Abstract
The proteins produced by the herpes simplex virus type 1 (HSV-1) genes UL15 and UL28 are believed to form part of the terminase enzyme, a protein complex essential for the cleavage of newly synthesized, concatameric herpesvirus DNA and the packaging of the resultant genome lengths into preformed capsids. This work describes the purification of recombinant forms of pUL15 and pUL28, which allowed the calculation of the average number of copies of each protein in A and B capsids and in capsids lacking the putative portal encoded by UL6. On average, 1.0 (±0.29 [standard deviation]) copies of pUL15 and 2.4 (±0.97) copies of pUL28 were present in B capsids, 1.2 (±0.72) copies of pUL15 and 1.5 (±0.86) copies of pUL28 were found in mutant capsids lacking the putative portal protein pUL6, and approximately 12.0 (±5.63) copies of pUL15 and 0.6 (±0.32) copies of pUL28 were present in each A capsid. These results suggest that the packaging machine is partly comprised of approximately 12 copies of pUL15, as found in A capsids, with wild-type B and mutant UL6(−) capsids containing an incomplete complement of cleavage and packaging proteins. These results are consistent with observations that B capsids form by default in the absence of packaging machinery in vitro and in vivo. In contrast, A capsids may be the result of initiated but aborted attempts at DNA packaging, resulting in the retention of at least part of the DNA packaging machinery.
During herpesvirus replication, branched concatemers of viral DNA are produced in the nucleus of an infected cell (47). Individual viral genomes are cleaved from the concatemer, and each is packaged into a preformed capsid (reviewed in references 9 and 26). This assembly step involves extensive interactions between the capsid, DNA, and the packaging machinery.
Four forms of herpesvirus intranuclear capsid have been described (22, 37). Procapsids, believed to be the precursors of all capsid types, are spherical structures containing an inner shell or scaffold consisting largely of the protein VP22a. During the packaging reaction, VP22a is cleaved by the UL26 protease, releasing it from the inside surface of the outer shell (19, 33, 45, 59). This dissociation of the scaffold coincides with a dramatic conformational change in the outer shell, which eventually forms a stable icosahedron (24, 57). Each procapsid is thereby converted into one of three angular capsid types. Type A capsids contain only the icosahedral shell, B capsids contain cleaved scaffold material within the outer shell, and C capsids contain packaged DNA in place of the cleaved scaffold (22).
All three mature forms of capsids contain outer shells consisting of 150 hexons and 12 pentons forming a T=16 structure (62). The pentons and hexons are made up of five and six copies, respectively, of VP5, the product of UL19; thus, each capsid contains 960 copies of VP5. Hexons and pentons are linked by triplexes which consist of one copy of VP19C (encoded by UL38) and two copies of VP23 (encoded by UL18) (14, 17, 22, 39, 66). Only C capsids exit the cell nucleus to become enveloped virions (46). One interpretation from previous studies of the different capsid types (35, 37, 52, 57) is that A capsids result from packaging reactions in which the DNA is inserted but not retained, whereas B capsids represent a mismatch in timing such that the formation of the impervious icosahedral shell precludes exit of the cleaved scaffold proteins.
Many DNA viruses use similar mechanisms to cleave genome-length DNA from concatemers and package it into preformed capsids. A model deduced from studies of double-stranded DNA bacteriophages predicts that in cells infected with herpesviruses, the newly synthesized viral DNA is transported to the empty procapsid by the terminase, an enzyme that specifically binds genomic ends and cleaves the DNA. The terminase, with bound DNA, is believed to attach to the portal protein which comprises a unique pore at one of the five fold axes of the capsid. The DNA is then translocated through the portal into the capsid using the ATPase and helicase activities of the terminase (reviewed in reference 12).
Six genes are known to be essential for the cleavage and packaging of the herpesvirus genome: UL6, UL15, UL17, UL28, UL32, and UL33. Deletion of any of these six genes precludes cleavage of viral DNA and results in the accumulation of mutant B capsids in the nuclei of infected cells (2, 3, 5, 6, 15, 31, 32, 41, 43, 49, 52, 55, 56, 60, 64).
There is increasing indirect evidence that the product of UL6 encodes the portal protein, and the UL15 and UL28 proteins comprise terminase subunits. The UL6 gene product (pUL6) has been localized to one vertex of the herpesvirus B capsid, where 14.8 ± 2.6 copies (mean ± standard deviation) of the protein were calculated to be present (38). Moreover, when purified from recombinant baculovirus-infected insect cells and solubilized in 1 M arginine, pUL6 forms rings with a mass corresponding to an oligomeric state of 12 (38), consistent with previously described bacteriophage portals or connector proteins (34, 53, 58). The UL15 gene shares limited homology with a nucleotide binding motif found in a number of bacteriophage terminase components, including gp17, the large subunit of the T4 bacteriophage (16, 36). In addition, both pUL15 and gp17 are susceptible to N-terminal cleavage (10, 48, 50). The UL15 protein is found in wild-type B capsids in three forms—a full-length 83-kDa protein and two N-terminally cleaved forms that migrate at 80 and 79 kDa. However, only the full-length form is detected in cells infected with viral mutants incompetent for DNA cleavage and packaging, suggesting that the N-terminal cleavage of pUL15 is linked to the cleavage and packaging process. The UL28 protein, when overexpressed and purified from Escherichia coli cells, has been shown to bind specifically to the pac1 sequence in herpes simplex virus type 1 (HSV-1) DNA, which is essential for the generation of correct genomic termini (4, 25). In addition, several studies have shown that pUL15 and pUL28 interact in vitro and in infected and uninfected cells (1, 11, 29, 30).
