Abstract
Bromoviral templates for plus-strand RNA synthesis are rich in A or U nucleotides in comparison to templates for minus-strand RNA synthesis. Previous studies demonstrated that plus-strand RNA synthesis by the brome mosaic virus (BMV) RNA replicase is more efficient if the template contains an A/U-rich template sequence near the initiation site (K. Sivakumaran and C. C. Kao, J. Virol. 73:6415-6423, 1999). These observations led us to examine the effects of nucleotide changes near the template's initiation site on the accumulation of BMV RNA3 genomic minus-strand, genomic plus-strand, and subgenomic RNAs in barley protoplasts transfected with wild-type and mutant BMV transcripts. Mutations in the template for minus-strand synthesis had only modest effects on BMV replication in barley protoplasts. Mutants with changes to the +3, +5, and +7 template nucleotides accumulated minus-strand RNA at levels similar to the the wild-type level. However, mutations at positions adjacent to the initiation cytidylate in the templates for genomic and subgenomic plus-strand RNA synthesis significantly decreased RNA accumulation. For example, changes at the third template nucleotide for plus-strand RNA3 synthesis resulted in RNA accumulation at between 18 and 24% of the wild-type level, and mutations in the third template nucleotide for subgenomic RNA4 resulted in accumulations at between 7 and 14% of the wild-type level. The effects of the mutations generally decreased as the mutations occurred further from the initiation nucleotide. These findings demonstrate that there are different requirements of the template sequence near the initiation nucleotide for BMV RNA accumulation in plant cells.
Efficient viral RNA synthesis requires specific recognition and proper interaction between the template RNA and the membrane-bound viral replicase (7, 12, 40, 41). This recognition process is likely to be quite complex because an RNA virus expresses different classes of RNAs at regulated levels and times (for review of viral RNA replication, see references 8 and 30). We study viral RNA replication with Brome mosaic virus (BMV) as a model plus-strand RNA virus.
BMV is a single-stranded, positive-sense RNA virus that belongs to the Bromoviridae family of plant viruses in the alphavirus-like superfamily (18, 23). The tripartite BMV genome is composed of RNAs designated RNA1 (3.2 kb), RNA2 (2.8 kb), and RNA3 (2.1 kb). Viral replication is dependent on BMV genomic RNAs 1 and 2, which encode the replication-associated proteins 1a and 2a, respectively. The dicistronic RNA3 encodes the 3a movement protein and coat protein, both of which are required for systemic infection in plants (5, 34, 38). Minus-strand RNA3 can also be transcribed to produce a subgenomic RNA4 (0.9 kb) that directs translation of the viral capsid (3).
Complete replication of the BMV genome requires three classes of RNA promoters that direct the BMV replicase to synthesize genomic minus-strand, genomic plus-strand, and subgenomic RNAs (Fig. 1A) (22, 23). A tRNA-like structure at the 3′ end of all positive-sense BMV RNAs directs the synthesis of the complementary minus-strand RNA (11, 14, 15). A second promoter at the 3′ end of the newly synthesized minus-strand RNA directs genomic plus-strand RNA synthesis (35). The third, subgenomic promoter is located within the minus-strand RNA3 (Fig. 1A) (1, 33, 44). During infection, plus-strand RNAs are maintained in a specific ratio, where RNA1 < RNA2 < RNA3 < RNA4, despite having nearly identical promoters for minus-strand RNA synthesis. Furthermore, plus-strand RNA is present in up to 100-fold molar excess over minus-strand RNA (32).
FIG. 1.
