Abstract
Both CD4+ and CD8+ T cells are required for clearance of the murine coronavirus mouse hepatitis virus (MHV) during acute infection. We investigated the effects of an epitope-specific CD8+ T-cell response on acute infection of MHV, strain A59, in the murine CNS. Mice with CD8+ T cells specific for gp33-41 (an H-2Db-restricted CD8+ T-cell epitope derived from lymphocytic choriomeningitis glycoprotein) were infected with a recombinant MHV-A59, also expressing gp33-41, as a fusion protein with enhanced green fluorescent protein (EGFP). By 5 days postinfection, these mice showed significantly (approximately 20-fold) lower titers of infectious virus in the brain compared to control mice. Furthermore mice with gp33-41-specific CD8+ cells exhibited much reduced levels of viral antigen in the brain as measured by immunohistochemistry using an antibody directed against viral nucleocapsid. More than 90% of the viruses recovered from brain lysates of such protected mice, at 5 days postinfection, had lost the ability to express EGFP and had deletions in their genomes encompassing EGFP and gp33-41. In addition, genomes of viruses from about half the plaques that retained the EGFP gene had mutations within the gp33-41 epitope. On the other hand, gp33-41-specific cells failed to protect perforin-deficient mice from infection by the recombinant MHV expressing gp33, indicating that perforin-mediated mechanisms were needed. Virus recovered from perforin-deficient mice did not exhibit loss of EGFP expression and the gp33-41 epitope. These observations suggest that the cytotoxic T-cell response to gp33-41 exerts a strong immune pressure that quickly selects epitope escape mutants to gp33-41.
Infection of mice with the murine coronavirus mouse hepatitis virus (MHV) provides a model for studying acute virus-induced neurological disease as well as providing a model for chronic demyelinating diseases, such as multiple sclerosis. Following intracranial (i.c.) inoculation, MHV-A59 replicates in the central nervous system (CNS) and causes acute encephalitis, which peaks at about 7 days postinfection (12); virus is cleared from the CNS by 7 to 14 days postinfection (27). Following clearance, surviving mice develop an immune-mediated demyelinating disease (12), which peaks at about 30 days postinfection (36).
Clearance of infectious MHV from the CNS requires multiple components of the immune response. Adoptive transfer experiments in combination with depletion experiments have demonstrated that both CD8+ and CD4+ T cells are critical for normal viral clearance (10, 16, 32-35, 38, 40). Furthermore, the peak of T lymphocyte infiltration into the CNS is coincident with falling titers of infectious virus in the CNS (39). While B cells are also recruited into the CNS, this occurs as virus replication is completed and the acute disease is resolving; neither B cells nor antibody is required for clearance of acute infection (13, 18).
The virus-specific CD8+ T-cell response to MHV has been studied in C57BL/6 (B6) mice. In B6 mice, two CD8+ T-cell epitopes have been identified within the structural proteins of MHV; both of these are within the spike protein (2). CD8+ T cells specific for the immunodominant S510-518 epitope (referred to here as S510) and the subdominant epitope S598-605 (referred to here as S598) have been detected in the CNS of mice infected with the JHM strain of MHV (MHV-JHM), a highly neurovirulent strain. A 52-amino-acid deletion in the region of the MHV-A59 spike surrounding S510 relative to the spike of MHV-JHM (15, 21) eliminates the S510 epitope in the MHV-A59 spike; thus, infection with MHV-A59 does not elicit a response to that epitope, but only to S598. While S598-specific CD8+ T cells have been viewed as an unimportant antiviral effector cells in vivo, at least in the context of infection with the JHM strain, response to this epitope is apparently sufficient to protect mice from MHV-A59 infection as it is the only identified CD8+ T-cell epitope in the MHV-A59 genome (16). Approximately 12% of the CD8+ cells in the CNS of MHV-A59-infected animals are specific for this epitope (as measured by an intracellular gamma interferon [IFN-γ] assay). This level of S598 epitope-specific CD8+ T cells is the same even when the S510 epitope is expressed (25). Furthermore, infection of β2-microglobulin-deficient mice (β2 M−/−) with MHV-A59 results in a highly lethal infection (7), suggesting that this epitope is important in controlling infection. (Alternatively, there may be as yet undefined epitopes in nonstructural proteins, which have not yet been investigated for CD8+ T epitopes.)
CD8+ T cells clear virus both by perforin-mediated mechanisms and through the actions of IFN-γ. Perforin-mediated cytolysis has been shown to play a role in controlling virus replication in astrocytes and microglia during acute infection, while IFN-γ is necessary for clearance from oligodendrocytes (14, 22). The mechanism of clearance from neurons has not yet been addressed. Mice deficient in either perforin or IFN-γ experience delayed clearance of MHV in the CNS (14, 22); however, IFN-γ is believed to play a more important role in viral clearance and protection from disease (16).
This study was designed to determine the ability of an epitope-specific CD8+ T-cell response to a specific epitope to protect animals from subsequent infection with a virus expressing that epitope. For this purpose we selected a recombinant murine coronavirus in which a strong CD8+ epitope from lymphocytic choriomeningitis virus (LCMV) glycoprotein, gp33-41 (referred to here as gp33) was expressed as a fusion protein with the enhanced green fluorescent protein (EGFP) in place of nonessential gene 4. To determine the ability of gp33-specific CD8+ T cells to provide protection, we challenged mice that had been previously immunized with a recombinant Listeria monocytogenes (rLm) also encoding the LCMV gp33 epitope (rLm-gp33). Immunization with rLm-gp33 did protect against virus replication and spread of MHV antigen in the CNS, in a perforin-dependent manner. However, despite the fact that gp33 epitope escape mutants were readily selected, virus was cleared with similar kinetics as in naive mice, that is, within the first week to 10 days postinfection.
MATERIALS AND METHODS
Mice, bacteria, and viruses.
Four-week-old C57BL/6 (B6) and perforin-deficient B6 (PKO) (9) mice were obtained from the National Cancer Institute and the Jackson Laboratory, respectively. rLm strains expressing the LCMV CD8+ (H-2Db) epitope gp33-41 (KAVYNFATC) or the (H-2Ld) epitope np118-126 (RPQASGVYM) were engineered as described previously (29); both epitopes are expressed as fusion proteins with dihydrofolate reductase. The strain expressing gp33-41 is called XFL703 and will be referred to here as rLm-gp33 (28). The strain expressing np118-126 is called XFL303 and will be referred to here as rLm-np118 (29). A recombinant MHV strain (SA59REGFP) expressing EGFP was derived from strain MHV-A59 by targeted recombination of the EGFP gene (catalogue no. 6085-1; Clontech) in place of the gene 4 sequence and has been described previously (5); for simplicity we will refer to this strain here as RA59-gfp (4, 8). Selection of the RA59-gfp/gp33, recombinant MHV-A59 expressing the LCMV gp33 epitope as a fusion protein with GFP is described in the next paragraph. fMHV (obtained from Paul Masters, New York State Department of Health, Albany) is a recombinant MHV-A59 which contains the ectodomain of the feline infectious peritonitis virus spike glycoprotein in place of that of the MHV spike (11). MHV stocks were prepared in 17Cl-1 cells and titrated by plaque assay in murine L2 cells (6).
