form related structural modules in TFIID and SAGA is not yet known.
EM studies showed that TFIID is a large flexible complex and that human and yeast TFIIDs have very similar structures (Andel et al. 1999; Leurent et al. 2002). TFIID is composed of three linked lobes termed A, B, and C, which form a “horseshoe”-shaped structure (Figure 10) (W. L. Liu et al. 2008; Papai et al. 2009). TFIID is assembled around two molecules of the WD-40 repeat-containing Taf5, which binds the other Taf HFD pairs to form a core crescent-shaped structure consisting of TFIID lobes B, C, and the lower part of lobe A (Cler et al. 2009). Although this subunit arrangement predicts a symmetric core domain built around the Taf5 dimer, localization studies and biochemical analysis suggest that the core is only pseudosymmetric.
Figure 10.
Organization of yeast TFIID. Numbers indicate the Taf subunit name and the three lobes observed in EM are shown. Adapted from Cler, E., G. Papai, P. Schultz, and I. Davidson, 2009, Recent advances in understanding the structure and function of general transcription factor TFIID. Cell. Mol. Life Sci. 66: 2123–2134; with kind permission from Springer Science+Business Media B.V.
Tafs 1, -2, and -7, along with TBP, bind to one lobe of the core domain, forming the complete TFIID complex. In this arrangement, TBP is positioned within the center of the horseshoe-shaped cleft. It is not yet clear how or if TBP interacts with DNA when TFIID binds DNA. TBP binding to TATA requires that TBP open the DNA minor groove, interacting closely with the hydrophobic surface of the groove through a complementary hydrophobic surface on the underside of TBP (J. L. Kim et al. 1993; Y. Kim 1993). Due to steric constraints, this binding mechanism is compatible only with the TATA sequence, and substitution of G-C at key positions within TATA causes a severe decrease in DNA-binding affinity (Patikoglou et al. 1999). This raises the question of how TFIID and TBP specifically interact with promoter DNA at TATA-less promoters. Initial in vitro experiments showed that a human TBP mutation that compromised TBP-TATA binding decreased TBP-driven transcription but not TFIID-driven transcription from a TATA-less promoter (Martinez et al. 1995). This suggests that if TBP interacts with DNA at a TATA-less promoter, then it does so by a different mechanism compared to TBP-TATA binding.
An additional complication for TBP-DNA binding within TFIID is due to the TAND (Taf1 N-terminal domain) domain of Taf1. TAND tightly binds the DNA-binding surface of TBP and is likely a major contributor to the stability of TBP within the TFIID complex (Liu et al. 1998), but the TAND must be removed from TBP to allow specific DNA binding. Repression by the TAND domain can be overcome by TFIIA, which competes with TAND for TBP binding (Kokubo et al. 1998). In one study, TFIID binding to the RPS5 promoter was observed only in the presence of TFIIA (Sanders et al. 2002), consistent with the competition of TFIIA and the TAND domain for TBP.
Higher eukaryotic Taf1 contains several domains with enzymatic activity: separate N- and C-terminal protein kinase activities, histone acetyl transferase activity, and double bromo and PhD domains, the latter two interacting with acetylated and methylated histones, respectively (Matangkasombut et al. 2004). Yeast Taf1 was reported to have histone acetyl transferases (HAT) activity, although much weaker than its human counterpart (Mizzen et al. 1996), and it is not clear whether this HAT activity is functionally important in yeast. The protein kinase activities and PhD domains are not conserved between human and yeast Taf1. A yeast factor containing a double bromodomain, Bdf1, is associated with TFIID and likely represents the acetyl lysine-binding activity functionally analogous to human Taf1 (Matangkasombut et al. 2000). It is thought that interaction of the Taf1 bromodomain with doubly acetylated histone H4 flanking the promoter stabilizes TFIID binding, contributing to the stability of the PIC.
Role of Tafs in gene expression
The yeast system has led the way in understanding the in vivo role of Tafs. Experiments in yeast have shown that all of the conserved Tafs are essential for growth (Reese et al. 1994; Poon et al. 1995). To examine the role of Tafs in expression, several Tafs were initially depleted from growing cells using temperature-sensitive mutations, expression of Tafs from repressible promoters, and/or controlled protein degradation. In surprising contrast to the prevailing view at the time, most genes assayed were expressed normally upon Taf depletion (Moqtaderi et al. 1996; Walker et al. 1996). These studies continued over the next several years until microarrays were used to systematically assay genome-wide expression upon depletion of all 14 individual Tafs (Shen et al. 2003).
Analysis of genome-wide Taf function revealed that there are two types of Pol II promoters: TFIID dependent and TFIID independent (Kuras et al. 2000; Li et al. 2000; Basehoar et al. 2004; Huisinga and Pugh 2004). In cells grown in rich media, 84% of the genes assayed required at least one Taf, and 16% of genes were completely Taf independent. TFIID and SAGA share a subset of Tafs, and the most broadly required Tafs are those shared by both coactivator complexes (Shen et al. 2003).
Some of the first ChIP studies were designed to examine the mechanism of TBP recruitment and Taf dependence. At most promoters, there is an excellent correlation between TBP binding and transcription, with most nonexpressed genes showing low levels of TBP crosslinked to promoters and an increase in TBP crosslinking upon gene activation (Kuras and Struhl 1999; Li et al. 1999). The Taf dependence of promoters also corresponds well to whether Tafs are recruited to promoters. TFIID Taf-dependent promoters have a high ratio of crosslinked TFIID Taf/TBP while TFIID Taf-independent promoters have a much lower ratio (Kuras et al. 2000; Li et al. 2000). An unexpected finding from these studies was that upon TBP depletion, TFIID Tafs were still crosslinked to regulatory regions upon gene induction, although recruitment of the remaining general factors and Pol II was abolished (Li et al. 2000). This result suggests that the TFIID Tafs are targeted by activator, which agrees with results from humans, Drosophila, and yeast that activators can interact with TFIID subunits.
Several studies have examined which promoter elements are involved in determining TFIID dependence by swapping UAS and core promoters from TFIID-dependent or -independent genes (Shen and Green 1997; Cheng et al. 2002; Li et al. 2002). These experiments have sometimes given conflicting results with TFIID Taf dependence tracking with either the UAS or the core promoter. The most recent and comprehensive study examined the RPS5 (TFIID-dependent) and GAL1 and ADH1 (TFIID-independent) promoters (Li et al. 2002). Measurement of transcription and TFIID recruitment at these chimeric promoters showed that efficient transcription requires compatible UAS and core promoter elements. For example, a TFIID-independent UAS, e.g., GAL1, works much better with a TFIID-independent promoter. This is presumably because Gal4 does not efficiently target TFIID, but rather the SAGA coactivator. In contrast, the RPS5 UAS could activate, although with a lowered efficiency, at a TFIID-independent core promoter.
SAGA
The coactivator SAGA modulates expression of many inducible genes that are generally distinct from genes regulated by TFIID (T. I. Lee et al. 2000; Huisinga and Pugh 2004). SAGA mutations alter expression of ∼10% of yeast genes, and these are usually TATA-containing, stress-regulated, and highly inducible. SAGA is targeted by gene-specific activators and functions through covalent modification of chromatin and by direct contact and recruitment of TBP. SAGA is a 1.8-mDa complex composed of 20 subunits (Baker and Grant 2007), is conserved in eukaryotes, and is orthologous to the mammalian coactivators TFTC, PCAF, and STAGA (Lee and Workman 2007). Like Mediator, SAGA was discovered in yeast using a combination of genetic and biochemical assays. Two SAGA subunits, Spt3 and Spt8, were discovered in a screen for suppression of Ty element insertions (Winston et al. 1984; Eisenmann et al. 1994). Several other subunits (Ada1, -2, -3) were found in a genetic screen for genes, which, when mutated, suppressed the toxic effects of an overexpressed transcription activator (Berger et al. 1992). A complex containing these and additional subunits was found in biochemical studies aimed at isolating HATs in yeast whole-cell extracts (Grant et al. 1997). Many of the yeast HAT complexes bind Ni Sepharose, and fractionation of these Ni-purified HATs led to isolation of the coactivator complexes SAGA and NuA4 containing the HATs Gcn5 and Esa1, respectively. SAGA preferentially acetylates histone H3 while NuA4 acetylates histone H4.
SAGA organization and TBP binding
Biochemical, genetic, structural, and recent mass spectrometry analysis has shown that SAGA is composed of at least five modules: (1) a core/Spt module important for SAGA integrity and TBP-binding containing Spt7, Ada1, Spt20, and the TBP-binding subunits Spt3 and Spt8 (Eisenmann et al. 1992; Grant et al. 1997, 1998a; Lee and Young 1998; Dudley et al. 1999; Sterner et al. 1999; Bhaumik and Green 2002; Wu and Winston 2002; Mohibullah and Hahn 2008; Lee et al. 2011); (2) a HAT module containing the HAT Gcn5, Sgf29, and the subunits Ada2 and Ada3, which modulate substrate specificity (Grant et al. 1999; Balasubramanian et al. 2002; M. Washburn and J. Workman, personal communication); (3) a four-subunit ubiquitin protease module (Rodriguez-Navarro et al. 2004; Ingvarsdottir et al. 2005; Lee et al. 2005; Shukla et al. 2006; Kohler et al. 2008); (4) Tra1, an activator-binding module (Brown et al. 2001); and (5) a Taf module (Lee et al. 2011). The subunits Ada1 and Taf12 are thought to dimerize via histone fold domains, and SAGA appears to contain two molecules of Ada1 located within distinct regions of the complex. EM studies suggest that most of the modules are located at separate positions within SAGA (Wu et al. 2004). The distribution of the Tra1, HAT, and TBP-binding modules to different regions suggests that SAGA is composed of submodules that physically associate via a central core. The SLIK complex regulates expression from a small set of genes and is closely related to SAGA (Pray-Grant et al. 2002). SLIK contains a proteolytically processed Spt7 subunit and the subunit Rtg1 in place of Spt8 (Wu and Winston 2002).
