Abstract
The human cytomegalovirus (HCMV) virion is comprised of a linear double-stranded DNA genome, proteinaceous capsid and tegument, and a lipid envelope containing virus-encoded glycoproteins. Of these components, the tegument is the least well defined in terms of both protein content and function. Several of the major tegument proteins are phosphoproteins (pp), including pp150, pp71, pp65, and pp28. pp28, encoded by the UL99 open reading frame (ORF), traffics to vacuole-like cytoplasmic structures and was shown recently to be essential for envelopment. To elucidate the UL99 amino acid sequences necessary for its trafficking and function in the HCMV replication cycle, two types of viral mutants were analyzed. Using a series of recombinant viruses expressing various UL99-green fluorescent protein fusions, we demonstrate that myristoylation at glycine 2 and an acidic cluster (AC; amino acids 44 to 57) are required for the punctate perinuclear and cytoplasmic (vacuole-like) localization observed for wild-type pp28. A second approach involving the generation of several UL99 deletion mutants indicated that at least the C-terminal two-thirds of this ORF is nonessential for viral growth. Furthermore, the data suggest that an N-terminal region of UL99 containing the AC is required for viral growth. Regarding virion incorporation or UL99-encoded proteins, we provide evidence that suggests that a hypophosphorylated form of pp28 is incorporated, myristoylation is required, and sequences within the first 57 amino acids are sufficient.
Human cytomegalovirus (HCMV), a betaherpesvirus, is an opportunistic pathogen that infects 50 to 90% of the human population by adulthood (39). In healthy adults, infection and viral latency are controlled largely by cell-mediated immunity, although there is a humoral response that may play a secondary role. However, when such immunity is not present, wanes, or is suppressed, the clinical effects of HCMV are realized. Thus, HCMV causes serious problems in congenitally infected infants, AIDS patients (especially in the pre-HAART [highly active antiretroviral therapy] era), and transplant patients.
The HCMV virion is comprised of an envelope and nucleocapsid, separated by the amorphous tegument comprised of numerous virus-encoded proteins (4, 34). Several of the major tegument proteins are phosphoproteins (pp), including pp150 (encoded by UL32 open reading frame [ORF]), pp71 (encoded by the UL82 ORF), pp65 (encoded by the UL83 ORF), and pp28 (encoded by the UL99 ORF). The functions of these proteins have not been completely elucidated. However, pp71 is a transactivator of immediate-early genes (30). Deletion of UL82 results in a virus that is severely impaired in culture (6). In contrast, deletion of UL83 does not significantly affect the growth of cultured virus (44). However, UL83-negative mutants do not make dense bodies, one of three viral particle (VP) types made by HCMV infection in culture (18, 34, 44). Mutagenesis of the phosphoprotein product of HCMV ORF UL32 in the context of recombinant HCMV has not been reported.
HCMV ORF UL99 encodes a 190-amino-acid, myristoylated tegument protein with an extremely unusual amino acid structure (Fig. 1A) (9). Unlike the other major phosphoproteins products of UL82 and UL83, which have early-late kinetics, the UL99 phosphoprotein is expressed with strict late kinetics (34). Sequences within 40 bases upstream of the UL99 coding region were identified to be sufficient for its expression with late kinetics (26, 28). The UL99 ORF is the 3′ most ORF in the UL93-UL99 transcription unit (55). The ORFs in this transcription unit each have separate promoters (with different expression kinetics) but share a common polyadenylation signal downstream of UL99. Thus, transcripts from these ORFs are overlapping. There is no obvious commonality in the function(s) of the genes encoded by this transcription unit, which include a kinase (UL97), an alkaline nuclease (UL98), a gene putatively involved in cleavage/packaging of progeny DNA (UL93), and several genes of unknown function (UL94 to UL96).
FIG. 1.
(A) Amino acid sequence of UL99 (9). Notable amino acid regions are indicated in boldface. (B to I) Subcellular localization of transiently expressed UL99-GFP fusion proteins. UL99-GFP fusion proteins were expressed transiently in HFF cells. (B and E) GFP; (C and F) UL99(1-190)-GFP; (D and G) UL99(1-190 [G2A])-GFP. Photos were taken at 1 day (B to D) and 2 days (E and G) posttransfection.
The function of the pp28 protein in the HCMV infectious cycle is also unknown, although recent evidence suggests that it has a role in envelopment (45). Britt and coworkers determined that pp28 localizes only to the cytoplasm (42). In the absence of other viral proteins, localization was to the endoplasmic reticulum (ER)-Golgi intermediate compartment (43). During infection, colocalization with pp150 in a Golgi/trans-Golgi network (TGN)-derived vacuole-like structures was reported (17, 42). We elucidated here the UL99 sequences required for proper trafficking and growth through the creation and examination of viral mutants. Using viruses expressing various UL99-green fluorescent protein (GFP) fusion proteins, our data indicate that in addition to the requirement for the myristoylation site (glycine 2), an acidic cluster (AC), comprising amino acids 44 and 57, is required for the punctate perinuclear and cytoplasmic (vacuole-like) localization observed for wild-type pp28 (17, 42). While the present study was in preparation, Silva et al. reported that the UL99 ORF is essential for viral growth through the use of a bacterial artificial chromosome (BAC)-mediated mutagenesis strategy (45). We extend this observation through the use of viral recombinants expressing a series of UL99 C-terminal deletions. The data from these mutants demonstrate that UL99 amino acids 58 to 190 are not essential for viral growth. Furthermore, UL99 amino acids 1 to 57 are sufficient for near wild-type growth kinetics in culture and virion incorporation of the protein. We also show that expression of UL99 as an early-late protein, rather than a strict late protein, has no effect on virus growth.
MATERIALS AND METHODS
DNA sequence.
The numbering system of Chee et al. (9) was used for the HCMV strain AD169 DNA sequence (GenBank accession number X17403).
Virus and cells.
HCMV strain AD169 was obtained from the American Type Culture Collection. All recombinant HCMV are derivatives of strain AD169. The origin and growth of human foreskin fibroblast (HFF) cells were described previously (20).
Recombinant mutant HCMV.
Construction and isolation of all HCMV recombinant mutants, including those expressing the β-glucuronidase and GFP reporter genes, were done according to the general method described previously (22). Plasmids used to make the mutants contained the reporter gene or gene of interest flanked by about 1.5 kb of viral “targeting” sequences to direct its homologous recombination with the HCMV genome. Typically, after cotransfection with HCMV wild-type strain AD169 genomic DNA, plaques containing reporter gene-expressing virus were picked and plaque purified. Plaques expression β-glucuronidase were identified by their blue color in the presence of overlay medium containing 125 μg of X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid)/ml. Plaques expressing GFP were identified by fluorescence microscopically. The proper genomic organization of each purified mutant was verified by DNA blot hybridization analysis (data not shown) as described previously (22). RV7150 is similar to RV134, except that it contains a simian virus 40 (SV40) promoter-driven β-glucuronidase gene in the intergenic region between US10 and US9 (22). RV17194 was derived from RV7150 after cotransfection with a plasmid containing the UL99 coding region and its flanking sequences (bases 144361 through 144996) under the control of a three-tetracycline-operator-modified US11 promoter (designated 3optx in reference 27). Proper recombination into the HCMV genome resulted in the simultaneous replacement of the US7 through US11 genes and β-glucuronidase. RV17194 was obtained after screening for β-glucuronidase-negative (white) plaques after X-Gluc overlay. Thus, RV17194 is diploid for UL99, having a second copy within the US6 gene family region. UL99 transcripts expressed from the US6 region of the genome utilize the polyadenylation signal that is downstream of US7, within the flanking sequences used to direct recombination. RV17044 expresses unfused GFP from the US9 to US10 intergenic region under the control of the SV40 promoter (19b). A complete listing and description of all HCMV used in this report are in Table 1.