Selection of RA59-gfp/gp33.
Complementary synthetic oligonucleotides (63 bases) encoding the first 9 amino acids of MHV open reading frame 4a (ORF4a) (including the initial AUG codon) followed by the 9 amino acids of gp33 were synthesized such that, when hybridized, the 5′ and 3′ ends of the double-stranded fragment contained sequences complementary to SalI- and BamHI-cleaved DNA ends, respectively (Table 1). The double-stranded fragment was ligated into SalI/BamHI-cleaved pEGFP (Clontech vector containing the EGFP gene) such that the ORF4a/gp33 fragment was adjacent to and in frame with the EGFP gene. The ORF4a/gp33/EGFP portion of the plasmids was sequenced using primers EGFP 530+ and EGFP 766− (Table 1) and the Taq dye terminator procedure according to the manufacturer's protocol (Taq DyeDeoxy Terminator Cycle Sequencing kit; Applied Biosystems). The fragment encoding ORF4a/gp33/EGFP was cleaved from this plasmid with SalI and NotI and inserted into pMH54-EGFP (5) to replace most of gene 4. The plasmid pMH54 and its use in targeted recombination have been described previously (11, 27). Briefly, pMH54 contains a T7 RNA polymerase promoter followed by a 9,139-nucleotide sequence (11); pMH54 encodes the 5′ end of the MHV genome fused in frame to codon 28 of the hemagglutinin esterase pseudogene, followed by the spike and the rest of the 3′ end of the MHV-A59 genome and finally a poly(A) tail. In the final pMH54-gp33/EGFPconstruct, the ATG for ORF4a is 50 nucleotides downstream of the gene 3/gene4 intergenic sequence. This is followed by the nine codons of ORF4a, the nine codons of gp33, seven intervening codons followed by the ATG for the EGFP gene, and the rest of the EGFP gene, such that ORF4a, gp33, and the EGFP protein are all in frame (Fig. 1). Recombination was carried out in feline FCWF cells between the donor RNA (transcribed from pMH54-containing the gp33/EGFP sequences) and the recipient virus, fMHV. The recombinant viruses were selected by two rounds of plaque purification on L2 cells (11, 27). Recombinant virus genomes were amplified and sequenced in the regions that were mutagenized, that is, from the 3′ end of the spike gene into the 5′ end of the EGFP coding region. Reverse transcriptase-mediated PCR (RT-PCR) amplification was carried out, using as templates cytoplasmic RNA extracted from virus-infected L2 cells and the primers FIJ81 and RIJ84 (Table 1).
TABLE 1.
Oligonucleotides used for cloning and sequencing
| Primer name | Primer sequencea (5′-3′) | Location |
|---|---|---|
| GP33F (SalI) | tcgacGATGGCCGTGTTGGTCCTAAGGCTACATAAGGCTGTCTACAATTTTGCCACCTGTCCG | Synthetic ORF4a/gp33 fragment |
| GP33R BamHI | gatccGGACAGGTGGCAAAATTGTAGACAGCCTTATGTAGCCTTAGGACCAACACGGCCATCG | Synthetic ORF4a/gp33 fragment |
| FIJ81 | GTATGGAGGACACCAGGACAG | 3′ end of spike gene |
| RIJ84 | CCATGCATCACTCACATGCC | 5′ end of ORF5a |
| EGFP530+ | CGGTAGGCGTGTACGGTGGGAGG | Upstream of multiple cloning site in pEGFP |
| EGFP 766− | GCTGAACTTGTGGCCGTTTACGTCG | Downstream of multiple cloning site in pEGFP |
Restriction sites are shown in lowercase type. The gp33-41 coding region is in boldface type. The ATG for ORF4a is underlined.
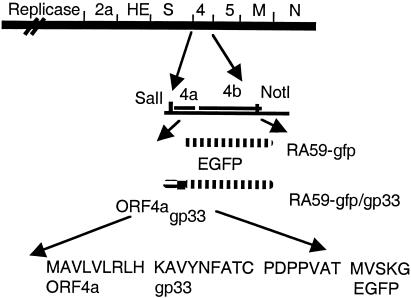
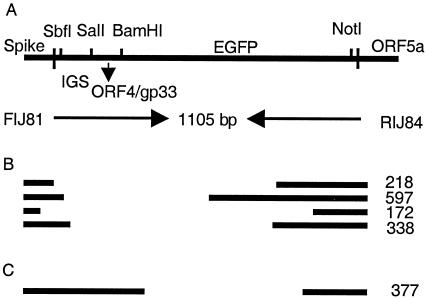
FIG. 1.
Recombinant viruses encoding EGFP and gp33. A schematic diagram of the genome of MHV-A59 is shown. Targeted recombination was used to replace gene 4 of MHV-A59 with the EGFP gene (RA59-gfp) or with a fragment of ORF4a, followed by gp33 and the EGFP gene (RA59-gfp/gp33), as described in the text. SalI and NotI sites were used to clone the gfp or ORF4a/gp33/gfp fragments into the pMH54 plasmids to generate synthetic RNA for recombination.
Inoculation of mice.
For immunization with rLm-gp33 or rLm-np118, mice were inoculated intraperitoneally (i.p.) with 104 CFU of rLm in 0.5 ml of phosphate-buffered saline (PBS). For i.c. inoculation of virus, mice were anesthetized with isoflurane and the amount of virus designated in each experiment was diluted in PBS containing 0.75% bovine serum albumin, and a total volume of 20 μl was injected into the left cerebral hemisphere. For i.p. inoculation, virus in PBS containing 0.75% bovine serum albumin, and a total volume of 500 μl was injected. Mock-infected controls were inoculated similarly but with an uninfected cell lysate at a comparable dilution.
Virus replication in mice.
For the measurement of virus replication in the brain and liver, at 5 days postinfection, mice were sacrificed and perfused with 10 ml of PBS, and brains and livers were removed. The left half of the brain and the liver were placed directly into 2 ml of isotonic saline with 0.167% gelatin (gel saline). The right half of the brain was used for histology and viral antigen staining as described below. All organs were weighed and stored frozen at −80°C until determination of virus titers. Organs were homogenized, and virus titers were determined by plaque assay on L2 cell monolayers (6).
Histology and immunohistochemistry.
For the analysis of inflammation and viral antigen expression, the right half of the brain harvested at 5 days postinfection (see above), was fixed in formalin overnight. Formalin-fixed tissue was embedded in paraffin and sectioned sagittally. Immunohistochemical analysis was performed by the avidin-biotin-immunoperoxidase technique (Vector Laboratories, Burlingame, Calif.) using 3,3′-diaminobenzidine as substrate and a 1:20 dilution of a monoclonal antibody (MAb) directed against the nucleocapsid protein (N) of MHV-JHM (MAb clone 1-16-1, kindly provided by Julian Leibowitz, Texas A & M University). Control slides from mice either uninfected or inoculated with rLm-gp33 but not with virus were incubated in parallel. All slides were read in a blinded manner.