Like Mediator, SAGA has a complex role in gene regulation since mutation of SAGA subunits can either increase or decrease gene expression. For example, genome-wide characterization of Gcn5-responsive genes reveals that Gcn5 functions as both a coactivator and a corepressor in S. cerevisiae and Schizosaccaromyces pombe (Xue-Franzen et al. 2010). Gene expression studies have also shown that Gcn5 and subunits of the SAGA TBP-binding module have opposing roles; S. cerevisiae SPT3 and GCN5 mutations were found to have opposite effects in transcriptional regulation of the HO and STE11 genes (Yu et al. 2003; Helmlinger et al. 2008), although it is possible that some of these phenotypes are due to indirect effects. SAGA can also repress basal expression of some genes in vitro and in vivo (Belotserkovskaya et al. 2000; Warfield et al. 2004).
Although both TFIID and SAGA share a subset of Tafs and function to recruit TBP, they have very different biochemical activities in TBP binding. TFIID stably binds TBP under certain conditions, while purified SAGA contains little TBP (Sterner et al. 1999; Sanders et al. 2002; Laprade et al. 2007). Genetic and biochemical studies implicate the subunits Spt3 and Spt8 in TBP binding. These Spt subunits genetically interact with TBP, and mutations in Spt3 can suppress TBP mutations (Eisenmann et al. 1992, 1994; Laprade et al. 2007). In one case, an Spt3 mutation was found to dramatically increase the amount of TBP copurifying with SAGA (Laprade et al. 2007). Structure modeling strongly suggests that the N- and C-terminal ends of Spt3 interact via a noncanonical histone fold domain, homologous to the Taf 11–13 structure (Birck et al. 1998). Mutations that result in an Spt3 phenotype lie primarily along one face of this model. (Eisenmann et al. 1992; Laprade et al. 2007). These mutations are presumably altered in their interaction with TBP because several can be suppressed in an allele-specific fashion by mutations in TBP. However, the interaction of SAGA and TBP is complex because mutations in Spt3 and Spt8 can have both positive and negative effects on gene expression (Yu et al. 2003; Helmlinger et al. 2008). Both Spt3 and Spt8 crosslink to TBP in PICs (Mohibullah and Hahn 2008), and mutations in Spt3 and Spt8 show defects in TBP recruitment in vivo (Dudley et al. 1999; Larschan and Winston 2001; Bhaumik and Green 2002; Barbaric et al. 2003; Mohibullah and Hahn 2008). Supporting the direct interaction of Spt3 and TBP is the finding that the site of TBP-Spt3 crosslinking lies very close to TBP mutations that suppress mutations in Spt3. However, the mechanism of TBP-Spt3 binding is not well understood because purified TBP and Spt3 do not interact (Madison and Winston 1997; Sterner et al. 1999; Sermwittayawong and Tan 2006).
Tra1 has multiple functions within SAGA and NuA4
The largest SAGA subunit is Tra1, a nearly 4000-residue essential protein that is shared with the NuA4 complex (Grant et al. 1998b; Saleh et al. 1998; Allard et al. 1999). Tra1 and its human homolog, TRRAP, are members of the PI3-related protein kinase family, but Tra1 and TRRAP have specifically lost kinase activity (Mutiu et al. 2007). Biochemical and genetic experiments showed that several activators, including Gcn4 and Gal4, interact with Tra1 and that this interaction is important for activated transcription of SAGA-dependent genes (Brown et al. 2001; Fishburn et al. 2005; Reeves and Hahn 2005). Tra1 is also likely responsible for activator recruitment of the NuA4 coactivator. Most of Tra1 is composed of short repeated motifs, including HEAT and TPR repeats, which are N-terminal to the PI3 kinase-like domain (Knutson and Hahn 2011). Systematic mutation of Tra1 has shown that about two-thirds of the protein is essential for growth. All lethal Tra1 mutations isolated to date abolish association of both SAGA and NuA4 subunits, showing that Tra1 uses identical regions to contact the subunits of both complexes. Nonlethal Tra1 mutations fall into three classes: (1) defective in activator-dependent promoter recruitment, (2) defective in HAT module recruitment, and (3) normal for HAT recruitment but defective for in vivo HAT activity. These latter two categories show that Tra1 is important for stability of the NuA4 and SAGA HAT modules as well as somehow being involved in activity or specificity of the HAT.
Although there has been significant progress on understanding function, there is not much known about how SAGA is regulated, organized, and interfaces with activators and the transcription machinery. Important areas for future work are (1) understanding the architecture of SAGA and the organization and functional relationships of the different SAGA modules; (2) understanding the mechanism of how SAGA interfaces with TBP, whether this binding is regulated, and if SAGA–TBP interaction is limiting at promoters; and (3) the mechanism of activator–Tra1 interaction.
Transcription Activation Mechanisms
Over the past 30 years, the yeast system has been used to answer a number of fundamental questions that are important for understanding gene control in all eukaryotes: (1) What mechanisms result in transcription stimulation? (2) How is formation of transcription preinitiation complexes regulated and what are the roles of coactivators in this process? (3) What is the nature of activation domains and what are their direct targets? and (4) How do activators interact with targets and are these interactions specific? Yeast has been an especially powerful system to dissect gene regulatory mechanisms and, in many cases, has led the way in understanding the fundamental mechanisms of transcriptional regulation.
In theory, transcription could be modulated by a number of different mechanisms (Hahn 1998; Keaveney and Struhl 1998) including (1) recruitment of coactivators and general transcription factors to promoters, (2) conformational changes in the transcription machinery leading to increased activity, (3) modification of chromatin structure by ATP-dependent remodelers or through covalent nucleosome modifications, and (4) by enhancing steps that occur after preinitiation complex formation. Each of these steps plays a role in eukaryotic regulation, although it is not yet clear if all of these mechanisms are used in yeast.
Activation by recruitment
Activation by recruitment is one of the best-studied regulatory mechanisms (Ptashne and Gann 2002), and there is overwhelming evidence that this is a major, but not the only, means of transcription stimulation in eukaryotes. Early support for the recruitment model was the finding in yeast of activation by “artificial recruitment” (Chatterjee and Struhl 1995; Klages and Strubin 1995; Xiao et al. 1995), where targeting an appropriate coactivator subunit or general transcription factor to a promoter by fusion to a DBD dramatically enhances transcription. The first artificial recruitment study enhanced transcription by fusion of TBP to the LexA DBD. These studies have been successfully repeated using Taf subunits, Mediator and SAGA subunits, and TFIIB (Gonzalez-Couto et al. 1997; Keaveney and Struhl 1998). Taken together, the artificial recruitment studies show that, under some circumstances, transcription can be enhanced by direct recruitment of the transcription machinery, but do not by themselves prove that natural activators work by recruitment. Similar conclusions were reached in a genetic study where a mutation (Gal11p) in the Mediator subunit Gal11/Med15 was found that created a new protein–protein interaction with the Gal4 dimerization domain and stimulated transcription in the absence of a Gal4 activation domain (Barberis et al. 1995; Hidalgo et al. 2001).
Interpretation of artificial recruitment experiments is not always straightforward because the ability of these protein fusions to activate is very dependent on the architecture of the reporter and whether the reporter is located on a plasmid or integrated into the chromosome, on the precise way in which the protein fusion is constructed, and on whether the fusion is highly overexpressed (Gaudreau et al. 1999; Cheng et al. 2004; Wang et al. 2010). A recent study found that fusion of a DBD to any of the three Mediator tail subunits could activate transcription (Wang et al. 2010). One consistent conclusion from these experiments is that the Mediator tail module, and Gal11/Med15 in particular, is an especially good target for stimulation by artificial recruitment. It is probably not a coincidence that Gal11/Med15 is a common target of several activation domains. Part of the variability in artificial recruitment experiments may be due to the absence of targeted chromatin modification and the differential requirement of promoters for chromatin remodeling. For example, Morse and colleagues have demonstrated that artificial recruitment does not activate transcription at promoters where a nucleosome blocks access to the promoter (Ryan et al. 2000).
The most conclusive evidence for the recruitment model are studies in yeast using ChIP to examine the level of factors at gene regulatory regions before and after gene activation (Kuras and Struhl 1999; Li et al. 1999). There are now numerous examples showing that the level of coactivators, chromatin remodelers, and the general transcription factors crosslinking to UAS and promoter regions significantly increase upon transcription stimulation (Green 2005; Weake and Workman 2010). Although there are a few exceptions to this finding, it seems like the recruitment mechanism is involved in transcription activation at nearly all Pol II-transcribed genes.
Other activation mechanisms
Activator-induced conformational change is another mechanism that has been proposed to contribute to transcription stimulation (Taatjes et al. 2002). The best evidence for this comes from mammalian systems, where activators binding to Mediator caused dramatic activator-specific changes in Mediator structure (Taatjes et al. 2002, 2004; Meyer et al. 2010). It is not yet proven that these conformational changes occur in functional transcription complexes, but once the mechanisms of these changes are understood, it should be possible to genetically manipulate Mediator conformation and test whether and how it contributes to activation. It will also be very informative to do similar EM studies with yeast Mediator to test if it undergoes conformational changes in response to activators. An additional possibility is that Mediator itself is the target of signaling pathways that directly modulate Mediator activity though covalent modification. Studies to examine Mediator modification upon activation of various signaling pathways should begin to reveal if this mechanism is important for gene regulation.
Numerous studies in many systems have demonstrated that chromatin modification and remodeling directed by transcription factors is a key mechanism for gene activation (Narlikar et al. 2002; Li et al. 2007; Weake and Workman 2010). An early example of the importance of chromatin remodeling in yeast was found at the yeast PHO5 promoter (Svaren and Horz 1997). In another example, it was first thought that nucleosome remodeling was unimportant in the mechanism of Gal4-mediated activation because transcription of genes such as GAL1,10 were not affected by mutation of the chromatin remodeler SWI/SNF. However, recent studies showed that SWI/SNF is important for the normal rapid induction kinetics of the GAL1,10 genes (Bryant et al. 2008). It has been argued that activation involving nucleosome remodeling is another example of the recruitment mechanism because known chromatin-modifying coactivators such as SWI/SNF and NuA4 are recruited to regulatory regions by direct interaction with activators (Ptashne and Gann 2002). However, these chromatin-modifying coactivators are not by themselves sufficient for activation since artificial recruitment of factors with only chromatin-remodeling activity has not been observed to stimulate transcription (Green 2005).