Initiation sequences for the three classes of RNAs produced from BMV RNA3. (A) Schematic of BMV RNA3 plus-strand (heavy line) and minus-strand (thin line) RNAs and the template sequence near the initiation cytidylate for the three RNAs that can be produced. Nontemplated nucleotides are in lowercase letters. The open reading frames in RNA3 are denoted by open boxes. The +1 marks the initiation nucleotide for each class of RNA. n denotes the nontemplated nucleotide at the 3′ end of minus-strand RNA3. (B) Bromoviral templates for plus-strand RNA3 and RNA4 synthesis but not minus-strand RNA3 are A/U-rich. The template for minus-strand RNA4 is not shown because it is identical to that from RNA3. +1 denotes the initiation nucleotide. G's and C's are in bold, and the percentages of G/C nucleotides are calculated from template nucleotides 2 to 9. The different viral species used were Broad bean mottle virus (BBMV), Cowpea chlorotic mottle virus (CCMV; two independently isolated and sequenced strains), and Spring beauty latent virus (SBLV).
Several factors in combination are likely to be responsible for the asymmetric accumulation of the BMV RNAs. In addition to the recognition of the core promoters, several regulatory sequences will affect RNA synthesis by the replicase. For example, the intercistronic sequence in RNA3 is required (36), in conjunction with the RNA termini, for amplification of RNA3 (16) and for the asymmetric replication of plus over minus strands (32). Other regulatory factors could include the ability of an RNA to compete for limited replicase, and the characteristics of RNA synthesis such as the processivity of the replicase enzyme, the frequency of initiation by the replicase, the ability of replicase to transition out of initiation, and the frequency of template switch (35, 46-49).
Examination of the template sequence from members of the Bromoviridae and other members of the bromoviruses revealed that the template sequences for plus-strand and minus-strand RNA synthesis tend to have dissimilar sequences near the initiation site (Fig. 1A). All of these RNAs initiate with a pyrimidine (a cytidylate for BMV), but the templates for plus-strand RNA synthesis tend to have an A/U-rich sequence following the initiation pyrimidine, while the minus-strand template does not (Fig. 1B) (9, 10, 46). In addition, an A and a U are invariant as the second and third template nucleotides in all of the bromoviruses. For genomic plus-strand RNA synthesis in vitro, changes of the template sequence near the initiation cytidylate generally had more detrimental effects, with changes to G or C generally having a more negative effect than changes to A or U (2, 46). These observations led us to examine the requirements of the template sequences near the initiation site for genomic minus-strand and plus-strand and subgenomic RNA synthesis in barley protoplasts, a natural host for BMV.
MATERIALS AND METHODS
Generation of mutations in BMV RNA3 template for genomic minus-strand, genomic plus-strand, and subgenomic RNAs.
The cDNA for BMV RNA3 in pB3TP8 (4) was used as the template to generate all of the site-directed mutants. Mutations on minus- and plus-strand template were generated with oligonucleotides containing the desired mutation either in antisense oligonucleotide or in an antisense oligonucleotide and Vent polymerase (New England Biolabs). In this PCR-based strategy, sense primers contained a T7 polymerase promoter, which allows in vitro transcription. Mutations in the subgenomic RNA template were generated with oligonucleotides and the QuickChange kit (Stratagene), followed by linearization of the plasmid with EcoRI and in vitro transcription. The presence of the mutations and the absence of spurious mutations were confirmed by DNA sequencing with the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Capped full-length transcripts were produced with the Message Machine kit as described by the manufacturer (Ambion Inc., Austin, Tex.). The template for RNA3 transcription is a PCR fragment generated by the Vent polymerase. It contains the T7 promoter and the cDNA sequence for RNA3. No additional 3′ nucleotides were intentionally added to this cDNA. Capped BMV wild-type RNA1 and RNA2 were generated from EcoRI-linearized pB1TP3 and pB2TP5, respectively (20). Capped transcripts were visually analyzed by agarose gel electrophoresis, followed by toluidine blue staining and quantification by spectroscopy.
Protoplast inoculations and analysis of progeny viral RNA.