Isolation and analysis of lymphocytes. (i) Isolation of mononuclear cells from the spleen and CNS.
Spleens were harvested from mice either at 5 or 7 days postinfection. Spleen cell suspensions were prepared by gently homogenizing spleens in a nylon bag (mesh opening, 64 μm) with a syringe plunger in RPMI 1640 medium supplemented with 1% fetal calf serum. Then, red blood cells were lysed with 0.83% NH4Cl. Mononuclear cells were prepared from the CNS as previously described by Phillips et al. (25) on day 7 after i.c. inoculation. Three to five brains were pooled per sample. In brief, animals were sacrificed and perfused with 10 ml of PBS, and a single-cell suspension from the brain was obtained by passing cells through a nylon mesh bag (mesh opening, 64 μm). Percoll (Pharmacia) was added to a final concentration of 30%, and the lysate was centrifuged at 1,300 × g for 30 min at 4°C. The cell pellet was resuspended, passed through a cell strainer (pore diameter, 70 μm; Becton Dickinson), and washed. The cells were then layered over 2 ml of Lympholyte-M (Cedarlane Laboratories) and centrifuged at 1,300 × g for 20 min at room temperature. Cells were removed from the interface, washed once, and counted.
(ii) Intracellular IFN-γ staining.
Intracellular IFN-γ expression in response to peptide stimulation was performed as previously described (19, 25). A total of 106 brain-derived monocytes or splenocytes per well were cultured for 5 h at 37°C in 200 μl of RPMI 1640 medium, supplemented with 5% fetal calf serum, 10 U of human recombinant interleukin-2, and brefeldin A (Golgiplug [1 μl/ml]; PharMingen) either with or without peptides. The peptides were used at a concentration of 1 μg/ml. Cells were then stained as described above for surface expression of CD8 and incubated overnight at 4°C. For intracellular IFN-γ staining, cells were then fixed and permeabilized using the Cytofix/Cytosperm kit (PharMingen) and stained with a fluorescein isothiocyanate-conjugated monoclonal rat anti-mouse IFN-γ antibody (clone XMG 1.2; PharMingen). Cells were washed and then suspended in PBS containing 2% paraformaldehyde and analyzed by FACSan flow cytometry (Becton-Dickinson).
Isolation of epitope escape mutants.
Viruses were plaqued from brain homogenates of infected animals sacrificed at 5 days postinfection as described above. Plaques were circled on the six-well culture dishes and then visualized by fluorescent microscopy. The number of green plaques was counted, and the percentage of fluorescent plaques was calculated. A selection of fluorescent and nonfluorescent plaques were picked, and viruses isolated from these plaques were used to infect L2 cells. Intracellular RNA from such infections was isolated. RT-PCR was used to amplify the region encoding gp33/gfp using primers FIJ81 and RIJ84 (Table 1). These amplified fragments were analyzed by agarose gel electrophoresis and in some cases sequenced using the same primers.
RESULTS
Selection of and immune response to recombinant MHV-A59 expressing the LCMV CD8+ epitope gp33.
We have selected a recombinant MHV-A59 (RA59) expressing a foreign CD8+ T-cell epitope, gp33, derived from the glycoprotein (or G protein) of LCMV. The gp33 epitope is expressed as a fusion protein, sandwiched between a 9-amino-acid fragment of the MHV ORF4a protein (37) and the EGFP (4). A schematic diagram of the genome of this virus (RA59-gfp/gp33) as well as a previously described recombinant MHV-A59 expressing the EGFP protein alone (RA59-gfp), is shown in Fig. 1. In both genomes the nonessential gene 4 is replaced by the foreign sequences. Selection of RA59-gfp was described previously (5). RA59-gfp/gp33 was selected using techniques similar to those used in the selection of RA59-gfp and is described in the Materials and Methods. Briefly, recombination occurred between fMHV and an RNA encoding, in place of gene 4, a recombinant protein containing 9 amino acids from ORF4a and 9 amino acids of gp33 fused in frame with the EGFP gene. Such recombinants were selected and plaque purified on murine L2 cells all as previously described (26) and were characterized further.
Cells infected with these viruses were notably less fluorescent than RA59-gfp, suggesting that the N-terminal addition to the EGFP protein may interfere with its function. Alternatively there may be less transcription or translation due to differences near the intergenic sequences (5, 20). RA59-gfp/gp33 (log10 50% lethal dose [LD50] = 5.6) displayed a similar level of virulence to RA59-gfp (log10 LD50 = 5.5). As previously published (5), RA59-gfp is somewhat attenuated relative to wild-type recombinant RA59 (log10 LD50 = 4); thus, both EGFP-expressing viruses display LD50 values approximately 10-fold higher than those displayed by wild-type RA59.
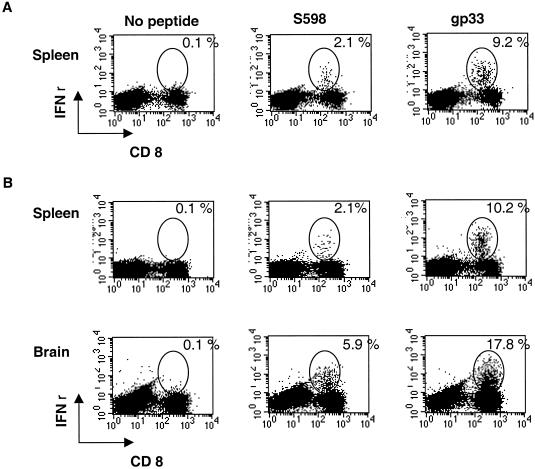
We quantified the epitope-specific CD8+ T-cell response to RA59-gfp/gp33 in the spleen and in the CNS after both i.c. and i.p. inoculation. Thus, 4-week-old B6 mice were inoculated either i.c. or i.p. and animals were sacrificed at day 7 postinfection (after i.c. inoculation) and on day 8 postinfection (after i.p. inoculation), the peak of inflammation for each route. Splenocytes were isolated from both sets of mice; CNS mononuclear cells were isolated from only those mice inoculated by the i.c. route. An intracellular IFN-γ assay was used to quantify the percentage of CD8+ T cells specific for the MHV spike protein CD8+ T-cell epitope S598 and for gp33 (19, 25). As shown in Fig. 2, S598 and gp33 epitope-specific CD8+ T cells are detected in both the spleen and the CNS. Response in the spleen is similar whether inoculation is carried out either i.c. or i.p.; following i.c. infection, the levels of CD8+ T cells specific for both epitopes are higher in the CNS, the site of viral replication, than in the spleen, as observed previously (3, 17).
FIG. 2.