TABLE 1.
Viruses used in this study
| Virusa | Descriptionb | Parent virus |
|---|---|---|
| AD169 | Laboratory parental wild-type strain | |
| RV7150* | β-Glucuronidase expressed from US9-US10 intergenic region | AD169 |
| RV17044† | GFP expressed from US6 region | AD169 |
| RV17179* | UL99 C-terminal deletion: aa 1 to 99 expressed | AD169 |
| RV17194‡ | Diploid for UL99: complete UL99 expressed from resident and US6 regions | RV7150 |
| RV18073* | UL99 C-terminal deletion: aa 1 to 99 expressed; complete UL99 expressed from US6 region | RV17194 |
| RV18074‡ | Pseudo-wild-type rescue virus | RV17179 |
| RV18196† | UL99(1-190)-GFP fusion expressed from US6 region | AD169 |
| RV18198† | UL99(aa 1-22)-GFP fusion expressed from US6 region | AD169 |
| RV18199† | UL99(1-100)-GFP fusion expressed from US6 region | AD169 |
| RV19012* | UL99 C-terminal deletion: aa 1 to 22 remain; complete UL99 expressed from US6 region | RV17194 |
| RV19023† | UL99(1-190 [G2A])-GFP fusion expressed from US6 region | AD169 |
| RV19024† | UL99(1-57)-GFP fusion expressed from US6 region | AD169 |
| RV19029* | UL99 C-terminal deletion: aa 1 to 57 expressed | AD169 |
| RV19030* | UL99 C-terminal deletion: aa 1 to 57 expressed; complete UL99 expressed from US6 region | RV17194 |
| RV19046* | UL99 C-terminal deletion: aa 1 to 43 remain; complete UL99 expressed from US6 region | RV17194 |
| RV19047† | UL99(1-43)-GFP fusion expressed from US6 region | AD169 |
*, isolation based on β-glucuronidase expression; †, isolation based on GFP expression; ‡, isolation based on lack of β-glucuronidase expression.
aa, amino acid(s).
UL99 deletion series plasmids.
A series of β-glucuronidase-positive HCMV mutants with deletions of the UL99 gene were derived from strain AD169 and RV17194. The general method for generation of plasmids used in this series of mutants is as follows. A TAA stop codon was placed after the designated amino acid, either amino acid 22, 43, or 57, by PCR mutagenesis. The fidelity of the resulting PCR fragment was verified by DNA sequencing and transferred into the recombination plasmid. Sequentially, the recombination plasmid contains: sequences upstream of UL99, including the UL99 promoter (HCMV bases 142993 to 144391); the UL99 coding region from amino acid 1 through the codon for the selected amino acid (HCMV bases 144392 to 144457 [amino acid 22], 144520 [amino acid 43], or 144562 [amino acid 57]); the inserted stop codon; an expression cassette containing the herpes simplex virus (HSV) type 1 tk polyadenylation signal (to terminate the truncated UL99 transcripts), the HSV gH promoter, β-glucuronidase, and the SV40 bidirectional polyadenylation signal; and UL99 downstream flanking sequences (HCMV bases 144929 through 146612). Note that although the downstream flanking sequences contain the coding region for the C-terminal 10 amino acids of UL99 (HCMV bases 144929 through 144961), signals to direct expression of this region are not present. The UL99 deletion after amino acid 99 was constructed in a slightly different fashion in that a stop codon was not engineered immediately after amino acid 99 (HCMV base 144688). Instead, the first encountered stop codon within the tk polyadenylation signal region was utilized, resulting in an additional 11 amino acids from that region being encoded after UL99 amino acid 99. Thus, after transfection into HFF cells and recombination into the HCMV genome (described above), the resident UL99 is truncated and the β-glucuronidase expression cassette is inserted.
UL99-GFP fusion protein series plasmids.
A series of UL99 fusions with enhanced GFP (EGFP; Clontech) were made by using standard cloning techniques and PCR-based mutagenesis. Sequentially, the relevant regions of plasmids for transient expression of UL99-EGFP contained the following: SV40 promoter, located 30 bases upstream of the UL99 coding region (HCMV bases 144362-144391); the UL99 coding region from amino acid 1 through the codon for the selected amino acid (HCMV bases 144392 to 144457 [amino acid 22], 144520 [amino acid 43], 144562 [amino acid 57], 144691 [amino acid 100], or 144961 [amino acid 190]); a linker encoding three glycine residues; the EGFP coding region; and the SV40 bidirectional polyadenylation signal. For recombination into the HCMV genome replacing the US6 family genes US9-US7, the SV40 promoter-regulated UL99-GFP fusions were inserted within the recombination plasmid and then transfected into HFF cells as described above. Sequentially, these plasmids contain US9 upstream flanking sequences (HCMV bases 200328 to 199021); the UL99-GFP gene fusion; and the US7 downstream flanking sequences (HCMV bases 196447 to 194741). The myristoylation-negative UL99-GFP fusion mutant was constructed by PCR-based mutagenesis changing the amino acid 2 glycine codon (GGT) to an alanine codon (GCT). All PCR mutagenesis products were confirmed by DNA sequencing.
Transient expression.
Uninfected cell analysis of UL99-GFP fusion protein expression was done by transient expression in HFF cells at 1 or 2 days posttransfection. The calcium chloride-DNA coprecipitation transfection technique was used, as described previously (27).
Antibodies.
Rabbit polyclonal antiserum reactive with abundant HCMV VP proteins was obtained after immunization of New Zealand rabbits with VPs from the media of infected cells, enriched by centrifugation through a 20% sorbitol cushion gradient (47). Anti-GFP monoclonal and polyclonal antisera were obtained from Clontech. Anti-pp28 monoclonal antibody is from ABI.
Protein analysis.
Radiolabeling, immunoprecipitation, and immunoblot techniques were done as described previously (22, 23). For radiolabeling experiments involving nonradioactive chases, the radioactive medium was removed and replaced with nonradioactive complete medium after two washes with phosphate-buffered saline; incubation was continued until the proper harvest time. In some cases, immunoprecipitates were treated with 800 U of lambda phosphatase (NEB) prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (51).
Fluorescence microscopy.
Transfected or infected HFF cells were examined live (i.e., unfixed) for expression and intracellular localization of UL99-GFP fusion proteins by standard fluorescence microscopy or fluorescence deconvolution microscopy, as indicated. Fluorescent-organelle-specific dyes were used, as indicated, according to the manufacturer's directions (Molecular Probes, Inc.): Hoechst 33342, a DNA stain, was used at 1 μg/ml; LysoTracker Red DND-99, a lysosome and acidic compartment stain, was used at 1 μM; and BODIPY TR ceramide, a Golgi stain, was used at 5 μM. Photographs were taken by using a Nikon fluorescence microscope and Image-Pro software (Media Cybernetics). Z-stacks were deconvoluted by using either 3D or Inverse Filter software, as indicated (AutoQuant Imaging, Inc.).
RESULTS
Transient expression of UL99-GFP fusion proteins.