Protoplasts were isolated from 5-day-old barley (Hordeum vulgare cv. Apex) primary leaves as described by Kroner et al. (28). A total of 106 cells were transfected with 0.5 μg of each desired combination of in vitro-capped transcript mixtures (RNA1, RNA2, and either wild-type or mutant RNA3) with polyethylene glycol. After inoculation, protoplasts were incubated at 25°C under lights for 21 h unless stated otherwise. Total RNA was extracted from protoplasts with a lysis buffer (final concentrations: 0.1 M glycine, 40 mM EDTA, 400 mM NaCl, 2% sodium dodecyl sulfate, 0.05% bentonite), and progeny RNA was analyzed by Northern blots. Equal quantities of total RNA samples, according to spectroscopic measurements, were denatured with glyoxal, electrophoresed in 1% agarose gels according to the protocol given by Sambrook and Russell (42), and transferred to nylon filters (Nytran; Schleicher & Schuell). Each blot was hybridized with radiolabeled strand-specific RNA probes designed to detect the 3′ 200 nucleotides of the BMV RNAs and the 18S rRNA as a control for the amount of RNA loaded in each lane.
Hybridization and washing were performed according to Ausubel et al. (6). Each blot was used to sequentially detect minus-strand and plus-strand progeny BMV RNAs and 18S rRNA, with the stripping of the bound probe in a low-salt buffer at 95°C between analyses. RNA accumulation was quantified with a Phosphorimager Datastorm analyzer. Autoradiograms were developed at desired exposure times, usually 8, 1, and 1 h for the minus-strand, plus-strand, and rRNAs, respectively. All of the experiments contained two independent transfections so as to determine whether there was much variation within each experiment. In most experiments, two additional transfections were performed and harvested at 21 h postinoculation to confirm the results of the first experiment. Where this was done, the four replicates were used to generate a mean and standard deviation for each mutant RNA that are summarized in Table 1.
TABLE 1.
Summary of the effects of the mutations tested for RNA accumulation in barley protoplasts
| Template | Construct (sequence) | % of wild-type levela
|
||
|---|---|---|---|---|
| Minus-strand RNA3 | Plus-strand RNA3 | Plus-strand RNA4 | ||
| Minus-strand RNA3 | Wild type (. . .uaaaagagacca3′) | 100 | 100 | 100 |
| M1,2ΔCA (. . .uaaaagagac--3′) | 59 ± 5 | 33 ± 1 | 36 ± 1 | |
| M1,2CC/GG (. . .uaaaagagaGGa3′) | 99 ± 4 | 83 ± 11 | 69 ± 9 | |
| M1C/G (. . .uaaaagagacGa3′) | 114 ± 1 | 108 ± 4 | 121 ± 4 | |
| M2C/G (. . .uaaaagagaGca3′) | 90 ± 14 | 109 ± 3 | 169 ± 7 | |
| M2C/A (. . .uaaaagagaAca3′) | 104 | 80 | 105 | |
| M2C/U (. . .uaaaagagaUca3′) | 85 | 53 | 71 | |
| M3A/C (. . .uaaaagagCcca3′) | 97 | NT | NT | |
| M3A/G (. . .uaaaagagGcca3′) | 56 | 43 | 51 | |
| M3A/U (. . .uaaaagagUcca3′) | 113 | 91 | 98 | |
| M5A/G (. . .uaaaagGgacca3′) | 86 | 63 | 74 | |