Epitope-specific CD8+ T-cell response of RA59-gfp/gp33-infected mice to gp33 and S598, as measured by intracellular IFN-γ expression. Four-week-old B6 mice were infected either i.p. with 104 PFU (A) or i.c. with 2 × 103 PFU (B). At either 8 (A) or 7 (B) days postinfection, mice were sacrificed, and mononuclear cells were harvested from the spleen and brain. IFN-γ assays were carried out to determine the percentage of CD8+ T cells specific for S598 or gp33 as indicated. Data shown are derived from pooled samples from three to five mice.
Epitope-specific CD8+ T cells confer protection against infection with RA59-gfp/gp33.
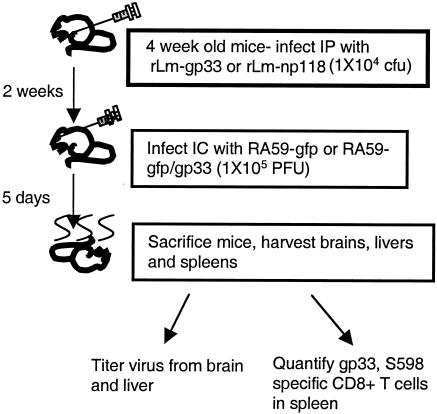
We wanted to determine the ability of the CD8+ T-cell response against a single epitope to protect mice from MHV replication and disease. The strategy was to carry out immunization with an rLm expressing gp33 (rLm-gp33) followed by challenge with RA59-gfp/gp33. (It had previously been shown that immunization with rLm expressing np118 epitope from LCVM could induce an epitope-specific memory response that was protective against LCMV infection (30)). Thus, 4-week-old B6 mice were inoculated i.p. with rLm-gp33 or, as a control, with rLm-np118 (rLm expressing an LCMV nucleocapsid epitope np118-126) (29). Two weeks later, mice were infected with either RA59gfp/gp33 or, as a control, RA59-gfp. At day 5, the peak of virus replication in the CNS, mice were sacrificed and the brains, livers, and spleens were harvested. The brains and livers were titrated for virus, and spleens were used for quantification of CD8+ T cells specific for gp33 and the MHV spike CD8+ T-cell epitope S598 (Fig. 3).
FIG. 3.
Schematic for protection experiment. Four-week-old mice were inoculated with rLm (104 CFU) expressing gp33 (rLm-gp33) or np118 (rLm-np118). Two weeks later mice were infected with 105 PFU RA59-gfp or RA59-gfp/gp33. Five days later mice were sacrificed. Virus was titrated from the brains and livers, and epitope-specific T cells were quantified from the spleens.
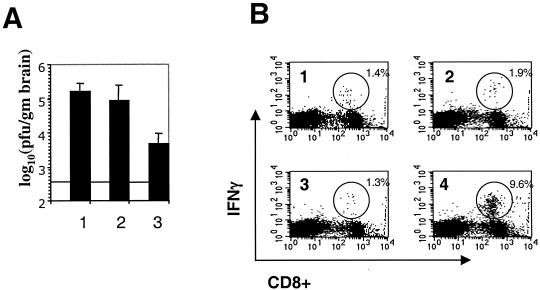
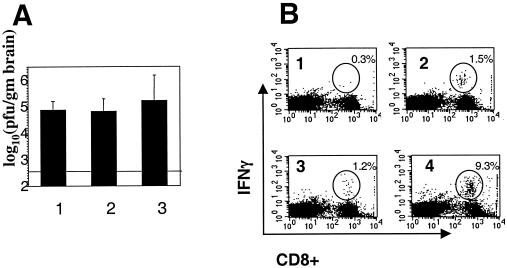
As shown in Fig. 4B, the immune response against gp33 following infection with RA59gfp/gp33 in animals that had been immunized with rLm-gp33 was greatly enhanced compared with that of mice inoculated with rLm-np118 or mice that were not inoculated with Listeria. Mice that had been immunized with rLm-gp33 and subsequently infected with RA59-gfp/gp33 had much higher levels (9.6% in the assay in Fig. 4B) of gp33 epitope-specific, IFN-γ positive CD8+ T cells than did animals inoculated with rLm-np118 (1.3% in the assay shown in Fig. 4B). Similar low levels of gp33 epitope-specific IFN-γ-positive CD8+ T cells were present in the spleens of mice inoculated with rLm-gp33 alone or RA59-gfp/gp33 alone. The percentage here for gp33 epitope-specific IFN-γ positive CD8+ T cells in the spleens of mice infected with RA59gfp/gp33 alone and sacrificed at day 5 (1.9% in the assay shown in Fig. 4B) was significantly lower than the value shown in Fig. 2 (10.2%). This is because the responses shown in Fig. 2 are from mice sacrificed at 7 days postinfection; CD8+ T-cell response in the spleen is maximal at day 7 and considerably higher than that at day 5. The percentage of S598 epitope-specific CD8+ T cells was the same (>1%) whether or not the virus expresses gp33.
FIG. 4.
Protection from recombinant MHV expressing gp33 by immunization with rLm expressing gp33. Four-week-old B6 mice were inoculated with rLm-gp33 or rLm-np118 or not inoculated. Two weeks later they were infected with virus, all as outlined in Fig. 3 legend. (A) Titers of virus in the brains of animals either not inoculated with rLm (lane 1) or inoculated with rLm-np118 (lane 2) or rLm-gp33 (lane 3). The values shown are averages from three experiments, a total of 13 mice inoculated with rLm-np118, 12 mice inoculated with rLm-gp33, and 5 mice that were not inoculated. The horizontal line shows the limit of detection for the assay. The titers from mice inoculated with rLm-gp33 were significantly less than those from mice inoculated with rLm-np118 (P < 0.001, Mann-Whitney U test). (B) Percentages of gp33-specific CD8+ T cells in the spleens of infected mice. Splenocytes were isolated from mice; the frequency of CD8+ T cells specific for the gp33 epitope was determined by intracellular IFN-γ staining. The numbers in the upper-right corners indicate the percentage of CD8+ T cells that are positive for the intracellular IFN-γ stain. Mice were inoculated with rLm-gp33 alone (quadrant 1); RA59-gfp/gp33 alone (quadrant 2); rLm-np118 followed 2 weeks later by RA59gfp/gp33 (quadrant 3); or rLm-gp33 followed 2 weeks later by RA59gfp/gp33 (quadrant 4). The immune response of every animal was examined; sometimes samples were pooled from two or three animals. Data shown are from one representative assay; similar results in all cases were obtained in assays from two more independent experiments.