Post-initiation mechanisms have most clearly been demonstrated in higher eukaryotes. The best-studied example is regulation of Pol II pausing, shortly after initiation (Buratowski 2009; Fuda et al. 2009). In genome-wide studies of mammalian and insect cells, it was found that pausing is a key and widely used mechanism of gene control and cell identity (Zeitlinger et al. 2007; Core et al. 2008; Nechaev et al. 2010). However, Pol II pausing in yeast has not been found to be a major mechanism of gene regulation. NELF, a key component of the metazoan regulatory circuit, is not conserved in yeast, and the yeast elongation factor Spt4–5 seems to have only a positive function in contrast to its metazoan counterparts. Second, studies mapping the distribution of Pol II along coding sequences have not found many instances where there is an abundance of Pol II confined to the gene 5′ end (Steinmetz et al. 2006).
Cooperativity between coactivators
It is clear from numerous studies in many systems that activator function ultimately results in the recruitment of a functional PIC to the promoter (Ptashne and Gann 2002; Green 2005). Measurement of factors crosslinked to promoters after induction showed that factors are recruited in an order specific to the gene under study (Cosma 2002). For example, at the yeast GAL1 promoter, the coactivator SAGA is initially recruited, followed shortly after by Mediator and subsequently by rapid binding of TBP, Pol II, and other general factors (Bryant and Ptashne 2003). Mediator recruitment appears blocked if SAGA is disrupted, suggesting that SAGA cooperatively recruits Mediator (Bhaumik et al. 2004). Variations of this recruitment mechanism are observed at other promoters. At several Gcn4-dependent promoters, SAGA and Mediator are recruited simultaneously in an interdependent fashion (Govind et al. 2005; Qiu et al. 2005). A common theme in these and other studies is that the targets of activators function cooperatively to generate an active PIC. There is little known about how different coactivators interact, and investigating the mechanism and function of coactivator cooperativity should reveal much about how signaling pathways converge to modulate transcription.
Activation domains
Activators are typically bipartite with separate DBD and activation domains. Most activation domains studied do not fold into well-ordered structures in the absence of a binding target, in contrast to the well-defined structure of DNA-binding motifs. For example, the activation domains of VP16, CREB, and Gcn4 all appear to be unstructured in the absence of a binding partner (Huth et al. 1997; Radhakrishnan et al. 1997; Uesugi et al. 1997; Dames et al. 2002; Freedman et al. 2002; Brzovic et al. 2011). In contrast to DNA-binding motifs, activation domain sequences are often not highly conserved (Martchenko et al. 2007). The mechanism of how activation domains specifically identify and bind their relevant multiple targets is a major unanswered question in the transcription field and a key for understanding the mechanism and specificity of many activators.
Acidic activation domains (enriched in acidic residues) are an important class that universally stimulate transcription in all eukaryotes tested (Ptashne and Gann 1990). However, as discussed below, the critical residues of these activation domains are typically hydrophobic while the function of the acidic residues is not yet clear. Originally recognized in yeast Gal4 and Gcn4, these activators encompass most of the well-characterized yeast activation domains and include strong mammalian and viral activators such as p53 and VP16. P53 contains two tandem activation domains, TAD1 and TAD2, and several structures containing the p53 activation domains have been determined (Kussie et al. 1996; Bochkareva et al. 2005; Di Lello et al. 2008; Feng et al. 2009). These structures all involve binding of one to two short α-helices to the target protein mediated primarily by hydrophobic interactions as well as some charged and polar interactions. While these structures are an important advance, the basis of activator-target specificity is not yet understood. Important questions yet to be understood include the following: (1) What are the common features of activator-binding domains? (2) How is activator-target specificity determined? and (3) How does the interaction of activators with these targets contribute to transcription activation?
Activator targets
A major question since the discovery of activation domains has been the identity of the relevant activator targets. Over the past 10 years, a combination of biochemical, genetic, and structural experiments has conclusively identified relevant targets for some mammalian and yeast activators (e.g., Stevens et al. 2002; Yang et al. 2004; Green 2005; Sampietro et al. 2006; Waters et al. 2006; Thakur et al. 2009; Herbig et al. 2010; Jedidi et al. 2010). In general, these targets are located in coactivator complexes and chromatin-remodeling or -modifying factors rather than in subunits of the general transcription factors. For example, protein crosslinkers positioned within the activation domains of Gcn4 and Gal4 have conclusively identified three common activator targets (Gal11, Tra1, and Taf12) that are subunits of four coactivator complexes (Mediator, SAGA, NuA4, and TFIID) (Fishburn et al. 2005; Reeves and Hahn 2005; Herbig et al. 2010). The activator–Tra1 interaction was also revealed by in vivo FRET analysis (Bhaumik et al. 2004) and by in vitro interaction and functional studies in vivo (Brown et al. 2001). Similarly, two subunits of the chromatin remodeler SWI/SNF interact with Gcn4 in functional assays (Prochasson et al. 2003). Recent studies indicate that Rap1, a yeast factor that participates in activation of most ribosomal protein genes as well as many other genes, functionally interacts with several subunits of TFIID (Layer et al. 2010; Papai et al. 2010).
Mechanism of Gcn4–Gal11 interaction
Activators interact with many of the same coactivator subunits, yet the sequences of the activation domains are not well conserved and the activator-binding subunits are not obviously related in sequence. This raises the question of how an activator can interact with multiple unrelated targets and whether activator–coactivator binding is specific. One of the best-studied examples of activator–target binding is that of the activator Gcn4 binding to the Mediator subunit Gal11/Med15 (Park et al. 2000; Herbig et al. 2010; Jedidi et al. 2010). Gal11 contains five conserved domains, four of which are involved in activator binding. Several studies have found that at least three of these Gal11 domains interact with both of the Gcn4 activation domains (residues 1–100 and 101–134) (Park et al. 2000; Majmudar et al. 2009; Herbig et al. 2010; Jedidi et al. 2010). Surprisingly, functional studies showed that each of these four Gal11 activator-binding domains contributes additively to activation by Gcn4 (Park et al. 2000; Herbig et al. 2010; Jedidi et al. 2010). NMR analysis has shown that the binding of Gcn4-Gal11 is unstable with a half-life of less than a millisecond (Brzovic et al. 2011). This property can explain why Gal11 has multiple activator-binding domains and Gcn4 has tandem activation domains that bind to multiple sites on Gal11 (Figure 11). The model proposed to explain activator–target binding in this system is that the two Gcn4 activation domains rapidly sample multiple activator-binding domains on Gal11. These rapidly cycling interactions are capable of allowing Gcn4 to recruit Mediator to gene regulatory regions in the absence of a stable protein–protein interaction.
Figure 11.
Model for Gcn4–Gal11 binding. Gcn4 contains tandem acidic activation domains and binds DNA as a dimer. Both activation domains contact at least three common activator-binding domains on Gal11/Med15, each of which contributes additively to activated transcription (Herbig et al. 2010). Activator–Gal11 binding has micromolar affinity and, for those sites measured, a half life of less than one millisecond (Jedidi et al. 2010; Brzovic et al. 2011). In this model, both Gcn4 activation domains rapidly sample the Gal11 activator-binding domains, and Mediator is recruited to the regulatory region without a stable high-affinity activator–target interaction. This binding mode can be scaled to increase Mediator recruitment by increasing the number of activator-binding sites at the promoter.
NMR structural analysis of the Gcn4 central activation domain bound to one Gal11 activator-binding domain (residues 158–238) has revealed much about the nature of activator–target binding and how activators can functionally interact with different unrelated coactivators (Brzovic et al. 2011). Upon binding to Gal11, about eight formerly unstructured Gcn4 residues form a helix that interacts with Gal11. The Gcn4–Gal11 protein interface is extremely simple and is purely hydrophobic, with no observed contribution from charged or polar interactions. Because of this simple interface, the Gcn4 backbone is highly flexible and is predicted by NMR to exist in multiple conformations, and, surprisingly, at least two of these conformations bind in approximately opposite orientations on Gal11. This activator–target complex is an example of a so-called “fuzzy complex” (Tompa and Fuxreiter 2008) where the structure of a protein complex cannot be described by a single conformational state. These properties probably explain how one class of activators interacts with multiple unrelated targets, and it is likely that this is a commonly used mechanism for activator–target interactions. Acidic residues in the activation domain could specifically interact with the coactivator target, contribute to a nonspecific long-range electrostatic attraction, or play no role. For the Gcn4 central activation domain, mutation of all 10 acidic residues to Ala had little effect, suggesting that, in this case, acidic residues do not play an important role (Brzovic et al. 2011). It is certainly possible that acidic residues in other activation domains play important functional roles, and it will be important to address this in future work.
Perspective
The S. cerevisiae system has made many invaluable and groundbreaking contributions to the understanding of gene control in eukaryotes. Although a few aspects of gene regulation occur only in higher eukaryotes, most of the fundamental mechanisms of transcriptional regulation have been conserved from yeast to humans. Because of the powerful combination of genetics, molecular biology, biochemistry, and genome-wide methods that can be utilized, the yeast system has often been at the forefront in discovering and understanding fundamental regulatory mechanisms. It is certain that in the next decade and beyond the yeast system will be at the forefront of fundamental discoveries in transcriptional regulation and serve as an excellent model for understanding regulatory mechanisms in other eukaryotes.