Sequence analysis revealed that the UL99-encoded protein has a very unusual amino acid composition: a glycine at position 2 followed closely by several cysteine residues, a very acidic stretch of amino acids, a lysine-rich region followed by a histidine-rich region, another acidic region, a proline-rich region, and a lysine/arginine-rich region (Fig. 1A) (9). Existing data have indicated that pp28 is myristolyated at glycine 2 and exists solely as a cytoplasmic protein (17, 42). Specifically, the UL99-encoded phosphoprotein localized to intracytoplasmic membranes, either ERGIC or TGN-like structure (42, 43). Furthermore, this intracytoplasmic membrane localization was dependent on the presence of the myristoylation (43). A major objective of our study was to determine the sequences required for proper subcellular localization of UL99. The strategy was to make deletions from the C terminus of UL99 and translationally fuse the remaining sequences with enhanced GFP. This strategy has proven to be useful to determine sequences necessary for trafficking of other herpesvirus proteins, including pseudorabies virus US9 glycoprotein, and the HSV type 1 (HSV-1) tegument proteins encoded by UL11 and UL49 (7, 8, 31). Fusion protein expression was under the control of the SV40 promoter and polyadenylation signal and was analyzed in a transient-expression assay (i.e., in the absence of other viral proteins) or an infected cell system (i.e., after incorporation into the HCMV genome). Cellular localization of fusion proteins was assessed by fluorescence in live cells to avoid possible alterations due to fixation.
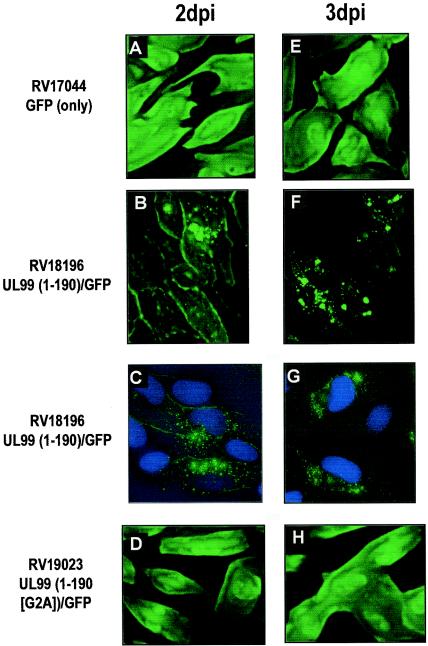
Initially, two fusions were made for control purposes: the full-length wild-type UL99 fused to GFP (UL99[1-190]/GFP) and the full-length myristoylation-negative mutant, UL99(1-190 [G2A])-GFP. In the latter construct, the codon encoding glycine at UL99 amino acid 2 was changed to an alanine codon, thereby eliminating the myristoylation consensus sequence. In transfected cells, Sanchez et al. demonstrated that (unfused) wild-type UL99 localized to intracytoplasmic vacuoles, whereas the corresponding mutant UL99 (G2A) was dispersed throughout the cell (43). Similar results were obtained with our analogous GFP fusion proteins in a transient-expression system (Fig. 1). Unfused GFP is distributed throughout the cell (Fig. 1B and E); UL99-GFP is excluded from the nucleus and accumulates in punctuate perinuclear and cytoplasmic structures (Fig. 1C and F). The myristoylation-negative mutant fusion protein is dispersed throughout the entire cell (i.e., cytoplasm and nucleus), although there is apparent concentration of this protein in the nucleus of the cell (Fig. 1D and G). After incorporation into the HCMV genome, these fusions were also examined in infected cells by fluorescence deconvolution microscopy; results were very similar (see below) to those obtained for experiments done in transfected cells. Thus, these control experiments have established that our systems replicate the data published previously (42, 43) and thereby establish the validity of the approach for analysis of additional fusion protein mutants.
Viral expression of UL99-GFP fusion proteins.
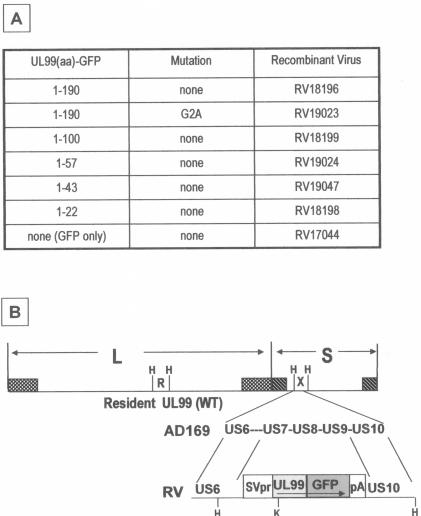
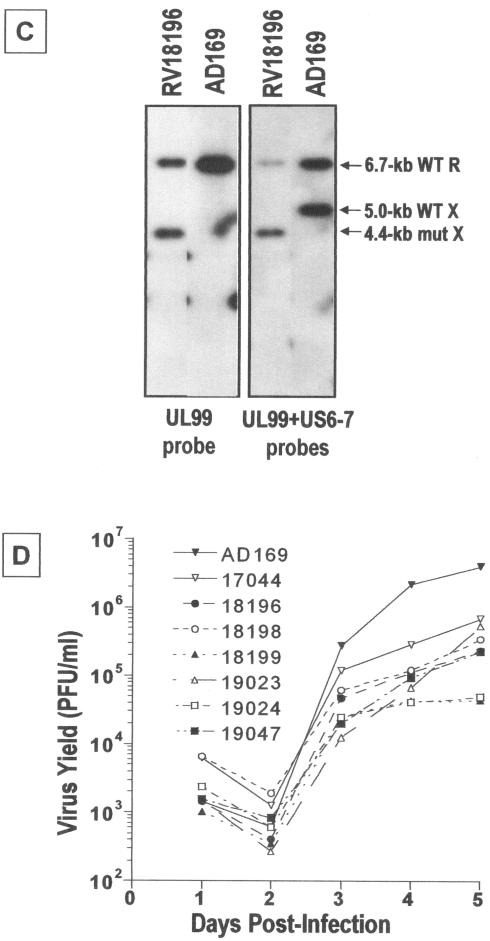
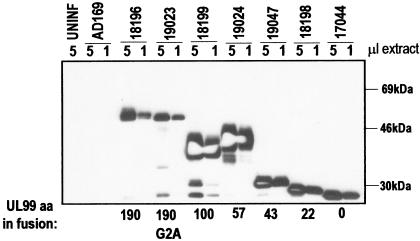
Trafficking of UL99 may be affected by the presence of other viral proteins, as has been suggested (42, 43). Therefore, the trafficking of UL99 fusion proteins was analyzed in the context of HFFs infected with recombinant HCMV, each expressing a UL99-GFP fusion protein from a series of sequential deletions (Fig. 2A). The UL99-GFP fusion cassette was inserted within the US6 glycoprotein family region of the HCMV genome, in place of the nonessential US7, US8, and US9 ORFs (Fig. 2B) (21). The correct arrangement of the inserted fusion cassette within this region of HCMV genome was confirmed by DNA blot analysis (Fig. 2C and data not shown); representative hybridization data for RV18196 are shown. A UL99-specific probe hybridizes with the wild-type 6.7-kb HindIII-R DNA fragment (containing the resident UL99 gene) in AD169 and RV18196. It also hybridizes with the altered 4.4-kb HindIII-X DNA fragment, containing the UL99-GFP fusion cassette, found in RV18196 only. A US6-7 intergenic region probe hybridizes to the wild-type 5.0-kb HindIII-X fragment in AD169, which is altered in RV18196. Note that in spite of the 1.9-kb insertion of the UL99-GFP fusion cassette, the mutated HindIII-X DNA fragment in RV18196 is ca. 0.6 kb smaller than the corresponding wild-type DNA fragment due to the designed deletion of 2.5-kb encompassing sequences of US7-US9, as result of homologous recombination directed by sequences flanking the fusion cassette (Fig. 2B). Additional hybridization experiments with KpnI-digested DNA, which cuts asymmetrically within the inserted UL99-GFP expression cassette (Fig. 2B), further confirmed that the insertions had recombined properly within the US6 glycoprotein family region (data not shown). Recombinant HCMV containing insertion of the UL99-GFP fusion cassettes within the HCMV genome were easily isolated and propagated. The growth kinetics of these viruses were similar to those of AD169 and RV17044, a virus that expresses unfused GFP, with burst occurring just after 2 days postinfection (dpi) (Fig. 2D). However, in some cases, the overall viral yield at late times postinfection were reduced by ∼1 log compared to RV17044. The integrity of the UL99-GFP fusion proteins expressed by these viruses was assessed by immunoblot analysis (Fig. 3). For most of the fusions, the major protein expressed was of the approximate anticipated size, with very little evidence of degradation. Note that the G2A myristoylation mutant fusion protein migrated slightly faster than its unmutated counterpart. An exception was the fusion protein expressed by RV19024 encoding UL99 amino acids 1 to 57 fused with GFP. The apparent molecular weight of this protein was larger than expected. Transient expression of this fusion, in the absence of other viral proteins, yielded a similar migration pattern, as did an independent reclone (data not shown). Furthermore, the fusion was shown to be correct by DNA sequencing. Thus, the aberrant migration of this protein in SDS-PAGE is due to the nature of this protein and not to mutation or incorrect cloning. The 1-57 fusion has a very acidic region of UL99 (amino acids 44 to 57) fused with GFP.