As shown in Fig. 4A, the mice previously immunized with rLm expressing the gp33 epitope have statistically significant lower titers of infectious virus in the brain at day 5 postinfection. There is an approximately 20-fold reduction of titer in mice immunized with the gp33 epitope but not in control mice inoculated with rLm-np118 or mice inoculated with virus only. The data in Fig. 4A represent animals pooled from three experiments, all with similar reductions in titer. Mice infected with RA59-gfp (n = 4 in each group) exhibited no difference in titers in the brain at day 5 whether they were previously inoculated with rLm-gp33 [average log10 (PFU per gram) = 4.62] or rLm-np118 [average log10 (PFU per gram) = 4.58]. Thus, there was a significant reduction in viral replication in mice immunized with rLm-gp33, indicating that the epitope-specific CD8+ T-cell response was able to protect against infection. Viral titers in the liver were very low whether the mice were immunized or not; this is likely because the mice were 6 weeks old by the time of inoculation. It was not possible to determine whether there were differences in the titers of virus from liver lysates of immunized and nonimmunized mice.
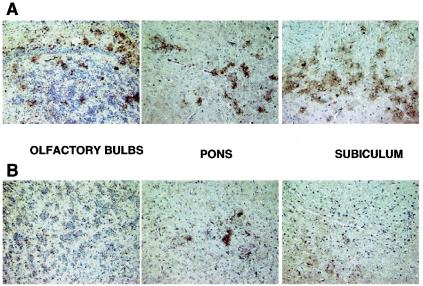
To complement the measurement of viral titers, we examined viral antigen spread in the brain. Thus, we used a MAb directed against MHV nucleocapsid protein and immunhistochemistry to detect viral antigen in various regions of the brain in sagittal tissue sections derived from immunized and nonimmunized mice. Figure 5 shows brain sections from mice, 5 days postinfection with RA59-gfp/gp33, that had been immunized with rLm-gp33 or mock immunized with rLm-np118 2 weeks prior to infection. (These data are representative of two sections from each of four mice.) Images are shown for three regions of the brain that represent common sights of viral antigen distribution, olfactory bulbs, subiculum, and pons-medulla. Consistent with the differences in viral titer (Fig. 4), there is a clear reduction in the amount of viral antigen expression in the brain of mice immunized with rLm-gp33 compared with nonimmunized mice.
FIG. 5.
Viral antigen expression in the brains of mice infected with RA59-gfp/gp33 after immunization with rLm-gp33 or rLm-np118. Mice were inoculated with rLm-np118 (A) or rLm-gp33 (B) and 2 weeks later were infected with RA59-gfp/gp33 as described in Fig. 3. Mice were sacrificed at day 5, and sagittal brain sections prepared as described in Materials and Methods. Representative sections (two sections from each of four mice) are shown for each of three regions of the brain.
Protection from MHV infection following immunization depends on perforin.
To begin to understand the mechanism of the protection, we investigated whether immunization with rLm-gp33 could protect against infection of perforin knockout (PKO) mice with RA59-gfp/gp33. Thus, following a scheme similar to that shown in Fig. 2, PKO mice were infected with RA59-gfp/gp33 without prior immunization or following immunization with either rLm-gp33 or rLm-np118. As shown in Fig. 6 protection was not achieved in the absence of perforin; there were no differences between the titers of virus in the brains in the mice that were immunized with rLm-gp33 or mock immunized with rLm-np118. These titers were similar to those detected in unprotected B6 mice. This lack of protection was not due to a lack of a gp33 epitope-specific CD8+ T-cell response, as demonstrated by the data in Fig. 6B. As measured by an intracellular IFN-γ assay, PKO mice immunized with rLm-gp33 had levels of spleen cells specific for gp33 (9.3% in the assay shown in Fig. 6) similar to levels found in immunized B6 mice (Fig. 4B and 6B). This demonstrates that perforin-mediated mechanisms play a role in protection from infection and disease.
FIG. 6.
Perforin-deficient (PKO) mice are not protected from RA59-gfp/gp33 after immunization with rLm-gp33. Four-week-old PKO mice were inoculated with rLm-gp33 or rLm-np118 or were not inoculated. Two weeks later they were infected with RA59-gfp/gp33, as in Fig. 4. (A) Titers of virus in the brains of animals either not inoculated with rLm (lane 1) or inoculated with rLm-np118 (lane 2) or rLm-gp33 (lane 3). The values shown are averages from two experiments, a total of five mice inoculated with rLm-np118, nine mice inoculated with rLm-gp33, and three mice not inoculated with rLm. The horizontal line shows the limit of detection for the assay. (B) Percentages of gp33-specific CD8+ T cells in the spleens of infected mice. Splenocytes were isolated from mice; the frequency of CD8+ T cells specific for the gp33 epitope was determined by intracellular IFN-γ staining. The numbers in the upper-right corners indicate the percentage of CD8+ T cells that are positive for the intracellular IFN-γ stain. Mice were inoculated with rLm-np118 alone (quadrant 1), RA59-gfp/gp33 alone (quadrant 2), rLm-np118 followed 2 weeks later by RA59gfp/gp33 (quadrant 3), or rLm-gp33 followed 2 weeks later by RA59gfp/gp33 (quadrant 4). The immune response of every animal was examined; sometimes samples were pooled from two or three animals. Data shown are from one representative assay; similar results in all cases were obtained from two independent experiments.
Selection of epitope escape mutants in mice immunized against rLm-gp33.
In determining the titers of virus from infected mice, we noticed that, under the fluorescent microscope, the majority of viral plaques obtained from the CNS of rLm-gp33 immunized mice were no longer fluorescent, suggesting that viruses no longer expressed EGFP activity. However, most of the plaques derived from the brains of nonimmunized mice were fluorescent. We reasoned that, in mice immunized with rLm-gp33, the strong epitope-specific response may cause a selection for virus lacking the gp33 epitope and that, since gp33 is expressed as a fusion protein with EGFP, there may be a loss of EGFP as well. Thus, we examined more quantitatively the percentage of fluorescent virus plaques isolated from the brains of such infected mice. Data from three independent experiments are shown in Table 2. More than 90% of the viral plaques from brains of animals inoculated with rLm-np118 and subsequently infected with RA59-gfp/gp33 expressing virus were fluorescent and thus retained the ability to express EGFP activity. On the other hand, only 15% of the plaques from the brains of mice that were protected by inoculation by rLm-gp33 were fluorescent. Lysates from the brains of animals infected with RA59-gfp contained similar percentages of EGFP expressing plaques whether or not they had been inoculated with rLm expressing gp33 (Table 2).
TABLE 2.
Percentage of virus retaining EGFP expression in mice vaccinated with rLm-np118 or rLm-gp33a
| Virus | Mouse strain | % of virus retaining EGFP expression in mice vaccinated withb:
|
|
|---|---|---|---|
| rLm-np118 | rLm-gp33 | ||
| RA59-gfp/gp33 | B6 | 93 (27/29) | 13 (3/24) |
| RA59-gfp/gp33 | B6 | 95 (54/57) | 2 (2/82) |
| RA59-gfp/gp33 | B6 | 94 (72/77) | 14 (5/36) |
| RA59-gfp/gp33 | PKO | 78 (31/40) | 89 (74/83) |
| RA59-gfp | B6 | 72 (38/53) | 90 (37/41) |
| RA59-gfp | B6 | 75 (40/53) | 71 (37/52) |
In each experiment plaques were isolated from four to seven mice. Three separate experiments were carried out with RA59-gfp/gp33 in B6 mice, two experiments were carried out with RA59-gfp in B6 mice, and one experiment was carried out in PKO mice with RA59-gfp/gp33.
