Abstract
The acylation mechanism of a prototypical serine protease-trypsin and its complete free energy reaction profile have been determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling.
Serine proteases catalyze the cleavage of peptide bond in two successive acylation and deacylation stages, and have long been paradigms of enzyme catalysis1–4 and subjects for drug development.5–8 In spite of a number of experimental9–14 and computational studies15–24, a few key mechanistic details remain controversial, in particular regarding the tetrahedral intermediate (TI) of the acylation process.1, 13, 25, 26
The essential functional unit of conventional serine proteases is the catalytic triad (Ser195, His57, Asp102 in the case of trypsin), and a two-step mechanism has generally been accepted for the acylation reaction: first His57 serves as a general base to abstract a proton from Ser195 to facilitate the formation of a metastable tetrahedral intermediate (TI); and then His57 serves as a general acid to protonate the leaving group for the breaking of the TI.1–3 Since serine is a very weak acid and it is expected that His57 needs to be ideally positioned to deprotonate it, one intriguing question is how the proton of His57 at the intermediate state transfers to the leaving group instead of re-protonating serine. A reaction-driven His-ring flip mechanism has been proposed as one solution of this dilemma,12, 13, 16 in which the ring of His57 is flipped at the TI to facilitate the protonation of the leaving group. However, recent crystal structural studies9, 14 provided evidences against it and suggested an economy of atomic motions during the acylation process. Meanwhile, based on the stereo-electronic consideration, it has been suggested that inversion of leaving nitrogen of the amine group takes place at the TI to favor the proton transfer to the leaving group and the cleavage of the C-N bond.11, 26, 27 To unambiguously elucidate this interesting mechanistic puzzle regarding the acylation tetrahedral intermediate has important implications in understanding serine protease substrate specificity as well as inhibition mechanisms, but is very difficult to be achieved by experimental means alone.
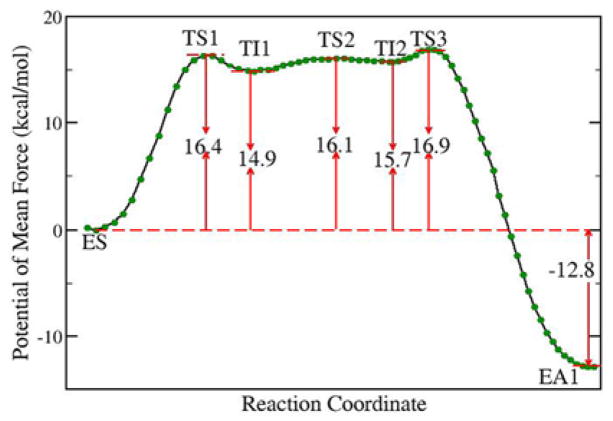
Herein our theoretical investigations center on Born-Oppenheimer ab initio QM/MM molecular dynamics with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions.28–34 It provides a first-principle description of chemical reaction and dynamics of the enzyme active site while properly taking account of interactions and fluctuations of the heterogeneous enzyme environment, and thus overcomes several inherent limitations in previous computational investigations of serine proteases. The initial structure of the substrate-enzyme complex was modeled based on the x-ray crystal structure of trypsin-inhibitor complex (PDB ID: 1MCT).35 The sequence of the substrate was taken from the inhibitor at the active site (Cys3-Pro4-Arg5-Ile6-Trp7-Met8), and the scissile bond is between Arg5 and Ile6. The QM sub-system, including the side chains of catalytic triad (His57, Asp102, and Ser195) and the scissile peptide portion of the substrate, was described at the B3LYP/6-31+G* level. The QM/MM boundary was treated by the pseudobond approach36–38 with the improved parameters39. Along the reaction path, 41 umbrella windows were employed. For each window, at least 30 ps B3LYP/6-31+G* QM/MM MD simulations have been carried out. The determined complete free energy reaction profile for the trypsin acylation reaction is shown in Fig. 1. The calculated overall activation barrier is 16.9 kcal/mol, in good agreement with the experimental value of 15~20 kcal/mol depending on substrates and experimental conditions.2
Fig. 1.
Free energy profile for the trypsin acylation reaction stage determined by B3LYP/6-31+G* QM/MM molecular dynamics simulations and umbrella sampling.