The association of pUL6, pUL15, and pUL28 with different types of capsids has supported the putative functions described above. The UL6 protein has been detected in procapsids, and similar amounts are present in B capsids and C capsids (41, 51, 65), indicating that it is an integral component of the capsid shell. The putative terminase components pUL15 and pUL28 are also present in procapsids (51); however, smaller amounts of these two proteins have been detected in C capsids than in B capsids (48, 54, 65). The reduction in the amount of pUL15 and pUL28 in C capsids is consistent with the behavior exhibited by bacteriophage terminase subunits that disengage from the capsid after packaging is complete. The amount of pUL15 has been reported to be smaller in capsids lacking pUL6 or pUL28 than in wild-type B capsids (50, 65), and recombinant forms of pUL15 and pUL28 have been shown to interact independently with each other and with pUL6 in vitro (61), suggesting that the ability of the UL15 protein to bind to B capsids is mediated through interactions with the UL6 and UL28 proteins.
In these studies, a stoichiometric approach was used to investigate the structure of the HSV packaging machine and to clarify the association of the putative terminase subunits with the presumed portal vertex. Histidine-tagged UL15 and UL28 proteins were purified and used as standards to calculate the number of copies of the UL15 and UL28 proteins in wild-type A and B capsids and in capsids lacking the UL6-encoded portal.
MATERIALS AND METHODS
Purification of the UL28 protein.
The UL28 open reading frame (ORF), excluding the stop codon and including a C-terminal six-histidine tag, was cloned into the pBakPAK8 vector (Clontech). Using the manufacturer's protocol, the DNA was used to generate a recombinant baculovirus that contained tagged UL28 under the control of the baculovirus polyhedron promoter. SF-21 insect cells were infected with the recombinant baculovirus at a multiplicity of infection of 5, and the cells were harvested 44 h later, pelleted by centrifugation, and stored at −20°C until use.
Approximately 3 × 107 cells were left on ice for 15 min to thaw and then resuspended in lysis buffer (50 mM Tris HCl [pH 8], 50 mM KCl, 10% glycerol, 5 mM β-mercaptoethanol) and EDTA-free Complete protease inhibitors (Roche Applied Science, Indianapolis, Ind.). The cells were lysed by Dounce homogenization, and the insoluble material was pelleted by centrifugation at 10,000 rpm in a JA-14 rotor for 20 min at 4°C. The pellet was resuspended in lysis buffer plus 6 M guanidine hydrochloride (GuHCl) and mixed slowly with a magnetic stirrer at 4°C for 90 min. After clarification at 12,000 rpm for 15 min in a JA-14 rotor at 4°C, the supernatant was collected and added to preequilibrated Ni-nitrilotriacetic acid beads (Qiagen) and the sample was rotated slowly at 4°C for 1 h. The beads were then washed extensively in lysis buffer and 6 M GuHCl. The GuHCl was then diluted by the addition of lysis buffer, and the beads with bound UL28 protein were washed sequentially in 20 mM imidazole and 50 mM imidazole and finally eluted three times with 0.75 ml of lysis buffer plus 1 M imidazole. The eluates were dialyzed extensively against storage buffer containing 50 mM Tris HCl (pH 8.0), 200 mM KCl, and protease inhibitors and stored at 4°C. The concentration of protein was calculated using a bicinchoninic acid (BCA) kit (Pierce Biotechnology Inc., Rockford, Ill.) and confirmed by electrophoretic separation on a denaturing polyacrylamide gel containing known amounts of bovine serum albumin (BSA) followed by fixation, staining with Coomassie blue, and densitometry.
Purification of the UL15 protein.
Plasmid pJB278 was cut with EcoRV and HindIII, allowing the isolation of a 2.2-kbp fragment containing the entire UL15 ORF tagged with six-histidine codons at the 3′ end. A kanamycin-resistant vector, pET30b (Novagen), was cut with NdeI and EcoRV and ligated to remove extraneous sequences in the vector. A blunt end was introduced into the multiple cloning site of this altered vector by cutting with BamHI and filling in with T4 polymerase. The vector was then cut with HindIII, and the 2.2-kbp EcoRV/HindIII fragment from plasmid pJB278 was inserted such that expression of the tagged UL15 ORF was driven by the T7 promoter within the vector. The plasmid was designated pJB279. Plasmid DNA was transfected into BL-21+ λDE3 codon plus bacteria (Stratagene) and grown in Luria-Bertani medium supplemented with kanamycin to an optical density of 0.6, and protein production was induced by adding 5 mM isopropyl-β-d-thiogalactoside (IPTG) and incubating for 2 h at 37°C. The cells were then harvested and frozen at −80°C. Cells from a 500-ml culture were resuspended in 100 ml of lysis buffer (50 mM Tris [pH 7.9 to 8], 50 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol) and EDTA-free Complete protease inhibitors, Dounce homogenized, and left on ice for 45 min. Insoluble material was pelleted by centrifugation at 8,000 rpm in a JA-14 rotor for 15 min at 4°C. The pellet was resuspended in 50 ml of lysis buffer, left on ice for 15 min, and centrifuged for 20 min in a JA-14 rotor at 10,000 rpm at 4°C. The pellet was resuspended in 50 ml of denaturation buffer (lysis buffer supplemented with 6 M GuHCl) and stirred at 4°C for 90 min. The solution was then centrifuged at 12,000 rpm for 15 min in a JA-14 rotor at 4°C, and the subsequent supernatant was passed through a 0.8-μm filter. The filtrate was added to preequilibrated Ni-nitrilotriacetic acid beads, and the sample was rotated slowly at 4°C for 1 h. The beads were washed three times in denaturation buffer with 1% Tween 20 and increasing amounts of imidazole, to a final concentration of 20 mM, to remove contaminants and then renatured by gradual dilution of the GuHCl. Protein was eluted from the beads by two separate elution steps using lysis buffer plus 1 M imidazole and dialysis against 50 mM Tris (pH 7.9 to 8), 100 mM NaCl, 50% glycerol, 1 mM dithiothreitol, 0.25 mM EDTA, 0.5% Tween 20, and 5 mM l-arginine. Proteins were stored at −20°C.