Literature Cited
- Agricola E., Verdone L., Xella B., Di Mauro E., Caserta M., 2004. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry 43: 8878–8884 [DOI] [PubMed] [Google Scholar]
- Agricola E., Verdone L., Di Mauro E., Caserta M., 2006. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol. Microbiol. 62: 1433–1446 [DOI] [PubMed] [Google Scholar]
- Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., et al. , 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., et al. , 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., et al. , 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing esa1p and the ATM-related cofactor tra1p. [In Process Citation] EMBO J. 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Horz W., 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5: 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andel F., III, Ladurner A. G., Inouye C., Tjian R., Nogales E., 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286: 2153–2156 [DOI] [PubMed] [Google Scholar]
- Andrau J. C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., et al. , 2006. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22: 179–192 [DOI] [PubMed] [Google Scholar]
- Badis G., Chan E. T., van Bakel H., Pena-Castillo L., Tillo D., et al. , 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa W., Torchia T. E., Hopper J. E., 1988. Yeast regulatory gene GAL3: carbon regulation; UASGal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol. Cell. Biol. 8: 3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. P., Grant P. A., 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26: 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A., Tan S., 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989–7995 [DOI] [PubMed] [Google Scholar]
- Bali M., Zhang B., Morano K. A., Michels C. A., 2003. The Hsp90 molecular chaperone complex regulates maltose induction and stability of the Saccharomyces MAL gene transcription activator Mal63p. J. Biol. Chem. 278: 47441–47448 [DOI] [PubMed] [Google Scholar]
- Barbaric S., Reinke H., Horz W., 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23: 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A., Pearlberg J., Simkovich N., Farrell S., Reinagel P., et al. , 1995. Contact with a compnent of the polymerase II holoenzyme suffices for gene activation. Cell 81: 359–368 [DOI] [PubMed] [Google Scholar]
- Basehoar A. D., Zanton S. J., Pugh B. F., 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Baumli S., Hoeppner S., Cramer P., 2005. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 280: 18171–18178 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Sterner D. E., Deng M., Sayre M. H., Lieberman P. M., et al. , 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20: 634–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L., Pina B., Silverman N., Marcus G. A., Agapite J., et al. , 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Liu C. L., Humphrey E. L., Perlstein E. O., Schreiber S. L., 2004. Global nucleosome occupancy in yeast. Genome Biol. 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J., Morillon A., 2009. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A., Workman C., Hollunder J., Radke D., Moller U., et al. , 2006. Integrated assessment and prediction of transcription factor binding. PLoS Comput. Biol. 2: e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22: 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Raha T., Aiello D. P., Green M. R., 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoite L. T., Yu Y., Stillman D. J., 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15: 2457–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick R., Young E. T., 2005. Yeast mediator and its role in transcriptional regulation. C. R. Biol. 328: 773–782 [DOI] [PubMed] [Google Scholar]
- Biddick R. K., Law G. L., Chin K. K., Young E. T., 2008a The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J. Biol. Chem. 283: 33101–33109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick R. K., Law G. L., Young E. T., 2008b Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS ONE 3: e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck C., Poch O., Romier C., Ruff M., Mengus G., et al. , 1998. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94: 239–249 [DOI] [PubMed] [Google Scholar]
- Bird A. J., Zhao H., Luo H., Jensen L. T., Srinivasan C., et al. , 2000. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 19: 3704–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund S., Gustafsson C. M., 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Blumberg H., Hartshorne T. A., Young E. T., 1988. Regulation of expression and activity of the yeast transcription factor ADR1. Mol. Cell. Biol. 8: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumental-Perry A., Li W., Simchen G., Mitchell A. P., 2002. Repression and activation domains of RME1p structurally overlap, but differ in genetic requirements. Mol. Biol. Cell 13: 1709–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E., Kaustov L., Ayed A., Yi G. S., Lu Y., et al. , 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. USA 102: 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm S., Frishman D., Mewes H. W., 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25: 2464–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. J., Ballou C., Fackenthal D. L., 1994. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol. Cell. Biol. 14: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. J., Carlson T., Fackenthal D. L., Paddock D., Storey K., et al. , 2000a Complex regulation of the yeast heat shock transcription factor. Mol. Biol. Cell 11: 1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. J., Chen D., Storey K., Tushan M., Lea K., 2000b Structural analysis of yeast HSF by site-specific crosslinking. J. Mol. Biol. 302: 581–592 [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., et al. , 2007. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Bourbon H. M., 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H. M., Aguilera A., Ansari A. Z., Asturias F. J., Berk A. J., et al. , 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Brickner J. H., 2009. Transcriptional memory at the nuclear periphery. Curr. Opin. Cell Biol. 21: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont P. A., Daugherty J. R., Cooper T. G., 1991. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A. S., Silver P. A., 1999. Nuclear transport HEATs up. Nat. Cell Biol. 1: E66–E67 [DOI] [PubMed] [Google Scholar]
- Brown C. E., Howe L., Sousa K., Alley S. C., Carrozza M. J., et al. , 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337 [DOI] [PubMed] [Google Scholar]
- Bryant G. O., Ptashne M., 2003. Independent recruitment in vivo by gal4 of two complexes required for transcription. Mol. Cell 11: 1301–1309 [DOI] [PubMed] [Google Scholar]
- Bryant G. O., Prabhu V., Floer M., Wang X., Spagna D., et al. , 2008. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 6: 2928–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P. S., Heikaus C. C., Kisselev L., Vernon R., Herbig E., et al. , 2011. The acidic transcription activator Gcn4 binds the Mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. W., Shore D., 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370–384 [DOI] [PubMed] [Google Scholar]
- Bulman A. L., Hubl S. T., Nelson H. C., 2001. The DNA-binding domain of yeast heat shock transcription factor independently regulates both the N- and C-terminal activation domains. J. Biol. Chem. 276: 40254–40262 [DOI] [PubMed] [Google Scholar]
- Buratowski S., 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., Guarente L., 1988. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334: 37–42 [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., Sharp P. A., 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561 [DOI] [PubMed] [Google Scholar]
- Bushnell D. A., Westover K. D., Davis R. E., Kornberg R. D., 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303: 983–988 [DOI] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G., 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Cai G., Imasaki T., Takagi Y., Asturias F. J., 2009. Mediator structural conservation and implications for the regulation mechanism. Structure 17: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Imasaki T., Yamada K., Cardelli F., Takagi Y., et al. , 2010. Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 17: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., et al. , 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Carter R., Drouin G., 2010. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 27: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S., Errede B., 1998. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 2884–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. W., Robert Liu F. G., Yu N., Sung H. M., Yang P., et al. , 2008. Roles of cis- and trans-changes in the regulatory evolution of genes in the gluconeogenic pathway in yeast. Mol. Biol. Evol. 25: 1863–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D., 1990. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol. Cell. Biol. 10: 2916–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Struhl K., 1995. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature 374: 820–821 [DOI] [PubMed] [Google Scholar]
- Chen A. C., Jawhari A., Fischer L., Buchen C., Tahir S., et al. , 2010. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. T., Hahn S., 2003. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12: 437–447 [DOI] [PubMed] [Google Scholar]
- Chen H.T., Hahn S., 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119: 169–180 [DOI] [PubMed] [Google Scholar]
- Chen H. T., Warfield L., Hahn S., 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 14: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lopes J. M., 2010. Multiple bHLH proteins regulate CIT2 expression in Saccharomyces cerevisiae. Yeast 27: 345–359 [DOI] [PubMed] [Google Scholar]
- Chen M., Lopes J. M., 2007. Multiple basic helix-loop-helix proteins regulate expression of the ENO1 gene of Saccharomyces cerevisiae. Eukaryot. Cell 6: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. X., Floer M., Ononaji P., Bryant G., Ptashne M., 2002. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 12: 1828–1832 [DOI] [PubMed] [Google Scholar]
- Cheng J. X., Gandolfi M., Ptashne M., 2004. Activation of the gal1 gene of yeast by pairs of ‘non-classical’ activators. Curr. Biol. 14: 1675–1679 [DOI] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L., 1989. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 56: 409–419 [DOI] [PubMed] [Google Scholar]
- Chua G., Morris Q. D., Sopko R., Robinson M. D., Ryan O., et al. , 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 103: 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cler E., Papai G., Schultz P., Davidson I., 2009. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell. Mol. Life Sci. 66: 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., et al. , 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76 [DOI] [PubMed] [Google Scholar]
- Company M., Errede B., 1988. A Ty1 cell-type-specific regulatory sequence is a recognition element for a constitutive binding factor. Mol. Cell. Biol. 8: 5299–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W., 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30: 250–255 [DOI] [PubMed] [Google Scholar]
- Cook W. J., Chase D., Audino D. C., Denis C. L., 1994. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol. Cell. Biol. 14: 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M. P., 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10: 227–236 [DOI] [PubMed] [Google Scholar]
- Cosma M. P., 2004. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 5: 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S., 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15: 2786–2796 [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Kornberg R. D., 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Cramer P., Armache K. J., Baumli S., Benkert S., Brueckner F., et al. , 2008. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37: 337–352 [DOI] [PubMed] [Google Scholar]
- D’Alessio J. A., Wright K. J., Tjian R., 2009. Shifting players and paradigms in cell-specific transcription. Mol. Cell 36: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dames S. A., Martinez-Yamout M., De Guzman R. N., Dyson H. J., Wright P. E., 2002. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA 99: 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J. K., Trumbly R. J., Dent S. Y., 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. A., Takagi Y., Kornberg R. D., Asturias F. A., 2002. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10: 409–415 [DOI] [PubMed] [Google Scholar]
- de Bruin D., Zaman Z., Liberatore R. A., Ptashne M., 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409: 109–113 [DOI] [PubMed] [Google Scholar]