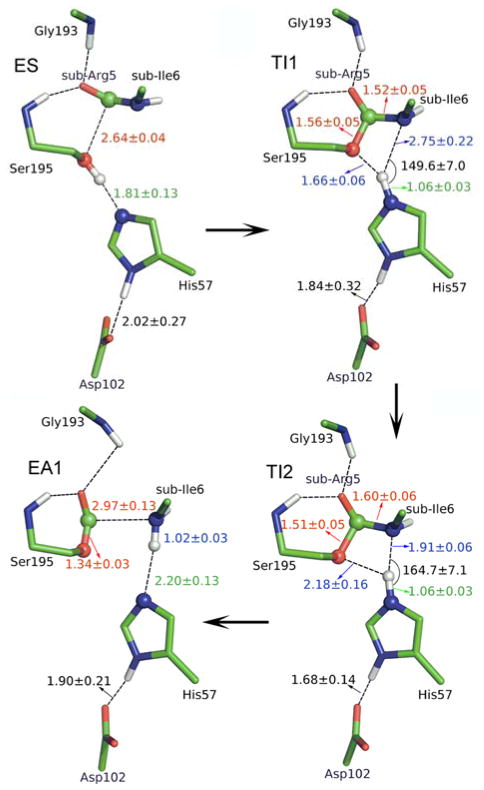
Our characterized acylation reaction proceeds in two chemical steps interspersed with one subtle His-ring reorientation step between two distinct TI configurations, as illustrated in Fig. 2. In the initial chemical step, His57 acts as a general base, facilitating the nucleophilic attack of Ser195 to the carbonyl carbon of the substrate Arg5 to form a metastable tetrahedral intermediate TI1.
Fig. 2.
Illustration of the structures for characterized acylation stage. ES, enzyme-substrate complex; TI1 and TI2 are two distinct tetrahedral intermediate configurations; EA1, acyl-enzyme.
At the TI1, we can see that the protonated His57 is indeed still close to OG atom of Ser195 and forms a stable hydrogen bond with an O…H distance at 1.66 ± 0.06 Å, while the proton is far from the leaving N of the scissile bond with an N…H distance at 2.75 ± 0.22 Å. In order to favor the forward progress of the reaction, our simulations indicate that the enzyme only needs a subtle re-orientation of the His57 ring to yield a second meta-stable tetrahedral intermediate TI2, in which the proton of His57 is well positioned to protonate the nitrogen of the leaving group, with an N…H distance at 1.91 ± 0.06 Å and a donor–hydrogen–acceptor angle at 164.7 ± 7.1°. Meanwhile, the subtle movement of His ring is accompanied with a slight change in the newly formed C-O bond and the C-N scissile bond. From TI1 to TI2, the distance for the newly formed C-O bond decreases from 1.56 ± 0.05 Å to 1.51 ± 0.05 Å, while the scissile bond increase from 1.52 ± 0.05 Å to 1.60 ± 0.06 Å. This indicates the C-O bond becomes a little stronger and the scissile bond C-N has elongated somewhat prior to the bond breaking. At TS3 state, the scissile bond is already extended to 1.81 ± 0.14 Å, with the distance between proton and N of His57 at 1.14 ± 0.05 Å, which means that the scissile bond breaks first before the proton transfer. Finally, the proton transfers to the leaving N atom, and the C–N bond breaks leading to a stable acyl-enzyme intermediate (EA1). We can see that the first chemical step is rate-determining, while the barriers for second and third steps are both quite small, only about 1.2 kcal/mol. Consistent with the determined free energy profile, unrestrained ab initio QM/MM MD simulations also indicate that both intermediates can be meta-stable for a few picoseconds. The free energy of the resulted acyl-enzyme (EA1) is 12.8kcal/mol lower than that of the enzyme-substrate (ES), indicating that the trypsin acylation process is exothermic and irreversible.
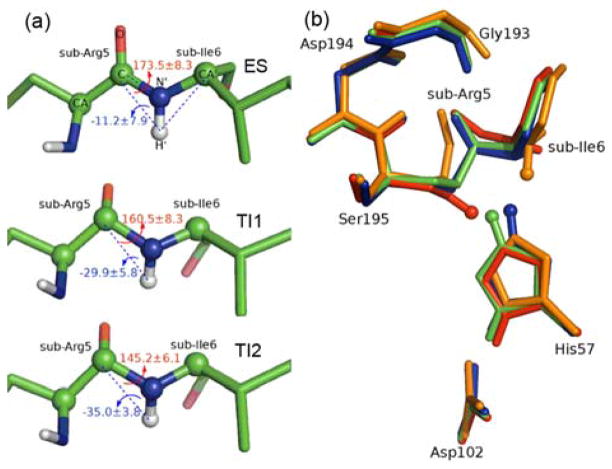
From our characterized reaction pathway, we can see that the His-ring flip mechanism is not needed to facilitate the protonation of the leaving group. Instead, the change between two distinct TI configurations only involves a subtle reorientation of the histidine ring. In order to examine other hypotheses that the nitrogen-inversion11, 26, 27 or rotation of the leaving group around C-N bond11, 19 takes place at the tetrahedral intermediate (TI) to favor the forward reaction, we have also calculated the improper angle τ and torsion angle ω (see Fig. 3a). Along the whole reaction path, the change of both torsion angles are quite small. The improper angle τ is always smaller than 0°, indicating that the scissile bond keeps in an S configuration and no nitrogen-inversion happens in the acylation process. For the ω angle, the difference between TI1 and TI2 is just about 15°, indicating a very subtle rotation of the scissile bond. Overall, these results support the recent hypothesis9, 14 that serine protease acylation proceeds with a remarkable economy of motion, which can be further demonstrated by a close superposition of four structures of trypsine active site along our characterized reaction pathway: ES, TI1, TI2 and EA1, as shown in Fig. 3b.