Plaques were recovered from the brains of mice that had been inoculated with rLm-np118 or rLm-gp33 and then infected with the viruses as indicated. Numbers in parentheses represent numbers of plaques expressing EGFP/total number of plaques.
We wanted to test our hypothesis that the gp33 epitope would also be disrupted in most of the viral genomes recovered from nonfluorescent plaques from the brains of protected animals. Thus, we analyzed the portions of the genomes encoding the ORF4a/gp33/EGFP fusion proteins from viruses isolated from both fluorescent and nonfluorescent plaques and from both protected and unprotected animals. RT-PCR was carried out to amplify a fragment encoding ORF4a/gp33/EGFP, using primers representing sequences at the 3′end of the spike gene and the 5′ end of ORF5 (FIJ81 and RIJ84, respectively [Table 1]); the genomes of viruses isolated from the nonfluorescent plaques contained deletions of various sizes; sequencing of these amplified fragments, (using the same primers) demonstrated deletions encompassing part or all of the EGFP coding region. In the case of viruses isolated from immunized, protected animals these deletions also included the gp33 coding region. All four of the nonfluorescent plaques (from individual animals) examined had gp33 deleted (Fig. 7B). In addition, of six genomes from fluorescent plaques (from six individual animals), in which there were no large deletions observed, three had mutations that would render the gp33 inactive (data not shown). These included a 7-base deletion; a 31-base insertion before the epitope, which would result in a frameshift and lack of gp33; and a finally a UGU-to-UGA mutation in the last codon of gp33, causing a translational termination within the gp33 epitope. (It is likely that, in the case of the last genome, translation reinitiates at the AUG initiation codon for EGFP, thus allowing this virus to produce fluorescent plaques.) Thus, the percentage of viral plaques containing virus in which gp33 is inactivated is even greater than the number negative for EGFP. In the case of unprotected animals, all the genomes derived from green plaques that were sequenced in this region (seven of seven from individual animals) had an (intact) gp33 epitope, as well as the EGFP gene. In the two sequenced genomes derived from nonfluorescent plaques, gp33 still remained intact despite the loss of EGFP activity, which was due to deletions within the EGFP gene. Figure 7C shows an example in which the gp33 epitope remains intact despite the EGFP deletion. These data suggest that immunization with the gp33 epitope results in a strong immune response and that during the stimulation of the gp33 response (after infection with virus expressing gp33), the immune response to gp33 exerts pressure to select viral epitope escape mutants. Interestingly, virus is cleared efficiently despite the epitope escape mutants, presumably due to the CD8+ T-cell response to the native viral epitope S598. As expected, selection of nonfluorescent variants does not occur when infection is carried out with RA59-gfp, which does not express gp33.
FIG. 7.
RT-PCR sequence analysis of the EGFP/gp33 region of the genomes of viruses isolated from the CNS of protected and unprotected mice. (A) Schematic diagram of a portion of the genome of RA59-gfp/gp33 including the intergenic sequence (IGS) preceding gene 4, a short region of ORF4a, gp33, and the EGFP gene. Genomic RNAs from viruses recovered from nonfluorescent plaques from brains lysates of mice that were inoculated with rLm-gp33 (B) or rLm-np118 (C) 2 weeks before RA59-gfp/gp33 infection were sequenced through this region using primers FIJ81 and RIJ84 as discussed in Materials and Methods and the text. All (four of four) genomes from protected mice had deletions in gp33 as well as in EGFP (B), while the genome from an unprotected animal had gp33 intact while EGFP was deleted (C).
The percentages of fluorescent plaques derived from brain lysates from PKO mice inoculated with rLm-gp33 or rLm-np118 were calculated as described above for B6 mice. Of plaques derived from rLm-np118-inoculated mice, 78% (31 of 40) were fluorescent and of those derived from rLm-gp33-inoculated mice, 89% (74 of 83) were fluorescent. Thus, consistent with the lack of protection in PKO mice, there was not an enrichment for viruses with deletions in the EGFP coding region and thus not for gp33. This strongly supports the notion that it is the immune pressure that drives the selection of epitope escape mutants.
DISCUSSION
Control of virus replication in the CNS of mice infected with the murine coronavirus MHV involves multiple aspects of the immune response. Both CD8+ and CD4+ T cells are required for clearance of virus during acute infection. The T-cell response to MHV infections clears virus through several mechanisms, including perforin-mediated cytolysis and secreting cytokines such as IFN-γ. In the case of astrocytes and microglia, clearance involves perforin but not Fas ligand or tumor necrosis factor alpha. IFN-γ is required for viral clearance from olgiodendrocytes (1, 14, 16, 23). The mechanism of clearance of virus from neurons is not known. In the CNS of perforin-deficient mice, viral clearance is delayed (14). IFN-γ-deficient mice exhibit delayed viral clearance despite the fact that perforin-mediated mechanisms remain intact; such mice exhibited increased clinical symptoms and mortality associated with persistent virus (22).
In this study we explored the role of an epitope-specific CD8+ T-cell response to a recombinant MHV in acute infection of the CNS. Thus, using an rLm (rLm-gp33) and a recombinant MHV-A59 (RA59-gfp/gp33), both expressing the same H-2Db-restricted CD8+ T-cell epitope derived from the LCMV glycoprotein, gp33, we demonstrated that the CD8+ T-cell response to a specific epitope can protect against virus replication and spread. Similar protection against recombinant coxsackie B3 virus (expressing LCMV epitope gp33) was achieved by immunization of mice with LCMV (31).
The T-cell response to MHV peaks at day 7 to 8 postinfection (day 7 after i.c. inoculation and day 8 after i.p. inoculation), at a time when infectious virus is nearly cleared (25, 39). At 7 to 8 days postinfection with RA59-gfp/gp33, as measured by an intracellular IFN-γ assay (19), approximately 2% of the CD8+ cells from the spleen were specific for the S598 MHV spike epitope and 9 to 10% were specific for the gp33 epitope, whether the virus was inoculated i.p. or i.c. Levels of specific cells were much higher in the CNS than in the spleen after i.c. inoculation: 5.9% for S598 and as high as 17.8% for gp33. Thus, the LCMV GP33 elicits a strong response when expressed from MHV-A59 genome, and epitope-specific T cells are recruited to and/or expanded in the CNS.