- Deckert J., Rodriguez Torres A. M., Simon J. T., Zitomer R. S., 1995. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6109–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Fontaine S. C., Chase D., Kemp B. E., Bemis L. T., 1992. ADR1c mutations enhance the ability of ADR1 to activate transcription by a mechanism that is independent of effects on cyclic AMP-dependent protein kinase phosphorylation of Ser-230. Mol. Cell. Biol. 12: 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lello P., Miller Jenkins L. M., Mas C., Langlois C., Malitskaya E., et al. , 2008. p53 and TFIIEalpha share a common binding site on the Tfb1/p62 subunit of TFIIH. Proc. Natl. Acad. Sci. USA 105: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mauro E., Verdone L., Chiappini B., Caserta M., 2002. In vivo changes of nucleosome positioning in the pretranscription state. J. Biol. Chem. 277: 7002–7009 [DOI] [PubMed] [Google Scholar]
- Dodou E., Treisman R., 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek K. M., Camier S., Young E. T., 1993. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol. Cell. Biol. 13: 4391–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A. J., Ebmeier C. C., Taatjes D. J., Espinosa J. M., 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17: 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech M., Coin F., Chipoulet J. M., Arai Y., Ohkuma Y., et al. , 2000. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell. Biol. 20: 8168–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon J. B., Liu D. R., 2007. Identification of eukaryotic promoter regulatory elements using nonhomologous random recombination. Nucleic Acids Res. 35: 5851–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna E., Bialkova A., Subik J., 2008. Transcriptional regulators of seven yeast species: comparative genome analysis. Review. Folia Microbiol. (Praha) 53: 275–287 [DOI] [PubMed] [Google Scholar]
- Drysdale C. M., Duenas E., Jackson B. M., Reusser U., Braus G. H., et al. , 1995. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol. 15: 1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker C. E., Simpson R. T., 2000. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J. 19: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A. M., Rougeulle C., Winston F., 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., Kalton H. M., Gaber R. F., 1998. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273: 18974–18978 [DOI] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R., 1991. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66: 563–576 [DOI] [PubMed] [Google Scholar]
- Eastmond D. L., Nelson H. C., 2006. Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J. Biol. Chem. 281: 32909–32921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner J., Chen H. T., Warfield L., Hahn S., 2010. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., 2009. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284: 18565–18569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Arndt K. M., Ricupero S. L., Rooney J. W., Winston F., 1992. SPT3 interacts with TFIID to allow normal transcription in S. cerevisiae. Genes Dev. 6: 1319–1331 [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Chapon C., Roberts S. M., Dollard C., Winston F., 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137: 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R., Tye B. K., 1991. Both activation and repression of a-mating-type-specific genes in yeast require transcription factor Mcm1. Proc. Natl. Acad. Sci. USA 88: 10966–10970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Teague M. A., Sherman F., 1984. Identification of regulatory regions within the Ty1 transposable element that regulate iso-2-cytochrome c production in the CYC7-H2 yeast mutant. Mol. Cell. Biol. 4: 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faitar S. L., Brodie S. A., Ponticelli A. S., 2001. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 21: 4427–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Struhl K., 2009. Where does mediator bind in vivo? PLoS ONE 4: e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Chou D. M., Struhl K., 2006. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 13: 117–120 [DOI] [PubMed] [Google Scholar]
- Fantino E., Marguet D., Lauquin G. J., 1992. Downstream activating sequence within the coding region of a yeast gene: specific binding in vitro of RAP1 protein. Mol. Gen. Genet. 236: 65–75 [DOI] [PubMed] [Google Scholar]
- Fascher K. D., Schmitz J., Horz W., 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 9: 2523–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W. J., Svejstrup J. Q., Henry N. L., Kornberg R. D., 1994. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Feller A., Dubois E., Ramos F., Pierard A., 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14: 6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Jenkins L. M., Durell S. R., Hayashi R., Mazur S. J., et al. , 2009. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure 17: 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L., Rodrigues-Pousada C., Struhl K., 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17: 6982–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T., Ptashne M., 1988. GAL4 activates transcription in Drosophila. Nature 332: 853–856 [DOI] [PubMed] [Google Scholar]
- Fishburn J., Mohibullah N., Hahn S., 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18: 369–378 [DOI] [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J., III, Sayre M. H., Tschochner H., Kornberg R. D., 1991. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438 [DOI] [PubMed] [Google Scholar]
- Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., et al. , 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman S. J., Sun Z. Y., Poy F., Kung A. L., Livingston D. M., et al. , 2002. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc. Natl. Acad. Sci. USA 99: 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Reynolds C., Schimmel P., 1989. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol. Cell. Biol. 9: 4056–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N. J., Ardehali M. B., Lis J. T., 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy D. J., Cassidy-Stone A., Llinas M., O’Rourke S. M., Herskowitz I., et al. , 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 18069–18074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo C., Flores C. L., 2008. Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Foat B. C., Bussemaker H. J., 2004. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics 5: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau L., Keaveney M., Nevado J., Zaman Z., Bryant G. O., et al. , 1999. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc. Natl. Acad. Sci. USA 96: 2668–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger S. R., Lorenzen K., Schreieck A., Hanecker P., Kostrewa D., et al. , 2010. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 39: 583–594 [DOI] [PubMed] [Google Scholar]
- Georges A. B., Benayoun B. A., Caburet S., Veitia R. A., 2010. Generic binding sites, generic DNA-binding domains: Where does specific promoter recognition come from? FASEB J. 24: 346–356 [DOI] [PubMed] [Google Scholar]
- Ghazy M. A., Brodie S. A., Ammerman M. L., Ziegler L. M., Ponticelli A. S., 2004. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol. Cell. Biol. 24: 10975–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C., Lis J. T., 1993. DNA melting on yeast RNA polymerase II promoters. Science 261: 759–762 [DOI] [PubMed] [Google Scholar]
- Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D., 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292: 1876–1882 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Couto E., Klages N., Strubin M., 1997. Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 94: 8036–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R., Hartemink A. J., Bulyk M. L., 2009. Distinguishing direct vs. indirect transcription factor-DNA interactions. Genome Res. 19: 2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G., 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 25: 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., et al. , 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (spt/Ada) complex. Genes Dev. 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grant P. A., Schieltz D., Pray-Grant M. G., Steger D. J., Reese J. C., et al. , 1998a A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53 [DOI] [PubMed] [Google Scholar]
- Grant P. A., Schieltz D., Pray-Grant M. G., Yates J. R., III, Workman J. L., 1998b The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2: 863–867 [DOI] [PubMed] [Google Scholar]
- Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., et al. , 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274: 5895–5900 [DOI] [PubMed] [Google Scholar]
- Gray W. M., Fassler J. S., 1996. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol. Cell. Biol. 16: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., 2005. Eukaryotic transcription activation: right on target. Mol. Cell 18: 399–402 [DOI] [PubMed] [Google Scholar]
- Gromoller A., Lehming N., 2000. Srb7p is essential for the activation of a subset of genes. FEBS Lett. 484: 48–54 [DOI] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N. L., Klapholz B., Bijma T., Boube M., et al. , 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Papachristoudis G., Altshuler R. C., Gerber G. K., Jaakkola T. S., et al. , 2010. Discovering homotypic binding events at high spatial resolution. Bioinformatics 26: 3028–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach A., Hon T., Zhang L., 1999. A new class of repression modules is critical for heme regulation of the yeast transcriptional activator Hap1. Mol. Cell. Biol. 19: 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. S., Hu Z., Thiele D. J., Iyer V. R., 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., 1998. Activation and the role of reinitiation in the control of transcription by RNA polymerase II. Cold Spring Harb. Symp. Quant. Biol. 63: 181–188 [DOI] [PubMed] [Google Scholar]
- Hahn S., 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11: 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Maurer P., Caesar S., Schlenstedt G., 2008. Classical NLS proteins from Saccharomyces cerevisiae. J. Mol. Biol. 379: 678–694 [DOI] [PubMed] [Google Scholar]
- Hall D. A., Zhu H., Zhu X., Royce T., Gerstein M., et al. , 2004. Regulation of gene expression by a metabolic enzyme. Science 306: 482–484 [DOI] [PubMed] [Google Scholar]
- Hampsey M., 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62: 465–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Grunstein M., 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M., 1988. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 7: 2221–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa N., Mizukami Y., Imazu H., Sakurai H., 2006. Mutated yeast heat shock transcription factor activates transcription independently of hyperphosphorylation. J. Biol. Chem. 281: 3936–3942 [DOI] [PubMed] [Google Scholar]
- Helmlinger D., Marguerat S., Villen J., Gygi S. P., Bahler J., et al. , 2008. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 22: 3184–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry N. L., Campbell A. M., Feaver W. J., Poon D., Weil P. A., et al. , 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8: 2868–2878 [DOI] [PubMed] [Google Scholar]
- Herbig A., Bird A. J., Swierczek S., McCall K., Mooney M., et al. , 2005. Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status. Mol. Microbiol. 57: 834–846 [DOI] [PubMed] [Google Scholar]
- Herbig E., Warfield L., Fish L., Fishburn J., Knutson B. A., et al. , 2010. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol. Cell. Biol. 30: 2376–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo P., Ansari A. Z., Schmidt P., Hare B., Simkovich N., et al. , 2001. Recruitment of the transcriptional machinery through GAL11P: structure and interactions of the GAL4 dimerization domain. Genes Dev. 15: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Hoeppner S., Baumli S., Cramer P., 2005. Structure of the mediator subunit cyclin C and its implications for CDK8 function. J. Mol. Biol. 350: 833–842 [DOI] [PubMed] [Google Scholar]
- Hoj A., Jakobsen B. K., 1994. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 13: 2617–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P. C., Pietz G., Fox C. A., 2001. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 15: 2445–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., et al. , 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Holstege F. C. P., Fiedler U., Timmers H. T. M., 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16: 7468–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T., Hach A., Lee H. C., Cheng T., Zhang L., 2000. Functional analysis of heme regulatory elements of the transcriptional activator Hap1. Biochem. Biophys. Res. Commun. 273: 584–591 [DOI] [PubMed] [Google Scholar]
- Hon T., Lee H. C., Hach A., Johnson J. L., Craig E. A., et al. , 2001. The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Mol. Cell. Biol. 21: 7923–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K., 1986. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46: 885–894 [DOI] [PubMed] [Google Scholar]
- Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., et al. , 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16: 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Yu J., Taylor J., Chinnaiyan A., Qin Z., 2010. On the detection and refinement of transcription factor binding sites using ChIP-Seq data. Nucleic Acids Res. 38: 2154–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Huth J. R., Bewley C. A., Jackson B. M., Hinnebusch A. G., Clore G. M., et al. , 1997. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6: 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K., Krogan N. J., Emre N. C., Wyce A., Thompson N. J., et al. , 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V., Struhl K., 1995a Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 15: 7059–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V., Struhl K., 1995b Poly (dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen B. K., Pelham H. R., 1991. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 10: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen B. K., Pelham H. R., 1988. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell. Biol. 8: 5040–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedidi I., Zhang F., Qiu H., Stahl S. J., Palmer I., et al. , 2010. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J. Biol. Chem. 285: 2438–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M. H., O’Rourke T. W., Reines D., 2008. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol. Cell. Biol. 28: 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Pugh B. F., 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Frey B. R., Evans M. L., Friel J. C., Hopper J. E., 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol. Cell. Biol. 29: 5604–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Carlson M., 1992. Regulation of carbon and phosphate utilization, pp. 193–281 Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae, edited by Jones E., Pringle J. R., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S., 1987. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell 50: 143–146 [DOI] [PubMed] [Google Scholar]
- Juan L. J., Walter P. P., Taylor I. C., Kingston R. E., Workman J. L., 1993. Nucleosome cores and histone H1 in the binding of GAL4 derivatives and the reactivation of transcription from nucleosome templates in vitro. Cold Spring Harb. Symp. Quant. Biol. 58: 213–223 [DOI] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., Levin D. E., 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T., Kadonaga J. T., 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacherovsky N., Tachibana C., Amos E., Fox D., III, Young E. T., 2008. Promoter binding by the Adr1 transcriptional activator may be regulated by phosphorylation in the DNA-binding region. PLoS ONE 3: e3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18: 5121–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K. H., Knochel W., Martinez D. E., 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14: 142–146 [PubMed] [Google Scholar]
- Kahana S., Pnueli L., Kainth P., Sassi H. E., Andrews B., et al. , 2010. Functional dissection of IME1 transcription using quantitative promoter-reporter screening. Genetics 186: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M., 1988. GAL4 activates gene expression in mammalian cells. Cell 52: 161–167 [DOI] [PubMed] [Google Scholar]
- Kang J. S., Kim S. H., Hwang M. S., Han S. J., Lee Y. C., et al. , 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003–42010 [DOI] [PubMed] [Google Scholar]
- Kanin E. I., Kipp R. T., Kung C., Slattery M., Viale A., et al. , 2007. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. USA 104: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., et al. , 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224: 111–171 [DOI] [PubMed] [Google Scholar]
- Keaveney M., Struhl K., 1998. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell 1: 917–924 [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Khaperskyy D. A., Ammerman M. L., Majovski R. C., Ponticelli A. S., 2008. Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol. Cell. Biol. 28: 3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K., 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365: 520–527 [DOI] [PubMed] [Google Scholar]
- Kim T. K., Ebright R. H., Reinberg D., 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B., 1993. Crystal structure of a yeast TBP/TATA-box complex. Nature 365: 512–520 [DOI] [PubMed] [Google Scholar]
- Klages N., Strubin M., 1995. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature 374: 822–823 [DOI] [PubMed] [Google Scholar]
- Knutson B. A., Hahn S., 2011. Domains of Tra1 important for Activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 31: 818–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Schneider M., Cabal G. G., Nehrbass U., Hurt E., 2008. Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10: 707–715 [DOI] [PubMed] [Google Scholar]
- Kohlhaw G. B., 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T., Takada R., Yamashita S., Gong D. W., Roeder R. G., et al. , 1993. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J. Biol. Chem. 268: 17554–17558 [PubMed] [Google Scholar]
- Kokubo T., Swanson M. J., Nishikawa J. I., Hinnebusch A. G., Nakatani Y., 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., O’Shea E. K., 2000. Nuclear transport and transcription. Curr. Opin. Cell Biol. 12: 355–360 [DOI] [PubMed] [Google Scholar]
- Koschubs T., Seizl M., Lariviere L., Kurth F., Baumli S., et al. , 2009. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J. 28: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschubs T., Lorenzen K., Baumli S., Sandstrom S., Heck A. J., et al. , 2010. Preparation and topology of the Mediator middle module. Nucleic Acids Res. 38: 3186–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., et al. , 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330 [DOI] [PubMed] [Google Scholar]
- Krishna S. S., Majumdar I., Grishin N. V., 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31: 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner J. N., Brow D. A., 2006. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 281: 14119–14128 [DOI] [PubMed] [Google Scholar]
- Kuehner J. N., Brow D. A., 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell 31: 201–211 [DOI] [PubMed] [Google Scholar]
- Kuras L., Struhl K., 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–613 [DOI] [PubMed] [Google Scholar]
- Kuras L., Kosa P., Mencia M., Struhl K., 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288: 1244–1248 [DOI] [PubMed] [Google Scholar]
- Kussie P., Gorina S., Marechal V., Elenbaas B., Moreau J., et al. , 1996. Structure of the MDM2 oncoprotien bound to the p53 tumor suppressor transactivation domain. Science 274: 948–953 [DOI] [PubMed] [Google Scholar]
- Lan C., Lee H. C., Tang S., Zhang L., 2004. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 279: 27607–27612 [DOI] [PubMed] [Google Scholar]
- Lane W. J., Darst S. A., 2010a Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 395: 671–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane W. J., Darst S. A., 2010b Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 395: 686–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade L., Rose D., Winston F., 2007. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177: 2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L., Geiger S., Hoeppner S., Rother S., Strasser K., et al. , 2006. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 13: 895–901 [DOI] [PubMed] [Google Scholar]
- Lariviere L., Seizl M., van Wageningen S., Rother S., van de Pasch L., et al. , 2008. Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev. 22: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F., 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., et al. , 2010. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 8: e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer J. H., Miller S. G., Weil P. A., 2010. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J. Biol. Chem. 285: 15489–15499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D., 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee H. C., Zhang L., 2009. A unique mechanism of chaperone action: heme regulation of Hap1 activity involves separate control of repression and activation. Protein Pept. Lett. 16: 642–649 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Workman J. L., 2007. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8: 284–295 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Florens L., Swanson S. K., Washburn M. P., Workman J. L., 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Sardiu M. E., Swanson S. K., Gilmore J. M., Torok M., et al. , 2011. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Chatterjee S., Struhl K., 2000. Genetic analysis of the role of pol II holoenzyme components in repression by the Cyc8-tup1 corepressor in yeast. Genetics 155: 1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Carlson T., Christian N., Lea K., Kedzie J., et al. , 2000. The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Mol. Biol. Cell 11: 1753–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. A., Jorgensen P., Bognar A. L., Peyraud C., Thomas D., et al. , 2010. Dissection of combinatorial control by the Met4 transcriptional complex. Mol. Biol. Cell 21: 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Young R. A., 1998. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12: 1398–1408 [DOI] [PubMed] [Google Scholar]
- Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., et al. , 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704 [DOI] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R. H., Davis R. W., et al. , 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Leurent C., Sanders S., Ruhlmann C., Mallouh V., Weil P. A., et al. , 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21: 3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li X. Y., Bhaumik S. R., Green M. R., 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288: 1242–1244 [DOI] [PubMed] [Google Scholar]
- Li X. Y., Virbasius A., Zhu X., Green M. R., 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399: 605–609 [DOI] [PubMed] [Google Scholar]
- Li X. Y., Bhaumik S. R., Zhu X., Li L., Shen W. C., et al. , 2002. Selective recruitment of TAFs by yeast upstream activating sequences: implications for eukaryotic promoter structure. Curr. Biol. 12: 1240–1244 [DOI] [PubMed] [Google Scholar]
- Liao S. M., Zhang J., Jeffery D. A., Koleske A. J., Thompson C. M., et al. , 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374: 193–196 [DOI] [PubMed] [Google Scholar]
- Liu D., Ishima R., Tong K. I., Bagby S., Kokubo T., et al. , 1998. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94: 573–583 [DOI] [PubMed] [Google Scholar]
- Liu W. L., Coleman R. A., Grob P., King D. S., Florens L., et al. , 2008. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol. Cell 29: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Vorontchikhina M., Wang Y. L., Faiola F., Martinez E., 2008. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol. Cell. Biol. 28: 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bushnell D. A., Wang D., Calero G., Kornberg R. D., 2010. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327: 206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kung C., Fishburn J., Ansari A. Z., Shokat K. M., et al. , 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the Scaffold complex. Mol. Cell. Biol. 24: 1721–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Warfield L., Zhang C., Luo J., Allen J., et al. , 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29: 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N. M., Austin S. E., Berman H. M., Thornton J. M., 2000. An overview of the structures of protein-DNA complexes. Genome Biol. 1: REVIEWS001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M., 1987a The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 50: 137–142 [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M., 1987b Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48: 847–853 [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M., 1988. Converting a eukaryotic transcriptional inhibitor into an activator. Cell 55: 443–446 [DOI] [PubMed] [Google Scholar]
- Ma J., Przibilla E., Hu J., Bogorad L., Ptashne M., 1988. Yeast activators stimulate plant gene expression. Nature 334: 631–633 [DOI] [PubMed] [Google Scholar]