Fig. 3.
a) improper angle τ and torsion angle ω for substrate at ES, TI1 and TI2 states. Improper angle τ: N′-C-H′-CA′ is employed to indicate the inversion of leaving N, while torsion angle ω: CA-C-N′-CA′ is defined to control the rotation of NH group around the scissile bond. b) Superposition of structures of active site region for ES (in red), TI1 (in green), TI2 (in blue) and EA1 (in orange) in stick mode. The HG atom of Ser195 is also presented in sphere mode.
For the hydrogen bond between His57 and Asp102, it becomes somewhat shorter at transition states and intermediates than at ES and EA1 states, indicating a stronger interaction due to the positive charge developing on His ring. However, throughout the reaction process, the average N-O distance between His57 and Asp102 is longer than 2.7 Å (see Table S1), indicating a normal hydrogen bond. Meanwhile, no spontaneous proton transfer between His57 and Asp102 has been observed in our ab initio QM/MM MD simulations. Thus our results provide further evidences against the charge-relay mechanism40, 41 and the LBHB mechanism.42–45, and support that the main catalytic role of Asp102 is to stabilize the positive charge on the imidazole ring of His57 at TS and TI states through electrostatic interactions.15, 17, 46
In summary, we have characterized the acylation reaction of trypsin in detail with a state-of-the-art computational approach. The simulation results indicate that the position of His57 ring only needs to be adjusted slightly during the reaction. The subtle re-orientation of the His ring at the tetrahedral intermediate serves as a bridge connecting two chemical steps: first His57 acts as a general base to accept a proton from Ser195, and then acts as a general acid to protonate the amine leaving group. This proton shuttle by the histidine proceeds with an economy of atomic motion, and does not need either the His-ring flip or the nitrogen-inversion of the amine leaving group.
Supplementary Material
Acknowledgments
We are grateful for the support of National Institute of Health (R01-GM079223), and National Science Foundation (CHE-CAREER-0448156, TeraGrid resources provided by NCSA and LONI). We also thank NYU-ITS for providing computational resources and Dr. Shenglong Wang for computing support.
Footnotes
Electronic Supplementary Information (ESI) available: [Computational details, Figures S1–S3, and Table S1]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Hedstrom L. Chem Rev (Washington, DC, U S) 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 2.Fersht A. A Guide to Enzyme Catalysis and Protein Folding. 2. W. H. Freeman and Company; New York: 1999. Structure and Mechanism in Protein Science. [Google Scholar]
- 3.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5. W. H. Freeman and company; New York: 2002. [Google Scholar]
- 4.Page MJ, Di Cera E. Cell Mol Life Sci. 2008;65:1220–1236. doi: 10.1007/s00018-008-7565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turk B. Nature Reviews Drug Discovery. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 6.Stoller JK, Aboussouan LS. Lancet. 2005;365:2225–2236. doi: 10.1016/S0140-6736(05)66781-5. [DOI] [PubMed] [Google Scholar]
- 7.Steinmetzer T, Sturzebecher J. Curr Med Chem. 2004;11:2297–2321. doi: 10.2174/0929867043364540. [DOI] [PubMed] [Google Scholar]
- 8.Leung D, Abbenante G, Fairlie DP. J Med Chem. 2000;43:305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- 9.Zakharova E, Horvath MP, Goldenberg DP. Proc Natl Acad Sci U S A. 2009;106:11034–11039. doi: 10.1073/pnas.0902463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrmann CN, Daugherty MD, Agard DA. J Am Chem Soc. 2006;128:9086–9102. doi: 10.1021/ja057721o. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Schofield CJ, Wilmouth RC. J Biol Chem. 2006;281:24024–24035. doi: 10.1074/jbc.M600495200. [DOI] [PubMed] [Google Scholar]