| M7A/G (. . .uaaaGgagacca3′) | 128 | 99 | 105 | |
| Plus-strand RNA3 | Wild type (3′cauuuuaugguu. . .) | 100 | 100 | 100 |
| P2A/C (3′ cCuuuuaugguu. . .) | 74 ± 5 | 10 ± 3 | 216 ± 14 | |
| P2A/U (3′ cUuuuuaugguu. . .) | 68 ± 2 | 9 ± 1 | 109 ± 18 | |
| P2A/G (3′ cGuuuuaugguu. . .) | 40 ± 4 | 6 ± 1 | 18 ± 2 | |
| P3U/C (3′ caCuuuaugguu. . .) | 52 ± 4 | 23 ± 4 | 10 ± 5 | |
| P3U/G (3′ caGuuuaugguu. . .) | 89 ± 8 | 18 ± 7 | 345 ± 28 | |
| P3U/A (3′ caAuuuaugguu. . .) | 90 ± 2 | 24 ± 4 | 358 ± 34 | |
| P4U/G (3′ cauGuuaugguu. . .) | 68 ± 4 | 62 ± 3 | 103 ± 2 | |
| P4U/A (3′ cauAuuaugguu. . .) | 66 ± 4 | 50 ± 3 | 154 ± 2 | |
| P4U/C (3′ cauCuuaugguu. . .) | 44 ± 3 | 35 ± 2 | 136 ± 2 | |
| P5U/G (3′ cauuGuaugguu. . .) | 106 ± 1 | 58 ± 4 | 99 ± 11 | |
| P5U/C (3′ cauuCuaugguu. . .) | 85 ± 1 | 50 ± 5 | 156 ± 6 | |
| P5U/A (3′ cauuAuaugguu. . .) | 72 ± 11 | 64 ± 3 | 128 ± 24 | |
| P7A/C (3′ cauuuuCugguu. . .) | 65 ± 5 | 50 ± 7 | 47 ± 11 | |
| P7A/U (3′ cauuuuUugguu. . .) | 61 ± 1 | 50 ± 7 | 55 ± 7 | |
| P11U/C (3′ cauuuuauggCu. . .) | 32 ± 7 | 32 ± 4 | 28 ± 2 | |
| P11U/A (3′ cauuuuauggAu. . .) | 48 ± 1 | 60 ± 4 | 46 ± 7 | |
| Subgenomic RNA | Wild type (3′. . .cauaauuau. . .) | 100 | 100 | 100 |
| S1C/U (3′. . .Uauaauuau. . .) | 48 ± 3 | 25 ± 3 | 1 ± 1 | |
| S2A/C (3′. . .cCuaauuau. . .) | 52 ± 2 | 28 ± 2 | 1 ± 1 | |
| S2A/G (3′. . .cGuaauuau. . .) | 69 ± 5 | 48 ± 8 | 8 ± 1 | |
| S2A/U (3′. . .cUuaauuau. . .) | 62 ± 3 | 56 ± 1 | 9 ± 1 | |
| S3U/C (3′. . .caCaauuau. . .) | 42 ± 2 | 42 ± 5 | 14 ± 1 | |
| S3U/G (3′. . .caGaauuau. . .) | 35 ± 1 | 36 ± 4 | 7 ± 1 | |
| S3U/A (3′. . .caAaauuau. . .) | 52 ± 3 | 72 ± 2 | 13 ± 1 | |
| S5A/C (3′. . .cauaCuuau. . .) | 37b | 33b | 16b | |
| S5A/G (3′. . .cauaGuuau. . .) | 46 ± 1 | 60 ± 1 | 48 ± 1 | |
| S5A/U (3′. . .cauaUuuau. . .) | 114 ± 9 | 155 ± 1 | 85 ± 8 | |
| S7U/C (3′. . .cauaauCau. . .) | 59 ± 1 | 59 ± 3 | 25 ± 3 | |
| S7U/A (3′. . .cauaauAau. . .) | 72 ± 1 | 81 ± 4 | 72 ± 6 | |
| S8A/C (3′. . .cauaauuCu. . .) | 31 ± 1 | 36 ± 2 | 25 ± 2 | |
| S8A/G (3′. . .cauaauuGu. . .) | 41 ± 2 | 34 ± 1 | 12 ± 1 | |
| S8A/U (3′. . .cauaauuUu. . .) | 70 ± 11 | 73 ± 10 | 66 ± 5 | |
| SIWT (3′. . .cauaauuaAAUUAu. . .) | 100 | 100 | 100 | |
| SI10′A/C (3′. . .cauaauuaACUUAu. . .) | 100 | 115 | 124 | |
| SI10′A/G (3′. . .cauaauuaAGUUAu. . .) | 75b | 86b | 63b | |
| SI10′A/U (3′. . .cauaauuaAUUUAu. . .) | 110b | 171b | 165b | |
| SI12′U/A (3′. . .cauaauuaAAUAAu. . .) | 101 | 86 | 97 | |
| SI12′U/C (3′. . .cauaauuaAAUCAu. . .) | 110 | 114 | 185 | |
| SI13′A/C (3′. . .cauaauuaAAUUCu. . .) | 143 | 170 | 253 | |
| SI13′A/G (3′. . .cauaauuaAAUUGu. . .) | 162 | 182 | 252 | |
| SI13′A/U (3′. . .cauaauuaAAUUUu. . .) | 134 | 171 | 199 | |
Numbers in bold denote that the mutation is in the template for a particular mode of synthesis. Where standard deviations are not shown, the data were from only two independently assayed samples. NT, not tested.