- MacPherson S., Larochelle M., Turcotte B., 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70: 583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J. M., Winston F., 1997. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TBP to confer promoter-specific transcriptional control in S. cerevisiae. Mol. Cell. Biol. 17: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmudar C. Y., Wang B., Lum J. K., Hakansson K., Mapp A. K., 2009. A high-resolution interaction map of three transcriptional activation domains with a key coactivator from photo-cross-linking and multiplexed mass spectrometry. Angew. Chem. Int. Ed. Engl. 48: 7021–7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Martchenko M., Levitin A., Whiteway M., 2007. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot. Cell 6: 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., Zhou Q., L’Etoile N. D., Oelgeschlager T., Berk A. J., et al. , 1995. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc. Natl. Acad. Sci. USA 92: 11864–11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O., Buratowski R. M., Swilling N. W., Buratowski S., 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14: 951–962 [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O., Auty R., Buratowski S., 2004. Structure and function of the TFIID complex. Adv. Protein Chem. 67: 67–92 [DOI] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G., 1980. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem. 255: 11992–11996 [PubMed] [Google Scholar]
- Maxon M. E., Herskowitz I., 2001. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. USA 98: 1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H. J., Brazas R. M., Yu Y., Nasmyth K., Stillman D. J., 1997. Long-range interactions at the HO promoter. Mol. Cell. Biol. 17: 2669–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Kingsman A. J., Kingsman S. M., 1987. A transcriptional activator is located in the coding region of the yeast PGK gene. Nucleic Acids Res. 15: 6243–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon B. B., Sarma N. J., Pasula S., Deminoff S. J., Willis K. A., et al. , 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Lin S. C., Bernecky C., Gao Y., Taatjes D. J., 2010. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17: 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Hahn S., 2006. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 13: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., et al. , 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA 103: 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T., Loch C. M., Li R., 2002. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 22: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C. A., Yang X. J., Kokubo T., Brownell J. E., Bannister A. J., et al. , 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Mohibullah N., Hahn S., 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22: 2994–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P. T., Mendes N. D., Teixeira M. C., d’Orey S., Tenreiro S., et al. , 2008. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 36: D132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai Y., Poon D., Weil P. A., Struhl K., 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383: 188–191 [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S., 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 73: 251–279 [DOI] [PubMed] [Google Scholar]
- Murakami K. S., Darst S. A., 2003. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13: 31–39 [DOI] [PubMed] [Google Scholar]
- Mutiu A. I., Hoke S. M., Genereaux J., Hannam C., MacKenzie K., et al. , 2007. Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1. Genetics 177: 151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E., 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Narlikar L., Gordan R., Hartemink A. J., 2007. A nucleosome-guided map of transcription factor binding sites in yeast. PLoS Comput. Biol. 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., et al. , 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Bruce C., Hart C., Leigh-Bell J., Gelperin D., et al. , 2009. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 23: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Sotelo J., Wiederrecht G., Okuda A., Parker C. S., 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62: 807–817 [DOI] [PubMed] [Google Scholar]
- Ostling J., Carlberg M., Ronne H., 1996. Functional domains in the Mig1 repressor. Mol. Cell. Biol. 16: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G., Tripathi M. K., Ruhlmann C., Werten S., Crucifix C., et al. , 2009. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 17: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G., Tripathi M. K., Ruhlmann C., Layer J. H., Weil P. A., et al. , 2010. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465: 956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Conlan R. S., Gounalaki N., Copf T., Tzamarias D., 2000. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem. 275: 8397–8403 [DOI] [PubMed] [Google Scholar]
- Park J., Park B., Jung K., Jang S., Yu K., et al. , 2008. CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res. 36: D562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., et al. , 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M. R., Jaehning J. A., 1992. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol. Cell. Biol. 12: 4981–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parua P. K., Ratnakumar S., Braun K. A., Dombek K. M., Arms E., et al. , 2010. 14–3-3 (Bmh) proteins inhibit transcription activation by Adr1 through direct binding to its regulatory domain. Mol. Cell. Biol. 30: 5273–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patikoglou G. A., Kim J. L., Sun L., Yang S. H., Kodadek T., et al. , 1999. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13: 3217–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C., 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75: 271–294 [DOI] [PubMed] [Google Scholar]
- Peng G., Hopper J. E., 2000. Evidence for Gal3p’s cytoplasmic location and Gal80p’s dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 5140–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Hopper J. E., 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. USA 99: 8548–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilauri V., Bewley M., Diep C., Hopper J., 2005. Gal80 dimerization and the yeast GAL gene switch. Genetics 169: 1903–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A., Reece R. J., 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 17: 4086–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polish J. A., Kim J. H., Johnston M., 2005. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 169: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D., Bai Y., Campbell A. M., Bjorkllund S., Kim Y.-J., et al. , 1995. Identification and characterization of a TFIID-like multiprotein complex for S. cerevisiae. Proc. Natl. Acad. Sci. USA 92: 8224–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M. G., Schieltz D., McMahon S. J., Wood J. M., Kennedy E. L., et al. , 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22: 8774–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochasson P., Neely K. E., Hassan A. H., Li B., Workman J. L., 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12: 983–990 [DOI] [PubMed] [Google Scholar]
- Ptashne M., 1988. How eukaryotic transcriptional activators work. Nature 335: 683–689 [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. F., 1990. Activators and targets. Nature 346: 329–331 [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A., 1997. Transcriptional activation by recruitment. Nature 386: 569–577 [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A., 2002. Genes and Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Qiu H., Hu C., Yoon S., Natarajan K., Swanson M. J., et al. , 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24: 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Zhang F., Hwang G. J., Swanson M. J., et al. , 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Hinnebusch A. G., 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I., Perez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., et al. , 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91: 741–752 [DOI] [PubMed] [Google Scholar]
- Ran F., Bali M., Michels C. A., 2008. Hsp90/Hsp70 chaperone machine regulation of the Saccharomyces MAL-activator as determined in vivo using noninducible and constitutive mutant alleles. Genetics 179: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F., Gadura N., Michels C. A., 2010. Hsp90 cochaperone Aha1 is a negative regulator of the Saccharomyces MAL activator and acts early in the chaperone activation pathway. J. Biol. Chem. 285: 13850–13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish J. A., Yudkovsky N., Hahn S., 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar S., Kacherovsky N., Arms E., Young E. T., 2009. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 182: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar S., Young E. T., 2010. Snf1 dependence of peroxisomal gene expression is mediated by Adr1. J. Biol. Chem. 285: 10703–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M., 1993. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science 261: 909–911 [DOI] [PubMed] [Google Scholar]
- Reese J. C., Apone L., Walker S. S., Griffin L. A., Green M. R., 1994. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature 371: 523–527 [DOI] [PubMed] [Google Scholar]
- Reeves W. M., Hahn S., 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25: 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. E., Evans K. J., Dyer N., Wernisch L., Ott S., 2010. Variable structure motifs for transcription factor binding sites. BMC Genomics 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A., Ebright R., Strick T. R., 2004. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc. Natl. Acad. Sci. USA 101: 4776–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K. A., Lopes J. M., 2000. SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28: 1499–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Fischer T., Luo M. J., Antunez O., Brettschneider S., et al. , 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Rothermel B. A., Thornton J. L., Butow R. A., 1997. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J. Biol. Chem. 272: 19801–19807 [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I., Mandel S., Robzyk K., Kassir Y., 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16: 2518–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Cox D., Williamson V. M., Young E. T., 1983. DNA sequences of two yeast promoter-up mutants. Nature 304: 652–654 [DOI] [PubMed] [Google Scholar]
- Ryan M. P., Stafford G. A., Yu L., Morse R. H., 2000. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 20: 5847–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Schieltz D., Ting N., McMahon S. B., Litchfield D. W., et al. , 1998. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J. Biol. Chem. 273: 26559–26565 [DOI] [PubMed] [Google Scholar]
- Sampietro J., Dahlberg C. L., Cho U. S., Hinds T. R., Kimelman D., et al. , 2006. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol. Cell 24: 293–300 [DOI] [PubMed] [Google Scholar]
- Sanders S. L., Garbett K. A., Weil P. A., 2002. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22: 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma N. J., Haley T. M., Barbara K. E., Buford T. D., Willis K. A., et al. , 2007. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 175: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C., 2000. The fungal GATA factors. Curr. Opin. Microbiol. 3: 126–131 [DOI] [PubMed] [Google Scholar]
- Schwank S., Ebbert R., Rautenstrauss K., Schweizer E., Schuller H. J., 1995. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 23: 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S., Abul-Hamd A. T., Cooper T. G., 2000. Roles of the Dal82p domains in allophanate/oxalurate-dependent gene expression in Saccharomyces cerevisiae. J. Biol. Chem. 275: 30886–30893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila A. C., Core L. J., Lis J. T., Sharp P. A., 2009. Divergent transcription: a new feature of active promoters. Cell Cycle 8: 2557–2564 [DOI] [PubMed] [Google Scholar]
- Sekinger E. A., Moqtaderi Z., Struhl K., 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Sellick C. A., Reece R. J., 2003. Modulation of transcription factor function by an amino acid: activation of Put3p by proline. EMBO J. 22: 5147–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick C. A., Reece R. J., 2005. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 30: 405–412 [DOI] [PubMed] [Google Scholar]
- Sentenac A., Hall B., 1982. Yeast nuclear RNA polymerases and their role in transcription, pp. 561–606 The Molecular Biology of the Yeast Saccharomyces, edited by Strathern J., Jones E., Broach J. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sermwittayawong D., Tan S., 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25: 3791–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.-C., Green M. R., 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90: 615–624 [DOI] [PubMed] [Google Scholar]
- Shen W.-C., Bhaumik S. R., Causton H. C., Simon I., Zhu X., et al. , 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22: 3395–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R., 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26: 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J., Breeden L., 1993. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Keegan L. P., Ptashne M., 1984. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc. Natl. Acad. Sci. USA 81: 5951–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Brent R., Ptashne M., 1986. DNA binding is not sufficient for nuclear localization of regulatory proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 4763–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P., Sadler I., Osborne M. A., 1989. Yeast proteins that recognize nuclear localization sequences. J. Cell Biol. 109: 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Barnett J., Hannett N., Harbison C. T., Rinaldi N. J., et al. , 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708 [DOI] [PubMed] [Google Scholar]