In order to examine the role of the CD8+ T-cell epitope-specific response, 4-week-old B6 mice were inoculated i.p. with rLm-gp33 or, as a control, rLm-np118 expressing an irrelevant epitope. Two weeks later they were inoculated i.c. with recombinant RA59-gfp/gp33. As shown in Fig. 4, by day 5 postinfection with RA59-gfp/gp33, approximately 10% of CD8+ T cells in the spleen were specific for gp33, considerably higher than the approximately 1 to 2% epitope-specific cells in the spleens of animals inoculated with rLm-gp33 alone or RA59-gfp/gp33 alone or in animals inoculated with rLm-np118 prior to viral infection. The 10% IFN-γ/CD8+ T cells in immunized mice represent the response to that epitope.
The increased number of epitope-specific CD8+ T cells, by day 5 postinfection, was able to limit viral replication during acute infection. There was an approximately 20-fold reduction in the titer of infectious virus in the brains of RA59gp/gp33-infected animals that had been immunized with rLm-gp33 (Fig. 4). Protection from MHV infection was not observed in perforin-deficient mice (PKO), as measured by viral titers in the CNS, indicating that perforin-dependent mechanisms are needed for protection.
We have observed in the past that spread of viral antigen in the CNS may be a more reliable indicator of virulence than titers of infectious virus (27). In these experiments, we observed a striking diminution in the amount of viral antigen in the brains from protected animals. Figure 5 shows sections from three regions of the brain: olfactory bulbs, pons, and subiculum. In all cases there is much less antigen in sections from protected mice than in sections from control mice; in some sections from protected mice there is little if any antigen (for example, the olfactory bulb in Fig. 5B).
In a previous study, vaccination of BALB/c mice was carried out with a recombinant vaccinia virus expressing MHV nucleocapsid protein (containing the immundominant H-2Ld CD8+ T-cell epitope). Mice infected with this recombinant vaccine were not protected from MHV-JHM, but cytotoxic T lymphocytes isolated from such vaccinia virus-infected animals and stimulated in vitro with epitope-specific peptide could protect when transferred to naive mice. In these studies (33), viral clearance was observed from astrocytes and microglia, cells expressing class I major histocompatibility complex molecules, consistent with the notion that perforin-mediated mechanisms play a role in protection from disease by CD8+ T cells. In a recent report (1), using mice deficient in both perforin and IFN-γ (PKO/GKO) it was shown, by the same group, that clearance by perforin requires the presence of IFN-γ, which plays an important role in upregulating major histocompatibility complex class I on astrocytes and microglia, a requirement for obtaining a good cytolytic T-cell response. In our study, the lack of clearance in PKO mice is not due to the lack of IFN-γ, since a normal IFN-γ response was observed in CD8+ T cells isolated from PKO mice (Fig. 6B). In the MHV-JHM studies referred to above, they also showed that transfer of CD8+ T cells from infected PKO mice (deficient only in perforin) into PKO/GKO mice can partially control replication; this suggests that in that model clearance may be more dependent on IFN-γ and may not require perforin. Although these results differ from ours, it is important to emphasize that in their model they use a variant of the highly neurotropic strain of MHV, JHM. This variant primarily infects glial cells, including oligodendrocytes, microglia, and astrocytes. Oligodendrocytes, specifically, require IFN-γ for virus clearance, which may account for the observation that cells from PKO mice can partially control virus replication. In contrast, our studies use the A59 strain of MHV, which primarily infects neurons and, to a lesser extent, astrocytes and microglia. The difference in virus tropism as well as strain of mouse may account for the differences in viral clearance requirements. It supports the notion that different cell types from the CNS require different components from the immune system for clearance of virus (16).
In the experiments described here, selection of escape mutants, in which gp33 has been inactivated either by deletion or mutation, were selected rapidly and with high frequency, in the CNS of infected mice. The presence of the EGFP gene allowed us to easily identify mutant viral genomes with deletions within EGFP and into gp33 epitope as well. More than 93% of the viruses recovered from plaques isolated from the CNS of protected mice were no longer fluorescent. Furthermore, of the minority of plaques that retained fluorescence, at least half of those had point mutations that inactivate the gp33 epitope. In unprotected animals, this phenomenon did not occur and the majority of virus retained EGFP and the gp33 epitope. These data suggest that there is a selective pressure to escape the CD8+ T-cell response to gp33. Selection of gp33 mutants did not occur in PKO mice; this is consistent with the observation that protection did not occur by immunization of such mice. This further supports the notions that gp33 mutants arise as a result of selective pressure and that perforin activity is an important immune mechanism for controlling MHV infection. There also was maintenance of EGFP expression in the majority of animals infected with RA59-gfp, whether they were immunized with rLm-gp33 or rLm-np118; this is as would be expected, because there was no selective pressure for deletion mutants.
Interestingly, in the liver EGFP deletions occur at high frequency for both the RA59-gfp- and RA59-gfp/gp33-infected whether or not immunization is carried out. This organ specific loss of EGFP expression has been observed by us in the liver (5) and by others in the heart (31) and may be due to toxicity of the EGFP in those organs.
Selection of escape mutants with amino acid substitutions within the S510 epitope of the spike protein of MHV-JHM has been described during the infection of suckling mice (24). Such mutants are more virulent than wild type when used to infect naive suckling mice. Selection of escape mutants has not been observed in weanling mice, even in B-cell-knockout mice in which virus persists in the CNS (18). Recombinant viruses that we have selected, in which the S510 epitope is inactivated by site-directed mutagenesis, are not more virulent than wild type (data not shown). Furthermore, recombinant viruses with mutations in the S598 epitope for the A59 spike were not viable (unpublished data). We suggest that these rapid and high-frequency deletions can occur in the model we describe here because the gp33 epitope is expressed as part of a nonessential protein rather than from within the spike protein. There are clearly constraints on changes in the spike protein structure. We are currently examining the likelihood of selection of escape mutants involving the natural spike epitopes after immunization with rLm encoding these epitopes.
Acknowledgments
This work was supported by NIH grants AI-47800 and AI-17418 (S.R.W.) and AI-45025 (H.S.). K. C. MacNamara was supported by NIH training grant T32-AI-07324.
We thank Julian Leibowitz for providing MAb clone 1-16-1.