Viruses and cells.
The G33 cell line is derived from Vero cells and contains HSV-1 DNA from UL6 to UL8 (41). G5 cells were transformed from Vero cells and contain HSV-1 DNA from UL16 to UL21 (18). Vero, G33, and G5 cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum and penicillin and streptomycin. The HSV-1(F) virus has been previously described (20), and the titer was determined on Vero cell monolayers. Virus CosUL6- was derived from HSV-1 strain 17 (HSV-1[17]) and contains a 4-bp insertion at a site corresponding to amino acid residue 381 (41). It was grown and the titer was determined on G33 cells. The K23Z virus contains a lacZ cassette in the UL18 ORF (18) and was grown and titers were determined on G5 cells. All CosUL6- and K23Z viral stocks were tested for revertant virus by titration on nonrescuing Vero cells. Only stocks with titers on rescuing cells greater than 10,000-fold over those obtained on Vero cells were used for further studies.
Capsid purification and analysis.
Capsids were purified as described previously with some modifications (42). Ten 175-cm2 flasks of confluent Vero cells were infected at a multiplicity of infection of 5, followed by a 16-h incubation at 37°C. The cells were then lysed in 1% Triton X and sonicated for 40 s at moderate power. After clarification for 15 min at 8,000 rpm at 4°C (7,227 × g) in a Sorvall Legend RT centrifuge, Heraeus rotor no. 3334, the supernatant was pelleted through a 6-ml 35% (wt/vol) sucrose cushion in a Beckman SW28.1 ultracentrifuge rotor for 1.5 h at 20,000 rpm. The pellet was then sonicated briefly and loaded on a 20-to-50% (wt/vol) continuous sucrose gradient and centrifuged for 1 h at 23,000 rpm in an SW41 rotor. The light-scattering bands were collected with a Pasteur pipette and pelleted at 30,000 rpm for 2 h in an SW41 rotor. Examination of the pelleted material by electron microscopy identified significant material inconsistent with capsid morphology (results not shown), so the resuspended material was further purified by centrifugation through a second continuous sucrose gradient. The capsids were collected again, pelleted, and resuspended in 50 μl of water (DNase-, RNase-, and protease-free; Acros) and stored at 4°C. Examination of the material by electron microscopy showed abundant capsids with minimal background material. All capsids were used within 24 h of purification. Where mentioned, the second sucrose gradient was fractioned using a fractioning device (Haake Buchler) beginning at the top of the gradient.
Protein quantification.
Capsids were resuspended in a buffer containing sodium dodecyl sulfate and β-mercaptoethanol, boiled for 3 min, and loaded on two denaturing 8% polyacrylamide gels that were run in parallel. BSA standards were included in one gel to allow calculation of the number of capsids present in the sample (see Results). The other gel contained either purified pUL15 or pUL28 to allow calculation of the amount of this protein in the capsid sample. The BSA-containing gel was stained with Coomassie blue and digitally scanned, and the band intensity was determined using Scion Image densitometry software for Windows. Initially, both the VP5 and VP19C protein bands were used to estimate the number of capsids present, and the data were compared. Similar results were obtained from both calculations, and the VP5 protein was used for all further calculations. The proteins on the second gel were transferred to either polyvinylidene difluoride (PVDF) or nitrocellulose paper and immunoblotted as previously described (8). Bands were visualized using either ECF (Amersham Bioscience) or ECL+ (Amersham Bioscience) development reagents, and chemiluminescence intensities were quantified using a Molecular Dynamics Storm Imager and associated ImageQuant software. The 83,000, 80,000, and 79,000 Mr pUL15 bands were included in the quantification.
The data were analyzed by a two-tailed t test, using Statistix 7 (Analytical Software, Tallahassee, Fla.).
Antibodies.
The UL15 protein was identified using either pUL15-specific rabbit polyclonal antisera, generated by immunization with an affinity-purified bacterial fusion protein (UL15-MBP) containing the malE gene product fused to the protein encoded by the majority of UL15 exon II (7), or rabbit polyclonal antisera recognizing the N-terminal 35 amino acids of pUL15 (50). The rabbit polyclonal antisera directed against the UL28 protein has been described previously (11).
RESULTS
Purification of pUL15 and pUL28.
The capsid shell components are present in more than 300 copies per capsid; however, the much lower copy numbers of pUL15 and pUL28 have precluded direct methods to quantify these proteins in samples of purified capsids. For example, bands specific for pUL15 and pUL28 are not readily distinguishable from ICP5 degradation products in profiles of Coomassie-stained electrophoretically separated capsid proteins (data not shown). Calculation of the amounts of these proteins in capsids therefore required more indirect methods. The strategy was to generate standard curves of purified pUL15 and pUL28 by subjecting known amounts of purified proteins to immunoblot analyses with specific antibodies. Thus, the intensity of a given band on an immunoblot would correspond to a known amount of purified protein, and the amounts in unknown samples (such as electrophoretically separated capsid proteins) could be calculated.