FIG. 2.
(A) Description of recombinant viruses expressing UL99-GFP fusion proteins. (B) Schematic representation of HCMV genome and the strategy used to construct recombinant viruses expressing UL99-GFP fusion proteins. The locations of the HindIII-R (containing the resident UL99 gene) and HindIII-X DNA fragments are shown. The UL99-GFP fusion protein expression cassettes recombined by homologous recombination within the HindIII-X DNA region of the HCMV genome in the orientation shown, replacing about 2.5 kb of the wild-type genome encompassing ORFs US7 through US9. An expanded view of the HindIII-X DNA region of the parental AD169 and recombinant RV series viruses is shown. The locations of relevant HindIII sites (H) and the asymmetrically located KpnI site (K) within UL99, used for other diagnostic DNA analyses (data not shown), are indicated. (C) DNA blot hybridization analysis of the genomic structure of RV18196. RV18196 and AD169 (parent) DNAs were digested with HindIII, electrophoresed, and hybridized with probes specific for either UL99 (left) and the HCMV US6-7 intergenic region (right). The right panel is the same blot as shown in the left panel, except that it was rehybridized with the US6 region probe, without striping the prior UL99 probe hybridization. The locations of the wild-type and mutant HindIII-X DNA fragments (5.0 and 4.4 kb, respectively), as well as the wild-type HindIII-R DNA fragment (6.7 kb), are indicated. (D) Single-cycle growth analysis of recombinant viruses expressing the UL99-GFP fusion proteins. HFF cells were infected, at a multiplicity of infection (MOI) of 1, with the indicated viruses. The total virus yield (intracellular plus extracellular) was harvested, and titers were determined daily.
FIG. 3.
Expression of UL99-GFP fusion proteins. HFF cells were infected with recombinant viruses (MOI = 1) expressing UL99-GFP fusion proteins. At 3 dpi, infected cell proteins were harvested and examined by immunoblot analysis with the GFP monoclonal antibody. The relative amounts of each extract used per lane are indicated.
Myristoylation mutant.
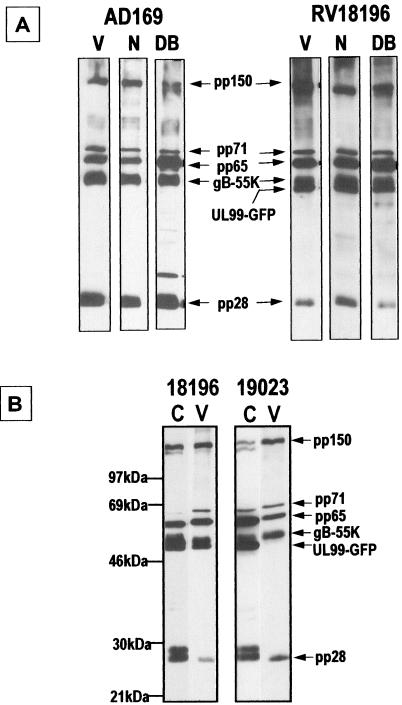
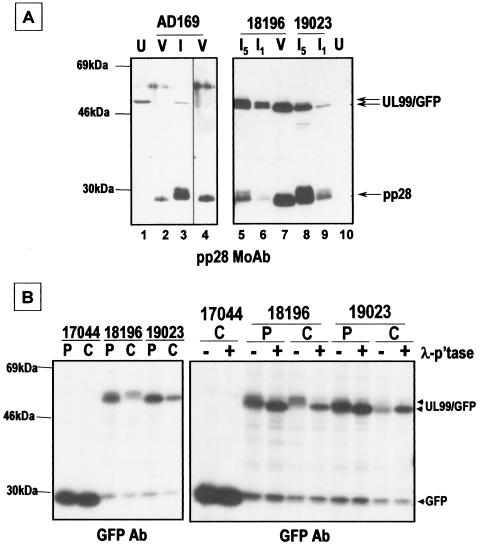
Trafficking of full-length UL99-GFP and its myristoylation site mutant (G2A) was assessed by deconvolution fluorescence microscopy. Infected HFF cells were examined at 2 and 3 dpi. RV17044, expressing unfused GFP, displays fluorescence throughout the cell (Fig. 4A and E). In contrast, RV18196, expressing UL99(1-190[WT])-GFP, displays plasma membrane and punctate perinuclear and cytoplasmic fluorescence at 2 dpi (Fig. 4B and C) but only punctate perinuclear and cytoplasmic fluorescence at 3 dpi (Fig. 4F and G). Expression of myristoylation mutant fusion protein UL99(1-190 [G2A])-GFP by RV19023 resulted in total cellular fluorescence identical to that of unfused GFP (Fig. 4D and H). These results indicate that myristoylation of glycine 2 is required for wild-type trafficking of UL99. The effect of myristoylation on virion incorporation was also investigated. Extracellular VPs were separated and enriched by sucrose density gradient centrifugation and then analyzed by immunoblotting. In parental AD169 particles, the 28-kDa UL99 protein (pp28) was detected in all three VPs (virions, noinfectious enveloped particles [NIEPs], and dense bodies [18]), as expected (Fig. 5A). Similarly, pp28 and the UL99(1-190)-GFP fusion protein was detected in all types of RV18196 VPs (Fig. 5A). However, when proteins present in RV18196- and RV19023-infected cells and virions were compared, the UL99-GFP fusion protein was detected at similar levels in cells infected by both viruses but was substantially reduced in RV19023 virions (Fig. 5B). Thus, myristoylation does not affect the apparent stability of the protein but does affect its propensity to be incorporated into virions, probably due to its reduced efficiency of association with cytoplasmic membranes involved in final envelopment (17, 43).
FIG. 4.
Cellular localization of UL99-GFP fusion proteins by fluorescence deconvolution microscopy. HFF cells were infected with the indicated recombinant virus (MOI = 1). At 2 or 3 dpi, Z-stacks of fluorescence photographs were taken and deconvoluted by using either three-dimensional filter (A, B, D to F, and H) or inverse filter (C and G) deconvolution software. Green fluorescence is GFP (all panels), and blue (Hoescht) fluorescence is nuclear (C and G only).
FIG. 5.