- Sloan J. S., Dombek K. M., Young E. T., 1999. Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J. Biol. Chem. 274: 37575–37582 [DOI] [PubMed] [Google Scholar]
- Smale S. T., Kadonaga J. T., 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449–479 [DOI] [PubMed] [Google Scholar]
- Smith H. E., Driscoll S. E., Sia R. A., Yuan H. E., Mitchell A. P., 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133: 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D., 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Smith R. L., Redd M. J., Johnson A. D., 1995. The tetratricopeptide repeats of Ssn6 interact with the homeo domain of alpha 2. Genes Dev. 9: 2903–2910 [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Sorger P. K., 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62: 793–805 [DOI] [PubMed] [Google Scholar]
- Stebbins J. L., Triezenberg S. J., 2004. Identification, mutational analysis, and coactivator requirements of two distinct transcriptional activation domains of the Saccharomyces cerevisiae Hap4 protein. Eukaryot. Cell 3: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., et al. , 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Sterner D. E., Grant P. A., Roberts S. M., Duggan L. J., Belotserkovskaya R., et al. , 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. L., Cantin G. T., Wang G., Shevchenko A., Berk A. J., 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758 [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Dorland S., Yu Y., 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics 136: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., 1985. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. USA 82: 8419–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., 1989. Molecular mechanisms of transcriptional regulation in yeast. Annu. Rev. Biochem. 58: 1051–1077 [DOI] [PubMed] [Google Scholar]
- Struhl K., 1995. Yeast transcriptional regulatory mechanisms. Annu. Rev. Genet. 29: 651–674 [DOI] [PubMed] [Google Scholar]
- Struhl K., 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- Sugihara F., Kasahara K., Kokubo T., 2011. Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 39: 59–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Fujimoto T., Fukuma M., Yano K. I., Sakurai H., Vonika A., et al. , 1996. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol. Cell. Biol. 16: 2504–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Horz W., 1997. Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22: 93–97 [DOI] [PubMed] [Google Scholar]
- Swamy K. B., Cho C. Y., Chiang S., Tsai Z. T., Tsai H. K., 2009. Impact of DNA-binding position variants on yeast gene expression. Nucleic Acids Res. 37: 6991–7001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes D. J., 2010. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes D. J., Naar A. M., Andel F., III, Nogales E., Tjian R., 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295: 1058–1062 [DOI] [PubMed] [Google Scholar]
- Taatjes D. J., Schneider-Poetsch T., Tjian R., 2004. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 11: 664–671 [DOI] [PubMed] [Google Scholar]
- Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., et al. , 2005. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 25: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Biddick R., Law G. L., Young E. T., 2007. A poised initiation complex is activated by SNF1. J. Biol. Chem. 282: 37308–37315 [DOI] [PubMed] [Google Scholar]
- Takagi Y., Calero G., Komori H., Brown J. A., Ehrensberger A. H., et al. , 2006. Head module control of mediator interactions. Mol. Cell 23: 355–364 [DOI] [PubMed] [Google Scholar]
- Tan K., Feizi H., Luo C., Fan S. H., Ravasi T., et al. , 2008. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc. Natl. Acad. Sci. USA 105: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Richmond T. J., 1998. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391: 660–666 [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., et al. , 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34: D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J. K., Arthanari H., Yang F., Chau K. H., Wagner G., et al. , 2009. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284: 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden J. B., Sellick C. A., Reece R. J., Holden H. M., 2007. Understanding a transcriptional paradigm at the molecular level. The structure of yeast Gal80p. J. Biol. Chem. 282: 1534–1538 [DOI] [PubMed] [Google Scholar]
- Thomas D., Jacquemin I., Surdin-Kerjan Y., 1992. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1719–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. C., Chiang C. M., 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Thompson C. M., Koleske A. J., Chao D. M., Young R. A., 1993. A multisubunit complex associated with TATA binding protein and the RNA polymerase II CTD in yeast. Cell 73: 1367–1375 [DOI] [PubMed] [Google Scholar]
- Tompa P., Fuxreiter M., 2008. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33: 2–8 [DOI] [PubMed] [Google Scholar]
- Tora L., 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16: 673–675 [DOI] [PubMed] [Google Scholar]
- Tornow J., Zeng X., Gao W., Santangelo G. M., 1993. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 12: 2431–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth-Petroczy A., Oldfield C. J., Simon I., Takagi Y., Dunker A. K., et al. , 2008. Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol. 4: e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A. E., Miller M. G., Raisner R. M., Johnson A. D., 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115: 389–399 [DOI] [PubMed] [Google Scholar]
- Tsong A. E., Tuch B. B., Li H., Johnson A. D., 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443: 415–420 [DOI] [PubMed] [Google Scholar]
- Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D., 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M., Nyanguile O., Lu H., Levine A. J., Verdine G. L., 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277: 1310–1313 [DOI] [PubMed] [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T., van Leenen D., et al. , 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522 [DOI] [PubMed] [Google Scholar]
- van Heusden G. P., Steensma H. Y., 2006. Yeast 14–3-3 proteins. Yeast 23: 159–171 [DOI] [PubMed] [Google Scholar]
- Verdone L., Wu J., van Riper K., Kacherovsky N., Vogelauer M., et al. , 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W. P., Yu Y., Takahata S., Kretschmann K. L., Lieb J. D., et al. , 2007. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 26: 4324–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. S., Reese J. C., Apone L. M., Green M. R., 1996. Transcription activation in cells lacking TAFIIs. Nature 383: 185–188 [DOI] [PubMed] [Google Scholar]
- Wandinger S. K., Richter K., Buchner J., 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283: 18473–18477 [DOI] [PubMed] [Google Scholar]
- Wang D., Hu Y., Zheng F., Zhou K., Kohlhaw G. B., 1997. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3p. J. Biol. Chem. 272: 19383–19392 [DOI] [PubMed] [Google Scholar]
- Wang D., Zheng F., Holmberg S., Kohlhaw G. B., 1999. Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J. Biol. Chem. 274: 19017–19024 [DOI] [PubMed] [Google Scholar]
- Wang W., Carey M., Gralla J. D., 1992. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science 255: 450–453 [DOI] [PubMed] [Google Scholar]
- Wang X., Muratani M., Tansey W. P., Ptashne M., 2010. Proteolytic instability and the action of nonclassical transcriptional activators. Curr. Biol. 20: 868–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Feng L. S., Matskevich V., Venkataraman K., Parasuram P., et al. , 2006. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J. Mol. Biol. 357: 1167–1183 [DOI] [PubMed] [Google Scholar]
- Warfield L., Ranish J. A., Hahn S., 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 18: 1022–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L., Yue B., Veverka V., Renshaw P., Bramham J., et al. , 2006. Structural diversity in p160/CREB-binding protein coactivator complexes. J. Biol. Chem. 281: 14787–14795 [DOI] [PubMed] [Google Scholar]
- Weake V. M., Workman J. L., 2010. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11: 426–437 [DOI] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P., 1988. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell 52: 169–178 [DOI] [PubMed] [Google Scholar]
- Werner F., Grohmann D., 2011. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 9: 85–98 [DOI] [PubMed] [Google Scholar]
- Wightman R., Bell R., Reece R. J., 2008. Localization and interaction of the proteins constituting the GAL genetic switch in Saccharomyces cerevisiae. Eukaryot. Cell 7: 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E., Dietze P., Karas H., Knuppel R., 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24: 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R., 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39: 675–682 [DOI] [PubMed] [Google Scholar]
- Wu J., Suka N., Carlson M., Grunstein M., 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7: 117–126 [DOI] [PubMed] [Google Scholar]
- Wu P. Y., Winston F., 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22: 5367–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. Y., Ruhlmann C., Winston F., Schultz P., 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15: 199–208 [DOI] [PubMed] [Google Scholar]
- Wyrick J. J., Holstege F. C., Jennings E. G., Causton H. C., Shore D., et al. , 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
- Xiao H., Friesen J. D., Lis J. T., 1995. Recruiting TATA-binding protein to a promoter: transcription activation without an upstream activator. Mol. Cell. Biol. 15: 5757–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Wang J., Shen Z., Zhu H., 2004. Enrichment of transcriptional regulatory sites in non-coding genomic region. Bioinformatics 20: 569–575 [DOI] [PubMed] [Google Scholar]
- Xue-Franzen Y., Johnsson A., Brodin D., Henriksson J., Burglin T. R., et al. , 2010. Genome-wide characterisation of the Gcn5 histone acetyltransferase in budding yeast during stress adaptation reveals evolutionarily conserved and diverged roles. BMC Genomics 11: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. B., 2002. How do 14–3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513: 53–57 [DOI] [PubMed] [Google Scholar]
- Yang F., DeBeaumont R., Zhou S., Naar A. M., 2004. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 101: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T., Saario J., Kacherovsky N., Chao A., Sloan J. S., et al. , 1998. Characterization of a p53-related activation domain in Adr1p that is sufficient for ADR1-dependent gene expression. J. Biol. Chem. 273: 32080–32087 [DOI] [PubMed] [Google Scholar]
- Young E. T., Kacherovsky N., Van Riper K., 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 277: 38095–38103 [DOI] [PubMed] [Google Scholar]
- Young E. T., Dombek K. M., Tachibana C., Ideker T., 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278: 26146–26158 [DOI] [PubMed] [Google Scholar]
- Yu Y., Eriksson P., Bhoite L. T., Stillman D. J., 2003. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 23: 1910–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G. C., Liu Y. J., Dion M. F., Slack M. D., Wu L. F., et al. , 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., et al. , 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Sumibcay L., Hinnebusch A. G., Swanson M. J., 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24: 6871–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hach A., Wang C., 1998. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol. 18: 3819–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Burkett T. J., Yamashita I., Garfinkel D. J., 1997. Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 4718–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S., 2005. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 33: 2838–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhao H., Mancera E., Steinmetz L. M., Snyder M., 2010. Genetic analysis of variation in transcription factor binding in yeast. Nature 464: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F., 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. USA 106: 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. M., Bai Y. L., Kohlhaw G. B., 1990. Yeast regulatory protein LEU3: a structure-function analysis. Nucleic Acids Res. 18: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Byers K. J., McCord R. P., Shi Z., Berger M. F., et al. , 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]