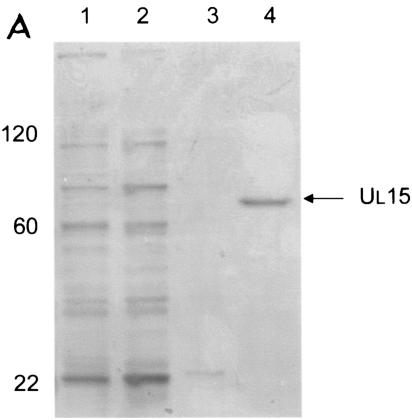
To purify pUL15, the UL15 ORF under the control of the T7 promoter was cloned in frame with a six-histidine tag such that the tag was inserted at the C terminus. Protein production was induced in E. coli by addition of IPTG to the culture medium, and induced protein was solubilized in GuHCl. The protein was then purified by affinity chromatography using Ni+ beads, followed by dilution of the GuHCl and eventual elution in imidazole (see Materials and Methods for details.) A representative purification is shown in Fig. 1A. Approximately 10 μg of protein was purified from 500 ml of an induced culture.
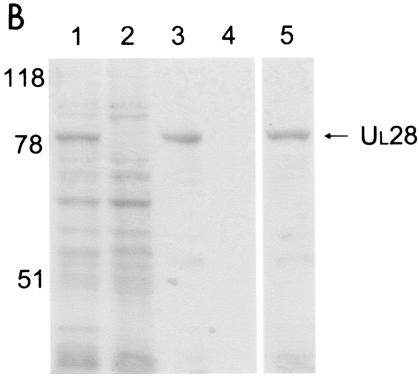
FIG. 1.
Scanned digital image of Coomassie-stained gels showing protein profiles from relevant steps of the purification of UL15 (A) and UL28 (B) proteins. (A) Lane 1, initial cell lysate; lane 2, supernatant after lysis; lane 3, supernatant from wash of insoluble material; lane 4, eluted protein after dialysis. (B) Lane 1, initial cell lysate; lane 2, supernatant after lysis; lane 3, Ni-nitrilotriacetic acid agarose beads after incubation with the insoluble, denatured protein fraction and extensive washing; lane 4, supernatant from wash of agarose beads; lane 5, eluted protein after dialysis. Molecular mass standards are indicated on the left in kilodaltons.
A similar strategy was used to purify the pUL28 protein, except that the C-terminally histidine-tagged protein was expressed in insect cells by infection with a recombinant baculovirus expressing the tagged protein. The recombinant protein was then purified by solubilization of inclusion bodies with GuHCl followed by affinity chromography on Ni+-containing beads and elution in imidazole. A representative purification is shown in Fig. 1B. The procedure yielded approximately 120 μg of purified protein per 2 × 107 insect cells.
Quantification of UL15 and UL28 in HSV-1(F) B capsids.
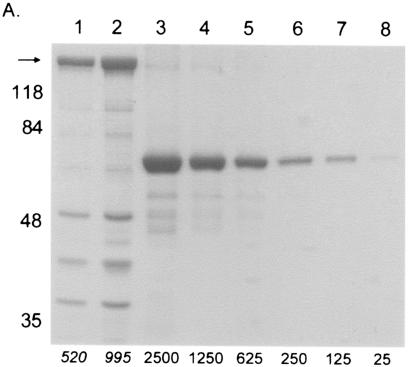
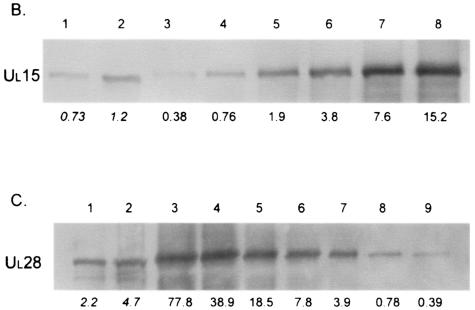
As viewed by examination of each capsid purification step by electron microscopy, it was necessary to use two successive sucrose gradients to generate highly purified B capsids. The number of capsids present in a given sample was then determined by calculating the amount of VP5 protein on a Coomassie-stained gel, using a standard curve of known amounts of BSA protein (Fig. 2A). The amount of UL15 or UL28 protein in a known amount of capsids was calculated from immunoblots by comparing the intensity of the band in a sample of the capsids to a standard curve of purified pUL15 or pUL28 (Fig. 2B and C).
FIG. 2.
Data used for calculating the average number of copies of pUL15 or pUL28 in capsids. (A) Coomassie-stained gel with B capsids (lanes 1 and 2) and BSA standards (lanes 3 to 8). The amount of BSA (in nanograms) loaded is indicated at the bottom of lanes 3 to 8. The R2 value of the graph produced by analysis of the data was 0.98, and the estimated amount of VP5 (arrow) is indicated in italics below lanes 1 and 2. The positions of molecular weight standards are indicated on the left. (B) Immunoblot of B capsids (lanes 1 and 2) and purified UL15 protein standards (lanes 3 to 8) probed with antisera directed against pUL15. The amount of pUL15 (in nanograms) is indicated below each lane. (C) Immunoblot probed with antisera directed against pUL28. B capsids are in lanes 1 and 2, and purified UL28 protein standards are in lanes 3 to 9. The amount of pUL28 (in nanograms) is indicated below each lane.