Immunoblot analysis of VPs and infected cell proteins with HCMV VP polyclonal antiserum. Extracellular VPs were separated by using a sucrose gradient. (A) AD169 and RV18196 extracellular VPs; (B) RV18196- and RV19023-infected cell and virion proteins. V, virions; N, NIEPs; DB, dense bodies; C, infected cell proteins.
During the course of these studies, it was observed that pp28 expressed in infected cells migrated diffusely in SDS-PAGE, whereas pp28 from VPs migrated as a more compact band (Fig. 6A). Similar observations were made for the UL99(1-190)-GFP protein encoded by RV18196. In the latter case, the faster-migrating form that was incorporated into virions comigrated with the nonmyristoylated form expressed in RV19023-infected cells. Since we have already shown that myristoylation is required for efficient incorporation into virions, it is unlikely that the faster-migrating form detected in RV18196 virions is not myristoylated. Pulse-chase radiolabeling experiments indicate that UL99(1-190)-GFP is posttranslationally modified to a slower-migrating form in the chase lane (Fig. 6B). A similar situation was reported previously for the UL97 gene product and was shown to be due to posttranslational phosphorylation (51). Treatment of the chase UL99-GFP immunoprecipitate with lambda phosphatase resulted in dephosphorylation and the conversion of the slower-migrating form to the faster form (Fig. 6B). As a control, the electrophoretic mobility of unfused GFP was not altered by phosphatase treatment, confirming its lack of phosphorylation and indicating that the phosphorylated residues of the fusion protein lie within the UL99 portion. Thus, the form of UL99(1-190)-GFP that is found in VPs is likely a hypophosphorylated form. By analogy, we hypothesize the same for wild-type (i.e., unfused) pp28. We also note that posttranslational modification, as detected by mobility shift in SDS-PAGE (i.e., in either pulse-chase or dephosphorylation experiments), was not observed for the nonmyristoylated UL99-GFP fusion encoded by RV19023 (Fig. 6B). However, phosphate radiolabeling experiments did indicate that it is phosphorylated (data not shown), although less than the UL99-GFP fusion encoded by RV18196.
FIG. 6.
(A) Immunoblot analysis of VPs and infected cell proteins with pp28 monoclonal antibody. U, uninfected cell proteins; I, infected cell proteins; V, virion proteins. The relative amounts of infected cell proteins are given (i.e., I5 has fivefold more infected cell protein than I1). Lane 4 is a fourfold-longer exposure of lane 2. (B) Pulse-chase radiolabeling-immunoprecipitation analysis of UL99-GFP fusion proteins. Infected cells (MOI = 1) were radiolabeled for 45 min with 200 μCi of 35S-labeled methionine-cysteine per ml at 20 h postinfection and then chased in nonradioactive medium for 4 h before the lysates were prepared. Immunoprecipitation was done with GFP polyclonal antibody. Lambda phosphatase treatment was done as indicated. P, pulse; C, chase.
C-terminal deletion mutants.
To determine motif(s) required for proper intracellular trafficking, localization of UL99 C-terminal deletion mutants fused to GFP were analyzed in the context of HFF cells infected with the appropriate recombinant HCMV at 2 and 3 dpi (Fig. 7). In cells infected with viruses expressing fusion proteins terminating at UL99 amino acids 100 and 57, mostly punctate perinuclear and cytoplasmic fluorescence, as well as some diffuse juxtanuclear fluorescence, was observed (Fig. 7A, B, E, and F), findings similar to that of the full-length UL99 wild-type fusion (Fig. 4C and G). In contrast, cells expressing fusion proteins terminating at UL99 amino acids 43 and 22 displayed mostly plasma membrane fluorescence and some diffuse juxtanuclear fluorescence but not punctate perinuclear and cytoplasmic fluorescence (Fig. 7C, D, G, and H). Thus, these data indicate that an important trafficking motif is within UL99 amino acids 44 through 57. This region contains a very acidic cluster of amino acids: 11 of 14 residues are either aspartate or glutamate (Fig. 1A). Differential staining with organelle-specific fluorescent probes was done to assess the intracellular localization of UL99 fusion proteins containing or lacking the AC (Fig. 8). BODIPY-TR ceramide was used to label the Golgi apparatus; LysoTracker-Red was used to label acidic compartments, including lysosomes, endosomes, and Golgi. RV19024-, RV19047-, and RV17044-infected HFF cells were examined as respresentatives of UL99-GFP fusion proteins containing the AC, or lacking the AC, and unfused GFP, respectively. The data indicate that a relatively small portion of both types of UL99 fusion proteins, either containing or lacking the AC, partially overlap with both the golgi and acidic compartments. Specifically, this overlap is limited to the diffuse juxtanuclear GFP fluorescence (red arrows in Fig. 8A and E). However, the AC-containing fusion proteins (i.e., those that display mostly punctate perinuclear and cytoplasmic fluorescence [i.e., vacuole-like structures]) do not overlap with either the Golgi or acidic compartments. Thus, the vacuole-like fluorescence does not represent accumulation of fusion protein in lysosomes since these structures are not labeled with LysoTracker. In contrast, we hypothesize that these structures represent sites of cytoplasmic (final) tegumentation or envelopment of capsids, similar to those observed and reported previously to which wild-type UL99-encoded protein localizes (17, 43). Further evidence suggestive of this is the colocalization of the Hoechst DNA stain to these structures (for examples, see yellow arrows in Fig. 8B). No such cytoplasmic DNA staining was observed in the cells infected with fusion proteins lacking the AC (Fig. 8F). As expected, there is no specific colocalization of the unfused GFP protein (Fig. 8I and J).
FIG. 7.
Cellular localization of UL99-GFP fusion proteins by fluorescence deconvolution microscopy. The experiment was done as described in the legend to Fig. 4, except that all Z-stacks were processed by using inverse filter deconvolution software.
FIG. 8.
Cellular localization of UL99-GFP fusion proteins by fluorescence deconvolution microscopy by using organelle markers. Recombinant virus-infected HFF cells were examined at 3 dpi by fluorescence deconvolution microscopy (inverse filter) as described in Fig. 4. RV19024 (A to D) and RV19047 (E to H) express truncated UL99-GFP fusions containing or lacking the AC, respectively. (I to J) RV17044 expresses unfused GFP. GFP fluorescence is green (A, C, E, G, and I); nuclear/DNA fluorescence is blue (A to J), organelle fluorescence is red and either specific for Golgi apparatus (B, F, and J) or for lysosomes and acidic compartments (D and H). Photos are in pairs (A-B, C-D, E-F, G-H, and I-J) showing the same field with different filters.
Region of UL99 required for viral replicative cycle.