REFERENCES
- 1.Bergmann, C. C., B. Parra, D. R. Hinton, R. Chandran, M. Morrison, and S. A. Stohlman. 2003. Perforin-mediated effector function within the central nervous system requires IFN-gamma-mediated MHC up-regulation. J. Immunol. 170:3204-3213. [DOI] [PubMed] [Google Scholar]
- 2.Castro, R. F., and S. Perlman. 1995. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J. Virol. 69:8127-8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro, R. F., and S. Perlman. 1996. Differential antigen recognition by T cells from the spleen and central nervous system of coronavirus-infected mice. Virology 222:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 5.Das Sarma, J., E. Scheen, S. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gombold, J. L., S. T. Hingley, and S. R. Weiss. 1993. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J. Virol. 67:4504-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gombold, J. L., R. Sutherland, E. Lavi, Y. Paterson, and S. R. Weiss. 1995. Mouse hepatitis virus-induced demyelination can occur in the absence of CD8+ T cells. Microb. Pathog. 18:211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 9.Kagi, D., B. Ledermann, K. Burki, P. Seiler, B. Odermatt, K. J. Olsen, E. R. Podack, R. M. Zinkernagel, and H. Hengartner. 1994. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369:31-37. [DOI] [PubMed] [Google Scholar]
- 10.Korner, H., A. Schliephake, J. Winter, F. Zimprich, H. Lassman, J. Sedgwick, S. G. Siddell, and H. Wege. 1991. Nucleocapsid or spike protein-specific CD4+ T lymphocytes protect against coronavirus induced encephalomyelitis in the absence of CD8+ cells. J. Immunol. 147:2317-2323. [PubMed] [Google Scholar]
- 11.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavi, E., D. H. Gilden, Z. Wroblewska, L. B. Rorke, and S. R. Weiss. 1984. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology 34:597-603. [DOI] [PubMed] [Google Scholar]
- 13.Lin, M. T., D. R. Hinton, N. W. Marten, C. C. Bergmann, and S. A. Stohlman. 1999. Antibody prevents virus reactivation within the central nervous system. J. Immunol. 162:7358-7368. [PubMed] [Google Scholar]
- 14.Lin, M. T., S. A. Stohlman, and D. R. Hinton. 1997. Mouse hepatitis virus is cleared from the central nervous systems of mice lacking perforin-mediated cytolysis. J. Virol. 71:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luytjes, W., L. Sturman, P. J. Bredenbeck, J. Charite, B. A. M. van der Zeijst, M. C. Horzinek, and W. J. M. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marten, N. W., S. A. Stohlman, and C. C. Bergmann. 2001. MHV infection of the CNS: mechanisms of immune-mediated control. Viral Immunol. 14:1-18. [DOI] [PubMed] [Google Scholar]
- 17.Marten, N. W., S. A. Stohlman, J. Zhou, and C. C. Bergmann. 2003. Kinetics of virus-specific CD8+-T-cell expansion and trafficking following central nervous system infection. J. Virol. 77:2775-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews, A. E., S. R. Weiss, M. J. Shlomchik, L. G. Hannum, J. L. Gombold, and Y. Paterson. 2001. Antibody is required for clearance of infectious murine hepatitis virus A59 from the central nervous system, but not the liver. J. Immunol. 167:5254-5263. [DOI] [PubMed] [Google Scholar]
- 19.Murali-Krishna, K., J. D. Altman, M. Suresh, D. J. Sourdive, A. J. Zajac, J. D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177-187. [DOI] [PubMed] [Google Scholar]
- 20.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 21.Parker, S. E., T. M. Gallagher, and M. J. Buchmeier. 1989. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoproteins of several strains of murine hepatitis virus. Virology 173:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra, B., D. R. Hinton, N. W. Marten, C. C. Bergmann, M. T. Lin, C. S. Yang, and S. A. Stohlman. 1999. IFN-gamma is required for viral clearance from central nervous system oligodendroglia. J. Immunol. 162:1641-1647. [PubMed] [Google Scholar]
- 23.Parra, B., M. T. Lin, S. A. Stohlman, C. C. Bergmann, R. Atkinson, and D. R. Hinton. 2000. Contributions of Fas-Fas ligand interactions to the pathogenesis of mouse hepatitis virus in the central nervous system. J. Virol. 74:2447-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pewe, L., G. F. Wu, E. M. Barnett, R. F. Castro, and S. Perlman. 1996. Cytotoxic T cell-resistant variants are selected in a virus-induced demyelinating disease. Immunity 5:253-262. [DOI] [PubMed] [Google Scholar]
- 25.Phillips, J. J., M. Chua, G. F. Rall, and S. R. Weiss. 2002. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation and virus-induced immunopathology in the central nervous system. Virology 301:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips, J. J., M. Chua, S. H. Seo, and S. R. Weiss. 2001. Multiple regions of the murine coronavirus spike glycoprotein influence neurovirulence. J. Neurovirol. 7:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.San Mateo, L. R., M. M. Chua, S. R. Weiss, and H. Shen. 2002. Perforin-mediated CTL cytolysis counteracts direct cell-cell spread of Listeria monocytogenes. J. Immunol. 169:5202-5208. [DOI] [PubMed] [Google Scholar]
- 29.Shen, H., J. F. Miller, X. Fan, D. Kolwyck, R. Ahmed, and J. T. Harty. 1998. Compartmentalization of bacterial antigens: differential effects on priming of CD8 T cells and protective immunity. Cell 92:535-545. [DOI] [PubMed] [Google Scholar]
- 30.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slifka, M. K., R. Pagarigan, I. Mena, R. Feuer, and J. L. Whitton. 2001. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells during picornavirus infection. J. Virol. 75:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stohlman, S. A., C. C. Bergmann, M. T. Lin, D. J. Cua, and D. R. Hinton. 1998. CTL effector function within the central nervous system requires CD4+ T cells. J. Immunol. 160:2896-2904. [PubMed] [Google Scholar]
- 33.Stohlman, S. A., C. C. Bergmann, R. C. van der Veen, and D. R. Hinton. 1995. Mouse hepatitis virus-specific cytotoxic T lymphocytes protect from lethal infection without eliminating virus from the central nervous system. J. Virol. 69:684-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stohlman, S. A., G. K. Matsushima, N. Casteel, and L. P. Weiner. 1986. In vivo effects of coronavirus-specific T cell clones: DTH inducer cells prevent a lethal infection but do not inhibit virus replication. J. Immunol. 136:3052-3056. [PubMed] [Google Scholar]
- 35.Sussman, M. A., R. A. Shubin, S. Kyuwa, and S. A. Stohlman. 1989. T-cell-mediated clearance of mouse hepatitis virus strain JHM from the central nervous system. J. Virol. 63:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland, R. M., M. M. Chua, E. Lavi, S. R. Weiss, and Y. Paterson. 1997. CD4+ and CD8+ T cells are not major effectors of mouse hepatitis virus A59-induced demyelinating disease. J. Neurovirol. 3:225-228. [DOI] [PubMed] [Google Scholar]
- 37.Weiss, S. R., P. W. Zoltick, and J. L. Leibowitz. 1993. The ns4 gene of mouse hepatitis virus (MHV), strain A59 contains two ORFs and thus differs from ns4 of the JHM and S strains. Arch. Virol. 129:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson, J. S., and S. A. Stohlman. 1990. Effective clearance of mouse hepatitis virus from the central nervous system requires both CD4+ and CD8+ T cells. J. Virol. 64:4589-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williamson, J. S. P., K. C. Sykes, and S. A. Stohlman. 1991. Characterization of brain-infiltrating mononuclear cells during infection with mouse hepatitis virus strain JHM. J. Immunol. 32:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi, K., N. Goto, S. Kyuwa, M. Hayami, and Y. Toyoda. 1991. Protection of mice from a lethal coronavirus infection in the central nervous system by adoptive transfer of virus-specific T cell clones. J. Neuroimmunol. 32:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]