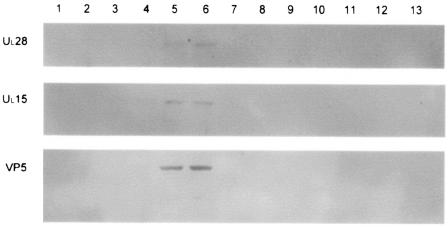
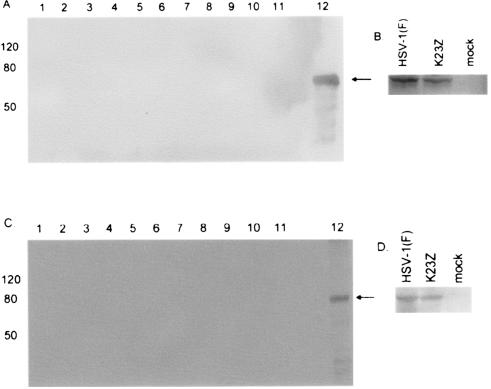
To ensure that pUL15 and pUL28 were associated with capsids and that background levels within the gradients did not affect the calculations, a representative second gradient was fractionated and each fraction was split between three separate gels that were then probed with antibodies directed against either VP5, pUL28, or pUL15 (Fig. 3). All three proteins were present only in fractions 5 and 6, the fractions corresponding to the light-scattering B capsid band seen on the sucrose gradient. As a further control, a capsid purification procedure was also carried out using lysates from cells infected with K23Z in parallel with HSV-1(F)-infected cells. The K23Z virus has a lacZ construct replacing most of the UL18 gene and is therefore unable to assemble capsids (18). No light-scattering bands were visible in the first K23Z sucrose gradient; however, the area corresponding to the A and B capsid band on the HSV-1(F) gradient was collected and run on a second sucrose gradient. This gradient was fractionated, the fractions were separated on a denaturing sodium dodecyl sulfate-polyacrylamide gel, and the proteins were transferred to PVDF. The membranes were then probed with antibodies directed against either pUL15 or pUL28, and the blot was intentionally overdeveloped (Fig. 4). No pUL15 or pUL28 was detected in any fraction; therefore, background levels of immunoreactivity as a consequence of pUL15 or pUL28 within the gradients would not be expected to contribute to immunoreactivity within capsid-containing fractions. Parenthetically, no UL15 or VP5 was detected when a nuclear lysate of K23Z-infected Vero cells was run on a single sucrose gradient, fractionated, and examined by immunoblotting (data not shown).
FIG. 3.
Immunoblots of B capsids purified through two sequential 20- to-50% sucrose gradients. The 14-ml gradient was fractionated from the top (fraction one), and the proteins present in each fraction were separated on an 8% denaturing polyacrylamide gel before being transferred to a PVDF membrane. The membrane was then probed with antisera against UL28, UL15, or VP5 and developed using the ECL+ method (see Materials and Methods). The image was generated using a Molecular Dynamics Storm PhosphorImager with chemiluminescence detection capability.
FIG. 4.
Nuclear lysate of Vero cells infected with the UL18 deletion virus K23Z, purified on two sequential 20- to-50% sucrose gradients. The 14-ml gradient was fractionated from top (lane 1) to bottom (lane 11), and the proteins present in each fraction were separated on an 8% denaturing polyacrylamide gel before being transferred to nitrocellulose membrane. The membrane was then probed with antisera against UL15 (A) or UL28 (C) and developed using the ECL+ method (see Materials and Methods). Lane 12 contains purified protein as a positive control (arrow). As a further control, lysates of HSV-1(F)-infected, K23Z-infected, or mock-infected cells were electrophoretically separated and reacted with antisera against UL15 (B) or UL28 (D) to confirm expression of the proteins in the cells infected with mutant virus.
Once it was confirmed that all of the pUL15 and pUL28 immunoreactivity was associated with capsids, the number of copies of each of the proteins present in HSV-1(F) B capsids was determined. A new preparation of capsids was used for each calculation. Assuming equal distribution of the proteins among all B capsids, each capsid was calculated to contain 1.0 copies of pUL15 and 2.4 copies of pUL28 (Table 1). Statistical analysis of these data revealed that each B capsid contained significantly more pUL28 than pUL15 (P = 0.03) (Table 1).
TABLE 1.
Summary of the amounts of pUL15 and pUL28 present in B, UL6(−), and A capsidsf
| Capsid type | UL28
|
UL15
|
||
|---|---|---|---|---|
| n | Amt (mean ± SD) | n | Amt (mean ± SD) | |
| B | 5 | 2.4 ± 0.97a,b | 4 | 1.0 ± 0.29a,c |
| UL6(−) | 3 | 1.5 ± 0.86 | 3 | 1.2 ± 0.72d |
| A | 2 | 0.6 ± 0.32b,e | 3 | 12.0 ± 5.63c,d,e |
More pUL28 than pUL15 was detected in B capsids; P = 0.03.
More pUL28 was detected in B capsids than in A capsids; P = 0.06.
Less pUL15 was detected in B capsids than in A capsids; P = 0.08.
Less pUL15 was detected in pUL6(−) capsids than in A capsids; P = 0.08.
Less pUL28 than pUL15 was detected in A capsids; P = 0.07.
Capsids were purified on two sequential sucrose gradients, and the amount of protein present in each capsid was calculated using a standard curve of purified protein (see Materials and Methods). Statistical analyses were calculated using a two-tailed t test, and significant differences are indicated.