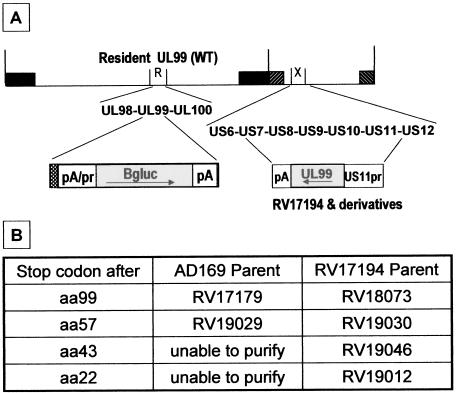
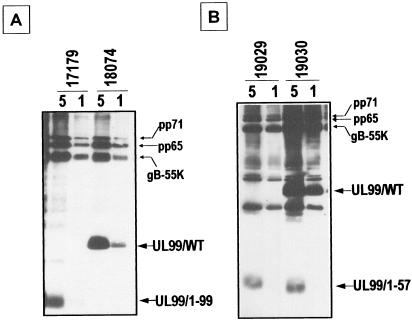
Deletion mutagenesis of UL99 was done to assess the requirement of UL99 in the HCMV infectious cycle. The strategy was to make a series of recombinant HCMVs that have sequential deletions from the C-terminal end of the resident UL99 gene. These deletions were made in the context of two parental strains: AD169 and, for positive control purposes, RV17194. RV17194 is diploid for UL99; it has a second copy of UL99 inserted within the nonessential US6 region of the HCMV genome, replacing the US11-US7 genes, and under control of the early-late US11 promoter (Fig. 9A). Thus, any deletion that results in a lethal mutation will not be able to be purified when made in the context of the AD169. However, the same deletion should be able to be purified when made in the context of the RV17194 parent, since there is a complementing wild-type UL99 expressed from the US6 region of the genome. The only exceptions are if the mutation at the resident UL99 is a dominant-negative lethal mutation or if it affects the expression of a neighboring ORF, such as the alkaline nuclease UL98 gene, whose C-terminal coding region overlaps the N-terminal 22-amino-acid coding region of UL99. Such dominant-negative lethal mutations did not occur. Deletions of the resident UL99 gene were done so that an in-frame stop codon, followed by the β-glucuronidase expression cassette, was inserted at the desired location. Since the polyadenylation signal for ORFs 93 to 99 is downstream of UL99 (55), the β-glucuronidase expression cassette has a second polyadenylation signal, located adjacent to the UL99 in-frame stop codon, to terminate upstream viral transcripts before the reporter gene (Fig. 9A). In this fashion, all RV17194-derived “positive control” mutants were obtained (and purified) with stop codons after resident UL99 amino acids 22, 43, 57, and 99 (Fig. 9B). However, only mutants with stop codons after amino acids 57 and 99 were able to be purified when we used the AD169 parent strain (Fig. 9B). Also, pseudo-wild-type virus (RV18074) was created by “rescuing” the UL99 mutation in RV17179 (a virus that had a stop codon and reporter cassette after UL99 amino acid 99) back to the wild type. Single cycle growth curves were generated for each virus; all grew with normal kinetics (burst just after 2 dpi), although overall viral yields were slightly (up to 1 log) lower than AD169 (Fig. 9C and D). Thus, the data indicate that a severely truncated UL99 protein (amino acids 1 to 57 present) is sufficient for near wild-type HCMV growth. Furthermore, UL99 amino acids 58 to 190 are nonessential, and their presence may only contribute a slight growth advantage to virus in cultured HFF cells. Recently, Silva et al. used BAC technology to demonstrate that UL99 is an essential gene (45). Compared to wild-type virus, their UL99 deletion mutant was severely impaired in HFF cells in a multicycle assay: no virus was detected, thus, the yield was reduced by at least 5 logs (45). Our results are in agreement and extend their data, suggesting that the AC, UL99 amino acids 44 to 57 that are required for proper intracellular trafficking, also imparts a function critical for viral growth. In immunoblot analyses, expression of the UL99 proteins truncated after amino acids 57 and 99 was detected in infected cells (data not shown) and virions (Fig. 10). Specifically, the ∼7-kDa UL99(1-57) protein was found in virions both in the presence (i.e., RV19030) and in the absence (i.e., RV19029) of wild-type pp28. Thus, sequence information within the first 57 amino acids is sufficient for virion incorporation.
FIG. 9.
(A) Schematic diagram of the HCMV genome showing the strategy for construction of deletion mutants of the resident UL99 gene by insertion of a β-glucuronidase expression cassette, with flanking UL99 in-frame stop codon (checkerboard box) and dual polyadenylation signals. The HSV tk polyadenylation signal adjacent to the UL99 in-frame stop codon terminates upstream viral transcripts from UL93-99 before the β-glucuronidase reporter gene. The resident UL99 is within the HindIII-R DNA fragment. A second copy of UL99, present in RV17194 and derivatives, was inserted within the HindIII-X DNA fragment, replacing US7 through US11, as shown. (B) Description of UL99 deletion mutant viruses. (C and D) Single-cycle growth analysis. HFF cells were infected at an MOI of 1; total virus was harvested daily, and titers were determined.
FIG. 10.
Immunoblot analysis of UL99 deletion mutant virion proteins with the HCMV VP antibody. Virions were enriched by sucrose gradient centrifugation from the extracellular medium of infected HFF cells. (A) RV17179 expresses the resident UL99 protein truncated after amino acid 99; RV18074 is a rescued derivative of RV17179 that expresses wild-type UL99. (B) RV19029 and RV19030 express UL99 proteins truncated after amino acid 57, either in the absence or in the presence of the wild-type UL99 protein, respectively.
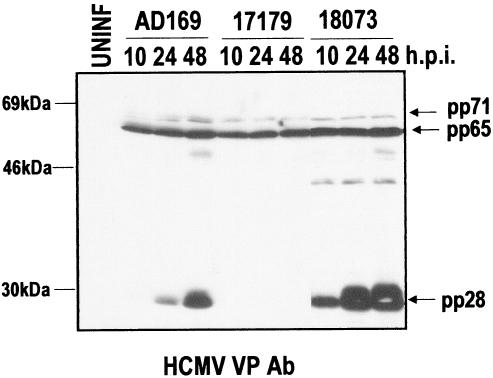
The fact that RV17194 and its derivatives (Table 1) are viable not only controls for resident UL99 mutations but also indicate that the wild-type strict late expression kinetics of UL99, as observed in AD169, is not required (Fig. 11). In RV17194-derived viruses, such as RV18073, UL99 is expressed with early-late kinetics, since it is under the control of the US11 promoter (Fig. 11). These data indicate that misregulation of UL99 is of no consequence, at least in a tissue culture system.
FIG. 11.
pp28 expression from AD169-, RV17179-, and RV18073-infected cells. HFF cells were infected at an MOI of 1; protein extracts were made at the indicated time (hours postinfection [h.p.i.]) and analyzed by immunoblotting with the HCMV VP antibody.
DISCUSSION
The tegument of herpesviruses is an amorphous collection of proteins that is the “cement” between the capsid and the viral envelope (33). Tegument proteins are found in all three types of HCMV VPs (virions, NIEPs, and dense bodies) (18) and many are phosphorylated (14, 18, 34, 41). In a detailed study, Baldick and Shenk (4) examined the protein composition of HCMV particles and identified at least nine proteins as probable abundant tegument components (4). The major HCMV tegument phosphoproteins are pp150, pp71, pp65, and pp28. Other, perhaps minor, phosphoprotein components of the tegument have been reported, including ppUL97 (51). Genetic or functional analysis of two of the most-studied phosphorylated tegument proteins has indicated the following: (i) pp71, encoded by UL82, is a transcriptional transactivator, acting early in the infectious cycle, is highly beneficial for viral growth in culture, and stimulates progression through the cell cycle (5, 6, 24, 30) and (ii) pp65, encoded by UL83, is a prominent target of the cellular immune system, is nonessential for growth in culture, is required for the production of dense bodies, and mediates accumulation and degradation of HLA class II in lysosomes (18, 25, 37, 44, 54). Functional analysis of the UL32 gene product, pp150, has not been reported. As reported recently, an initial genetic analysis of the pp28 tegument protein, a true late product of the UL99 ORF, indicates that this protein is indeed essential for completion of the HCMV replication cycle (45). Our genetic studies of UL99 here extend this observation and indicate the following: (i) proper intracellular trafficking of UL99-encoded protein requires myristoylation and an AC; (ii) over two-thirds of UL99 is nonessential; and (iii) the amino-terminal one-third of UL99, amino acids 1 to 57 containing the AC, is sufficient for virus growth at near wild-type levels. Our data also suggest that a truncated protein containing UL99 amino acids 1 to 43 does not support viral growth and UL99 proteins incorporated into virions are a hypophosphorylated subset of those found in infected cells.