- 12.Haddad KC, Sudmeier JL, Bachovchin DA, Bachovchin WW. Proc Natl Acad Sci U S A. 2005;102:1006–1011. doi: 10.1073/pnas.0409279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ash EL, Sudmeier JL, Day RM, Vincent M, Torchilin EV, Haddad KC, Bradshaw EM, Sanford DG, Bachovchin WW. Proc Natl Acad Sci U S A. 2000;97:10371–10376. doi: 10.1073/pnas.97.19.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radisky ES, Lee JM, Lu CJK, Koshland DE. Proc Natl Acad Sci U S A. 2006;103:6835–6840. doi: 10.1073/pnas.0601910103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warshel A, Naray-Szabo G, Sussman F, Hwang JK. Biochemistry. 1989;28:3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- 16.Scheiner S. J Phys Chem B. 2008;112:6837–6846. doi: 10.1021/jp710617w. [DOI] [PubMed] [Google Scholar]
- 17.Ishida T. Biochemistry. 2006;45:5413–5420. doi: 10.1021/bi051515b. [DOI] [PubMed] [Google Scholar]
- 18.Ishida T, Kato S. J Am Chem Soc. 2004;126:7111–7118. doi: 10.1021/ja030405u. [DOI] [PubMed] [Google Scholar]
- 19.Ishida T, Kato S. J Am Chem Soc. 2003;125:12035–12048. doi: 10.1021/ja021369m. [DOI] [PubMed] [Google Scholar]
- 20.Topf M, Richards WG. J Am Chem Soc. 2004;126:14631–14641. doi: 10.1021/ja047010a. [DOI] [PubMed] [Google Scholar]
- 21.Topf M, Varnai P, Richards WG. Theor Chem Acc. 2001;106:146–151. [Google Scholar]
- 22.Topf M, Varnai P, Richards WG. J Am Chem Soc. 2002;124:14780–14788. doi: 10.1021/ja026219q. [DOI] [PubMed] [Google Scholar]
- 23.Perakyla M, Kollman PA. J Am Chem Soc. 2000;122:3436–3444. [Google Scholar]
- 24.Nemukhin AV, Grigorenko BL, Rogov AV, Topol IA, Burt SK. Theor Chem Acc. 2004;111:36–48. [Google Scholar]
- 25.Polgar L. Cell Mol Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bizzozero SA, Dutler H. Bioorg Chem. 1981;10:46–62. [Google Scholar]
- 27.Dutler H, Bizzozero SA. Acc Chem Res. 1989;22:322–327. [Google Scholar]
- 28.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke Z, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Hu P, Zhang Y. J Phys Chem B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Wang S, Zhang Y. J Phys Chem B. 2010;114:8817–8825. doi: 10.1021/jp104258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu R, Wang S, Zhou N, Cao Z, Zhang Y. J Am Chem Soc. 2010;132:9471–9479. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang QC, Liu SP, Tang YQ. J Mol Biol. 1993;229:1022–1036. doi: 10.1006/jmbi.1993.1102. [DOI] [PubMed] [Google Scholar]
- 36.Zhang YK. Theor Chem Acc. 2006;116:43–50. [Google Scholar]
- 37.Zhang YK, Lee TS, Yang WT. J Chem Phys. 1999;110:46–54. [Google Scholar]
- 38.Zhang YK, Liu HY, Yang WT. J Chem Phys. 2000;112:3483–3492. [Google Scholar]
- 39.Zhang YK. J Chem Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 40.Blow DM, Birktoft JJ, Hartley BS. Nature. 1969;221:337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 41.Blow DM. Trends Biochem Sci. 1997;22:405–408. doi: 10.1016/s0968-0004(97)01115-8. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Westler WM, Cleland WW, Markley JL, Frey PA. Proc Natl Acad Sci U S A. 1998;95:14664–14668. doi: 10.1073/pnas.95.25.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fodor K, Harmat V, Neutze R, Szilagyi L, Graf L, Katona G. Biochemistry. 2006;45:2114–2121. doi: 10.1021/bi0517133. [DOI] [PubMed] [Google Scholar]
- 44.Guthrie JP. Chem Biol. 1996;3:163–170. doi: 10.1016/s1074-5521(96)90258-6. [DOI] [PubMed] [Google Scholar]
- 45.Perrin CL, Nielson JB. Annu Rev Phys Chem. 1997;48:511–544. doi: 10.1146/annurev.physchem.48.1.511. [DOI] [PubMed] [Google Scholar]
- 46.Warshel A, Papazyan A, Kollman PA. Science. 1995;269:102–104. doi: 10.1126/science.7661987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.