The amount of pUL15 and pUL28 in pUL6(−) capsids is similar to the amount present in HSV-1(F) B capsids.
To determine whether the presence or absence of the putative portal encoded by pUL6 influenced the capsid association of pUL15 and pUL28, the number of copies of pUL15 and pUL28 in pUL6(−) capsids was calculated using the same method as described above. An average of 1.5 copies of pUL28 and 1.2 copies of pUL15 were present in pUL6(−) capsids. No statistical difference was detected when comparing the amount of either pUL15 or pUL28 in UL6(−) and wild-type B capsids (P > 0.1) (Table 1).
There is significantly more pUL15 and less pUL28 in A capsids than in B capsids.
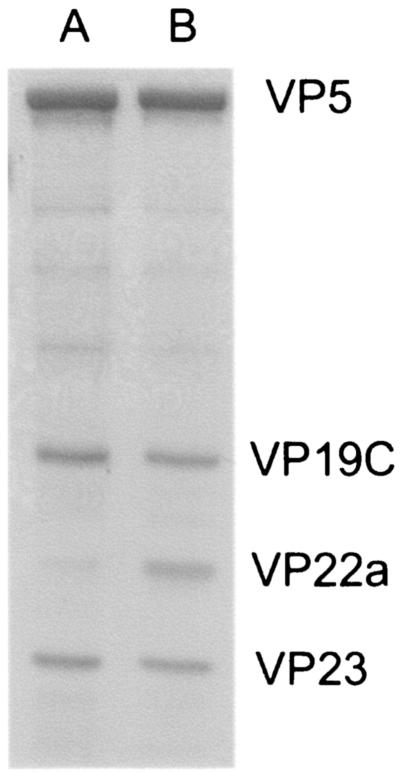
The association of any of the six cleavage and packaging proteins with wild-type A capsids has not been reported previously. Type A capsids were purified from approximately 4 × 108 HSV-1(F)-infected cells, associated proteins were electrophoretically separated on an 8% polyacrylamide gel, and the gel was stained with Coomassie brilliant blue. Lanes containing denatured A capsid proteins contained markedly reduced levels of scaffold proteins (VP22a) relative to other capsid proteins, confirming that the purified A capsids did not contain substantial numbers of B capsids (Fig. 5).
FIG. 5.
Coomassie-stained gel of A and B capsids purified as described in Materials and Methods. The positions of the capsid shell proteins are indicated on the right. Note the very small amount of VP22a present in the A capsid sample compared to that in B capsids.
The number of copies of pUL15 and pUL28 per A capsid was then determined. An average of 12.0 copies of pUL15 per capsid was detected, significantly more than amounts detected in wild-type B capsids and pUL6(−) capsids (P = 0.08 for both comparisons) (Table 1). In contrast, the level of pUL28 in A capsid preparations was near the lowest limit of detection, as assessed by comparison with the standard curve. Consequently, immunoreactivity that corresponded to an intensity within the standard curve was only obtained on two occasions. For these experiments, an average of 0.6 copies of pUL28 per capsid was calculated, which was significantly less than the amount of pUL28 present in B capsids (P = 0.06). Since three further experiments failed to reveal sufficient pUL28 to allow calculation from comparison with the standard curve, the 0.6 copies calculated likely represents the higher end of the range of levels of pUL28 in A capsids.
Cleaved pUL15 is present in A capsids.
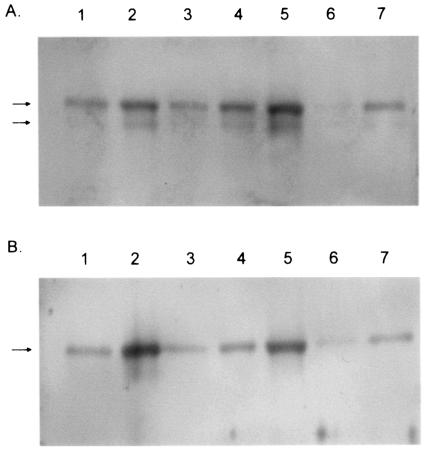
The UL15 protein is proteolytically cleaved at the N terminus in close association with the maturation of genomic DNA. The cleavage shortens the 83-kDa full-length UL15 protein to approximately 80,000 and 79,000 Mr (48, 50). To determine whether the pUL15 was proteolytically cleaved in A capsids, A, B, and pUL6(−) capsids were purified on two successive sucrose gradients as detailed above. Capsid-associated proteins were electrophoretically separated on 8% polyacrylamide gels, transferred to a PVDF membrane, and probed with antisera directed against either the entire exon II of the UL15 protein or the N-terminal 35 amino acids of the protein. Figure 6 shows that both the full-length and cleaved form of pUL15 were present in A capsids and wild-type B capsids but, as has been reported previously, only the full-length form was present in pUL6(−) capsids (48). A sample from the same A capsid preparation was separated on another gel and stained with Coomassie blue to confirm the lack of scaffold protein VP22a and, hence, the absence of contamination by B capsids (data not shown).
FIG. 6.
Digital image of immunoblot probed with antisera directed against the portion of the UL15 protein encoded by the entire exon II of the gene (A) or the N-terminal 35 amino acids (B). Lanes 1 and 2, A capsids; lanes 3, 4, and 5, B capsids; lanes 6 and 7, UL6(−) capsids. All capsids were purified through two successive sucrose gradients (see Materials and Methods). The 83,000 and 80,000 apparent Mr forms of pUL15 are indicated with arrows.