The genetic manipulation of UL99 within recombinant viruses was confounded by the fact that the UL99 N-terminal coding region (i.e., first 22 amino acids) is overlapped by the C-terminal coding region of the upstream UL98 ORF that encodes an alkaline exonuclease (9). By analogy with its HSV homolog UL12, mutation of UL98 may cause a near lethal phenotype, similar to UL12 mutants, since the HSV protein is involved in processing progeny DNA replication intermediates and recombination (32, 40, 53). HCMV UL98 has been shown to complement a HSV UL12-deficient mutant (12). Furthermore, ORFs upstream of UL99 share a common polyadenylation signal located just downstream of UL99 (55). Thus, the creation of UL99 mutations could affect the expression of the upstream ORFs. Our mutagenesis strategy to define essential/nonessential domains of UL99 has addressed these concerns so that the proper interpretations could be made. Specifically, (i) an alternate polyadenylation signal was present on the upstream side of our inserted reporter cassette; (ii) the coding region of alkaline nuclease UL98 was not disturbed; and (iii) two parent viruses were used, including a positive control virus diploid for UL99, so that the possible creation of an otherwise lethal or dominant-negative mutant by expression of a truncated UL99 protein would be identified. All UL99 deletion mutants were obtained with the diploid parent (RV17194) when deletions of the resident UL99 gene were made after codons for amino acids 22 and 43, even though these mutations were unable to be purified with the wild-type (i.e., haploid) parent virus (AD169). Mutations resulting in deletions after UL99 amino acids 57 and 99 were obtained with both parental strains and grew with near-wild-type kinetics (Fig. 9C and D). The interpretation of the data is that the AC, amino acids 44 to 57, is required for virus growth. These data are in agreement with, and extend, observations made recently using a BAC-mediated HCMV mutagenesis strategy that demonstrated that a UL99 mutant truncated after amino acid 22 does not grow in the absence of a complementing cell line (45).
Some herpesvirus tegument proteins are believed to be involved both in entry (i.e., cytoplasm to nucleus transport after fusion) and egress (33). Very little is known regarding the entry process. Meredith and coworkers propose that herpesvirus tegument kinases phosphorylate other tegument proteins shortly after infection to facilitate dissociation from the capsid (35, 36). Recently, it was shown that cellular p180, an ER-resident protein, binds pUL48, a HCMV tegument protein that, in turn, binds tightly to capsid proteins (38). This interaction may be required for successful capsid migration to the nucleus early during infection and for efficient egress from the nucleus to the site of virion assembly in the cytoplasm at late times (38). Another tegument protein, ppUL97, a protein kinase, was recently been shown to be required for efficient egress from the nucleus (29). Regarding HCMV and other herpesviruses, substantial evidence has indicated that the site of virion assembly (i.e., final tegumentation and envelopment) is at a intracytoplasmic membrane of a Golgi- or TGN-derived intracytoplasmic vacuole (13, 15-17, 33, 42, 46, 49). In fact, it has been shown that pp28, pp71, pp150, and major HCMV glycoproteins, such as gB, accumulate at these vacuolar membranes (17, 42). Thus, the evidence suggests that progeny capsids interact with, or acquire, tegument proteins at two different sites, one within the nucleus and later at a intracytoplasmic membrane. Tegument proteins pp65 and pUL48 localize to the nucleus and may associate with capsids there. In contrast, the major phosphoproteins pp150, pp71, and pp28 localize exclusively to intracytoplasmic vesicles (17, 42). Our data demonstrate that an UL99 AC comprised of mostly aspartate and glutamate residues (i.e., amino acids 44 to 57; Fig. 2) is required for proper intracellular trafficking to the vacuolar-like structures, as well as function. In our trafficking studies, the indicator UL99-GFP fusion was expressed from recombinant viruses that also expressed wild-type pp28. Thus, the fact that the same region was identified as being required for proper intracellular trafficking and viral growth is not due to the absolute requirement that the fusion proteins be functional. Instead, we hypothesize that proper intracellular trafficking and the function of pp28 are interlinked.
HCMV UL99 has positional homology and regional amino acid similarity with HSV UL11 (9). UL11 encodes a 96-amino-acid phosphoprotein that is both myristoylated and palmitylated (31). Interestingly, the amino acid similarity between HSV UL11 and HCMV UL99 resides within the N-terminal half of the latter protein. Similar to UL99, UL11 is myristoylated at glycine 2, has several cysteine residues within the first 13 amino acids, and contains an aspartate- and glutamate-rich AC in the region of amino acid 40. UL11 binds to the cytoplasmic face of intracytoplasmic membranes and is recycled between the plasma membrane and Golgi/TGN via PACS-1 (for phosphofurin AC sorting protein) (2, 31). As with other herpesvirus proteins that utilize PACS-1 for this function, including HCMV gB (10, 19, 50), pseudorabies virus US9 (7), and varicella-zoster virus gE (1), UL11 trafficking is dependent on the AC (31). Furthermore, casein kinase 2 (CK2)-dependent phosphorylation/phosphatase 2A dephosphorylation in the area of the AC is also usually involved in trafficking between the plasma membrane and Golgi/TGN cycling loops (48). We speculate that the apparent hypophosphorylated subset of UL99 protein that is incorporated into virions may arise due to the phosphorylation and dephosphorylation associated with this trafficking (Fig. 6). Consistent with this are our observations that, unlike the wild-type fusion, the UL99-GFP myristoylation mutant does not traffic to intracytoplasmic vesicle-like structures, is not efficiently incorporated into virions, and does not have hyperphosphorylated forms (Fig. 4 to 6). Furthermore, it was reported that the HSV UL49-encoded tegument protein VP22 traffics to membranes of TGN-derived acidic compartments and that a hypoposphorylated VP22 form is preferentially packaged into virions (8, 35).
Studies of an HSV UL11 deletion series have indicated that information for trafficking is contained with the first 49 amino acids of UL11, which are analogous to the first 57 amino acids of UL99 that we identified herein (31). UL99-GFP fusion proteins that are truncated after UL99 amino acids 57 and 100 are similar to a wild-type fusion localizing to the plasma membrane, as well as to punctate perinuclear and cytoplasmic vacuole-like structures at 2 dpi. However, by 3 dpi, these fusions localized to the latter areas only. Conversely, fusions truncated after UL99 amino acids 22 and 43 mostly localized to the plasma membrane through 3 dpi. Serine residues at 41 and 42 are potentially phosphorylated since they are within consensus CK2 sites (S/TxxD/E). Truncations after amino acid 43 would not only remove the AC but also a portion of both CK2 sites. Thus, UL99 truncations after amino acids 22 and 43, although they retain the myristoylation sequence, localize to but are not efficiently retrieved from the plasma membrane due to the absence of the AC and CK2 phosphorylation sites. HSV UL11 fusions in which the AC has been either deleted or mutated behave similarly (31).