DISCUSSION
Purified recombinant UL15 and UL28 proteins were used as standards to calculate an average of 1.0 (±0.29) copies of pUL15 and 2.4 (±0.97) copies of pUL28 per B capsid, substantially less than the 15 copies of pUL6 (38) or 42 copies of pUL25 (40) previously reported in B capsids. Similar amounts of pUL15 and pUL28 were detected in capsids lacking the putative portal protein pUL6 (Table 1). In contrast, highly purified A capsids were found to contain an average of 12.0 (±5.6) copies of the UL15 protein per capsid, a significantly larger amount than was found in either wild-type B capsids or pUL6(−) capsids. Only 0.6 (±0.32) copies of pUL28 were detected per A capsid on average, significantly less than the amount of pUL28 present in wild-type B capsids. Extensive efforts were made to quantify pUL15 and pUL28 in C capsids, but the instability of these capsids in successive sucrose gradients and the relatively low levels of pUL15 and pUL28 within them precluded accurate calculations.
In many double-stranded DNA viruses, including bacteriophages λ (63) and φ29 (28), the portal vertex of the capsid represents the docking site for the terminase enzyme. When extrapolated to the HSV system, a prediction of this hypothesis is that pUL6(−) capsids should lack the putative terminase components pUL15 and pUL28. There have been previous reports of smaller amounts of pUL15 in UL6(−) capsids than in wild-type B capsids (65), and our preliminary data seemed to support this observation (50). However, once quantitative analyses were applied, no significant difference was detected between the amounts of pUL15 or pUL28 in the UL6(−) capsids and those in wild-type B capsids (P > 0.1) (Table 1). Thus, small amounts of both pUL15 and pUL28 are able to associate with B-like capsids in the presence or absence of the UL6-encoded portal protein. This discrepancy between present and previous studies in estimating the relative amounts of pUL15 in B and UL6(−) capsids may stem from the fact that in this report capsids were purified through two, rather than one, sucrose gradients and extensive efforts were taken to separate A capsids from B capsids.
Type A capsids are thought to be the result of an aborted cleavage and packaging reaction and are widely assumed to have at least engaged the packaging machinery (37, 52, 57). The presence of the N-terminally cleaved form of pUL15 in A capsids (Fig. 6) is consistent with this hypothesis. It is therefore possible that A capsids bear 12 copies of pUL15 as a remnant of the functional DNA packaging complex docked with the capsid. If these subunits were to form a complex in conjunction with the portal vertex, such a structure would be reminiscent of the proposed DNA translocation machinery assembled by the bacteriophages φ29 and SPP1 (23, 27, 53). In these models the oligomeric ATPase subunit of the terminase is present at the portal vertex, possibly in 6- or 12-fold symmetry, and participates in the packaging of the DNA into the capsid. However, it must also be acknowledged that the method described here for estimating the copy number of both pUL15 and pUL28 in A and B capsids is indirect and therefore provides more accurate relative, rather than absolute, data.
The consistently small amounts of pUL15 present in both wild-type B capsids and pUL6(−) capsids do not exclude the possibility that the terminase docks with the capsid via interaction with the portal protein. Rather, the data suggest that neither B capsids nor UL6(−) capsids contain the fully assembled pUL15 structure, therefore adding to the increasing amount of indirect evidence suggesting that B capsids are dead-end by-products rather than intermediates in capsid development. For example, several lines of evidence indicate that proteolytic cleavage and expulsion of the scaffold protein are tightly linked with entry of viral DNA (13, 21, 35, 44). This implies that the relatively stable B capsids, with a rigid, icosahedral shell and cleaved but retained scaffold protein, are unlikely to be able to package DNA. Further, there is in vitro evidence that, over time, procapsids automatically progress to B-like capsids in the absence of any packaging machinery (37), suggesting that B capsids form by default if the cleavage and packaging reaction is delayed. Thus, while wild-type B capsids may be relevant for structural studies of the portal vertex, they are unlikely to be useful for investigations into the portal vertex-terminase complex.
The disparity between the stoichiometry of pUL15 and pUL28 in A and B capsids was unexpected and may indicate that the interaction between these proteins changes during DNA packaging. Thus, different stages of the packaging reaction may require different stoichiometries of, and interactions between, terminase subunits as recently suggested for bacteriophage T4 (10). An alternative explanation is that many copies of pUL28 may also comprise part of the cleavage and packaging machinery but that these are lost either during the capsid purification procedure or during A capsid morphogenesis, where a greater affinity for DNA than for capsids or pUL15 (4) causes the protein to be lost when the DNA with which it is associated aberrantly exits the capsid.
Both pUL15 and pUL28 have been detected in the procapsid (51), a precursor to A, B, and C capsids, suggesting that the cleavage and packaging machinery associates with capsids at an early stage. Quantification of the amounts of pUL15 and pUL28 in procapsids would likely shed further light on the overall structure of the packaging machine and allude to the possible functions of these proteins.
Acknowledgments
These studies were supported by R01 grants GM50740 and AI52341 from the National Institutes of Health.
We gratefully acknowledge the technical assistance of Jarek Okulicz-Kozaryn, Elizabeth Wills for electron microscopy, and Hollis Erb for statistical advice. We also thank P. Desai and S. Person for the K23Z virus, A. Patel for the UL6 deletion virus, and G. Cohen and R. Eisenberg for the VP5 antibody.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Addison, C., F. J. Rixon, J. W. Palfreyman, M. Ohara, and V. G. Preston. 1984. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 3.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type-1 UL28 gene-product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 4.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene-product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 6.Baines, J. D., C. Cunningham, D. Nalwanga, and A. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines, J. D., A. P. W. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus type 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus type 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines, J. D., and S. K. Weller. Cleavage and packaging of herpes simplex virus 1 DNA. In C. E. Catalano (ed.), Bacteriophage genome packaging, in press. Landes Press, Georgetown, Tex.
- 10.Baumann, R. G., and L. W. Black. 2003. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 278:4618-4627. [DOI] [PubMed] [Google Scholar]
- 11.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, G. H., M. Ponce de leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. Eisenburg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham, C., and A. J. Davison. 1993. A cosmid based system for constructing mutants of herpes simplex virus type-1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 16.Davison, A. J. 1992. Channel Catfish Virus—a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 17.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding 2 capsid proteins (VP24 and VP26) of herpes simplex virus type-1. J. Gen. Virol. 73:2709-2713. [DOI] [PubMed] [Google Scholar]
- 18.Desai, P., N. A. Deluca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type-1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diianni, C. L., D. A. Drier, I. C. Deckman, P. J. Mccann, F. Y. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley. 1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence-analysis of autoproteolytic cleavage products. J. Biol. Chem. 268:2048-2051. [PubMed] [Google Scholar]
- 20.Ejercito, P. M., and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 21.Gao, M., L. MatusickKumar, W. Hurlburt, S. F. Ditusa, W. W. Newcomb, J. C. Brown, P. J. Mccann, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Ruth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi 29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 24.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 25.Hodge, P. D., and N. D. Stow. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 27.Ibarra, B., J. R. Caston, O. Llorca, M. Valle, J. M. Valpuesta, and J. L. Carrascosa. 2000. Topology of the components of the DNA packaging machinery in the phage phi 29 prohead. J. Mol. Biol. 298:807-815. [DOI] [PubMed] [Google Scholar]
- 28.Ibarra, B., J. M. Valpuesta, and J. L. Carrascosa. 2001. Purification and functional characterization of p16, the ATPase of the bacteriophage phi 29 packaging machinery. Nucleic Acids Res. 29:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koslowski, K. M., P. R. Shaver, J. T. Casey, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koslowski, K. M., P. R. Shaver, X. Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 32.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, F. Y., and B. Roizman. 1993. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus-1 UL26 protein. J. Virol. 67:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lurz, R., E. V. Orlova, D. Gunther, P. Dube, A. Droge, F. Weise, M. van Heel, and P. Tavares. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310:1027-1037. [DOI] [PubMed] [Google Scholar]
- 35.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell, M. S., S. Matsuzaki, S. Imai, and V. B. Rao. 2002. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30:4009-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 38.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid—molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 40.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 42.Perdue, M. L., M. C. Kemp, C. C. Randall, and D. J. O'Callaghan. 1974. Studies of the molecular anatomy of the L-M strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology 59:201-216. [DOI] [PubMed] [Google Scholar]
- 43.Poon, A. P. W., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of the herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston, V. G., F. J. Rixon, I. M. McDougall, M. McGregor, and M. F. Alkobaisi. 1992. Processing of the herpes-simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology 186:87-98. [DOI] [PubMed] [Google Scholar]
- 46.Roizman, B., and D. Furlong. 1974. Comprehensive virology. Plenum Press, New York, N.Y.
- 47.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2213-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 48.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmon, B., D. Nalwanga, Y. Fan, and J. D. Baines. 1999. Proteolytic cleavage of the amino terminus of the UL15 gene product of herpes simplex virus type 1 is coupled with maturation of viral DNA into unit-length genomes. J. Virol. 73:8338-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman, G., and S. L. Bachenheimer. 1987. DNA processing in temperature sensitive morphogenetic mutants of HSV-1. Virology 158:427-430. [DOI] [PubMed] [Google Scholar]
- 53.Simpson, A. A., Y. Z. Tao, P. G. Leiman, M. O. Badasso, Y. N. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi 29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taus, N. S., and J. D. Baines. 1998. Herpes simplex virus 1 DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology 252:443-449. [DOI] [PubMed] [Google Scholar]
- 55.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 56.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type-1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 58.Valpuesta, J. M., N. Sousa, I. Barthelemy, J. J. Fernandez, H. Fujisawa, B. Ibarra, and J. L. Carrascosa. 2000. Structural analysis of the bacteriophage T3 head-to-tail connector. J. Struct. Biol. 131:146-155. [DOI] [PubMed] [Google Scholar]
- 59.Weinheimer, S. P., P. J. Mccann, D. R. Oboyle, J. T. Stevens, B. A. Boyd, D. A. Drier, G. A. Yamanaka, C. L. Diianni, I. C. Deckman, and M. G. Cordingley. 1993. Autoproteolysis of herpes simplex virus type-1 protease releases an active catalytic domain found in intermediate capsid particles. J. Virol. 67:5813-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weller, S. K., E. P. Carmichael, D. P. Aschman, D. J. Goldstein, and P. A. Schaffer. 1987. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology 161:198-210. [DOI] [PubMed] [Google Scholar]
- 61.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wildy, P., W. C. Russell, and R. W. Horne. 1960. The morphology of herpes virus. Virology 12:204-222. [DOI] [PubMed] [Google Scholar]
- 63.Yeo, A., and M. Feiss. 1995. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage-lambda, with the portal protein of the prohead. J. Mol. Biol. 245:141-150. [DOI] [PubMed] [Google Scholar]
- 64.Yu, D., A. K. Sheaffer, D. J. Tenney, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 71:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A capsid determined from 400-Kv spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]