It is not known whether the portion of HSV UL11 encompassing amino acids 1 to 49 is sufficient for virus growth. However, an HSV-1 mutant deleted of most of UL11 is severely defective, resulting in a 3- to 4-log reduction in yield and time-delayed appearance of extracellular virons (3). In that study, the absence of UL11 protein caused the accumulation of naked nucleocapsids in the cytoplasm. Our data extend the previously known genome position and amino acid similarities between HSV-1 UL11 and HCMV UL99, suggesting that there may be functional homologies between the two proteins as well. Stretches of highly charged amino acids are likely to be exposed on the surface of folded proteins and serve as sites of interaction with other proteins. Thus, UL99 may contribute to the cytoplasmic assembly of complete virions by providing a surface(s) for the interaction of the cytoplasmic domain of virion glycoproteins and/or other tegument proteins, thereby facilitating their maturation and transport to the extracellular space. Consistent with this view is the recent report that a severely defective UL99 deletion mutant virus accumulated tegumented, nonenveloped capsids in the cytoplasm (45). We note that coronavirus, an enveloped positive-strand RNA virus, expresses a small essential protein, E, that has an acidic region near its C terminus and may provide function similar in virus assembly (11, 52) as UL11 and UL99 proteins do for their respective herpesviruses.
Lastly, UL99-encoded pp28 is expressed with true late kinetics and has been studied as the prototypical gene for this class in promoter studies (26, 28). It was not known whether such restricted expression was required or, alternatively, whether misregulation is deleterious. We created recombinant mutants whereby UL99 was expressed with early-late kinetics rather than with true late kinetics (Fig. 10). Some of these mutants also expressed functional UL99 protein from the resident gene and promoter, whereas others did not. Early-late expression kinetics did not affect the growth of any of these viruses in cultured cells. This observation is consistent with data from other recombinant viruses made in our laboratory that contain substantial promoter alterations regulating other essential or highly beneficial genes, including UL54 polymerase, UL97 kinase, and UL98 alkaline exonuclease (19a).
In summary, our data indicate that the C-terminal two-thirds of the 190-amino-acid UL99-encoded tegument protein is nonessential for completion of the HCMV infectious cycle. A mutation deleting ∼90% of this ORF could not be isolated, unless a complementing UL99 gene was present, a finding in agreement with recent data indicating that UL99 is essential (45). Conversely, we also show that, at most, the N-terminal one-third of UL99, encompassing the myristoylation site and AC, is sufficient for viral growth and incorporation into VPs. The myristoylation site and AC are also required for proper intracytoplasmic vesicular localization. The HCMV UL99-encoded protein, especially its N-terminal one-third, appears to be very similar to HSV UL11-encoded protein. These data provide the basis for future studies to more precisely define the UL99 sequences necessary for the essential function(s) of pp28 and to further assess its functional similarity with HSV UL11.
Acknowledgments
We thank John O'Connell, Joe Baldick, Hua Zhu, Luz Hermida-Matsumoto, and Jeanette Fairhurst for support and helpful suggestions.
REFERENCES
- 1.Alconada, A., U. Bauer, and B. Hoflack. 1996. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 15:6096-6110. [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus UL11 proteins are associated with cytoplasmic and nuclear membranes and nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., and T. Shenk. 2000. UL82 virion protein activates expression of immediate-early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau, A. D., T. Del Rio, E. J. Wolfe, and L. W. Enquist. 1999. Intracellular trafficking and localization of the pseudorabies virus US9 type II envelope protein to host and viral membranes. J. Virol. 73:4372-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 membrane protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 10.Fish, K. N., C. Soderberg-Naucler, and J. A. Nelson. 1998. Steady-state plasma membrane expression of human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J. Virol. 72:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, M., B. J. Robertson, P. J. McCann, D. R. O'Boyle, S. K. Weller, W. W. Newcomb, J. C. Brown, and S. P. Weinheimer. 1998. Functional conservations of the alkaline nuclease of herpes simplex type 1 and human cytomegalovirus. Virology 249:460-470. [DOI] [PubMed] [Google Scholar]
- 13.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, W. 1983. Protein counterparts of human and simian cytomegaloviruses. Virology 128:391-406. [DOI] [PubMed] [Google Scholar]
- 15.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granzow, H., F. Weiland, A. Jons, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a non-infectious virion-like particle released from the cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis, M. A., K. N. Fish, C. Soderberg-Naucler, D. N. Streblow, H. L. Meyers, G. Thomas, and J. A. Nelson. 2002. Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J. Virol. 76:5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Jarvis, M. A., T. R. Jones, D. D. Drummond, P. P. Smith, W. J. Britt, J. A. Nelson, and C. J. Baldick. 2004. Phosphorylation of human cytomegalovirus glycoprotein B (gB) at the acidic cluster casein kinase 2 site (Ser900) is required for localization of gB to the trans-Golgi network and efficient virus replication. J. Virol. 78:285-293. [DOI] [PMC free article] [PubMed]
- 19b.Jones, T. R., S.-W. Lee, S. V. Johann, V. Razinkov, R. J. Visalli, B. Feld, J. D. Bloom, and J. O’Connell. 2004. Specific inhibition of human cytomegalovirus glycoprotein B-mediated fusion by a novel thiourea small molecule. J. Virol. 78:1289-1300. [DOI] [PMC free article] [PubMed]
- 20.Jones, T. R., and V. P. Muzithras. 1991. Fine mapping of transcripts expressed from the US6 gene family of human cytomegalovirus strain AD169. J. Virol. 65:2024-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T. R., V. P. Muzithras, and Y. Gluzman. 1991. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 65:5860-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, T. R., L. Sun, G. A. Bebernitz, V. P. Muzithras, H. J. Kim, S. H. Johnston, and E. Z. Baum. 1994. Proteolytic activity of human cytomegalovirus UL80 protease cleavage site mutants. J. Virol. 68:3742-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalejta, R. F., and T. Shenk. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virol. 77:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T-cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709-1716. [DOI] [PubMed] [Google Scholar]
- 26.Kerry, J. A., M. A. Priddy, C. P. Kohler, T. L. Stanley, D. Weber, T. R. Jones, and R. M. Stenberg. 1997. Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J. Virol. 71:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, H.-J., C. Gatz, W. Hillen, and T. R. Jones. 1995. Tetracycline repressor-regulated gene repression in recombinant human cytomegalovirus. J. Virol. 69:2565-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, C. P., M. Carter, J. A. Kerry, V. P. Muzithras, T. R. Jones, and R. M. Stenberg. 1994. Use of recombinant virus to assess human cytomegalovirus early and late promoters in the context of the viral genome. J. Virol. 68:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krosky, P. M., M.-C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loomis, J. S., B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2674. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 35.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 36.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odeberg, J., B. Plachter, L. Branden, and C. Soderberg-Naucler. 2003. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood 101:4870-4877. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa-Goto, K., S. Irie, A. Omori, Y. Miura, H. Katano, H. Hasegawa, T. Kurata, T. Sata, and Y. Arao. 2002. An endoplasmic reticulum protein, p180, is highly expressed in human cytomegalovirus-permissive cells and interacts with the tegument protein encoded by UL48. J. Virol. 76:2350-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pass, R. 2001. Cytomegalovirus, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 40.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roby, C., and W. Gibson. 1986. Characterization of the phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59:714-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva, M. C., Q-C Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stinski, M. F. 1976. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J. Virol. 19:594-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tooze, J. M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 50.Tugizov, S., E. Maidji, J. Xiao, and L. Pereira. 1999. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J. Virol. 73:8677-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Zeijl, M., J. Fairhurst, E. Z. Baum, L. Sun, and T. R. Jones. 1997. Expression and posttranslational modification of human cytomegalovirus UL97 phosphotransferase. Virology 231:72-80. [DOI] [PubMed] [Google Scholar]
- 52.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]
- 54.Wills, M., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and P. J. G. Sissons. 1996. The human cytotoxic-T-lymphocyte (CTL) response to cytomegaloviorus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wing, B. A., and E. S. Huang. 1995. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J. Virol. 69:1521-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]