These values were adjusted to normalize for the amount of rRNA in these samples.
For many of the key mutations, we also examined the effects of the mutations in another time course experiment. Five independently transfected protoplast suspensions were pooled and then aliquoted for incubation under lights at 25°C. Total RNAs were harvested at 0, 3, 10, 18, and 24 h with lysis buffer and stored at −70°C until the entire set of RNAs was ready for further processing as described above. Because the time of harvesting was different than in other transfections, we did not add the results of the time courses to the calculation of the means and standard deviations.
Examination of RNA stability.
To measure RNA stability, 32P-labeled capped transcripts of BMV RNA3 were inoculated into barley protoplasts along with RNA1 and RNA2 as described above. Total RNA was extracted over a 1-h period, denatured with glyoxal, and electrophoresed in 1% agarose gels. The gels were dried and quantified directly with a Phosphorimager.
In vitro translation.
In vitro transcripts of wild-type or mutant RNAs were used for translation with a rabbit reticulocyte lysate kit according to manufacturer's instructions (Ambion Inc.) in the presence of l-[35S]methionine. Protein samples were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (final concentrations: 0.5 M Tris-HCl, pH 6.8, 10% [wt/vol] glycerol, 10% β-mercaptoethanol, 2% sodium dodecyl sulfate, 0.01% bromophenol blue), denatured at 100°C for 5 min, separated by SDS-12% PAGE according to Laemmli (29), and detected by autoradiography.
Plant plaque assays.
Chenopodium quinoa plants were kept in the dark for 18 h prior to inoculation and mechanically inoculated with approximately 50 μl of a mixture of viral transcripts of the wild type or desired mutants along with wild-type RNA1 and RNA2 (each at 5 μg/ml) per inoculated leaf. The inoculated plants were kept in a growth chamber at 25°C and 12-h light-dark regimens for observation.
RESULTS
Nomenclature.
The large number of mutations affecting different portions of BMV RNA3 require clearly defined names. All the mutations that we make will be described as being in the template nucleotide even though the nucleotide in the complementary strand will also be affected. The template nucleotides will be named numerically, with the initiation nucleotide being +1. Mutants with changes in the template for plus-strand RNA3 synthesis will have names preceded by a P, followed by the position of the nucleotide mutated and the identities of the original and mutated bases. For example, a change of the second nucleotide A at the 3′ end of minus-strand RNA3 to a uridylate will be named P2A/U. Mutants affecting the template for minus-strand RNA3 synthesis (3′ end of RNA3) will be designated by the letter M. Mutations in the subgenomic template will be designated by the letter S.
Genomic minus-strand RNA3 accumulation.
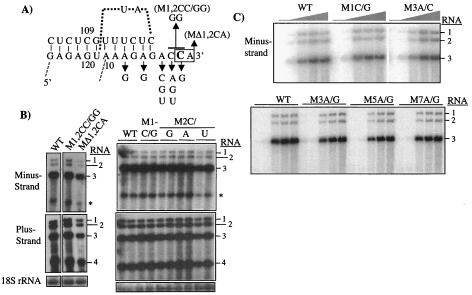
An examination of how the template sequence may affect minus-strand BMV RNA3 accumulation needs to take into account several features of the 3′ tRNA-like structure. First, there is the potential for the repair of the canonical CCA sequence that is also the initiation site, possibly with a cellular ATP(CTP):tRNA nucleotidyl transferase (37). Second, changes in the template sequence need to retain an RNA pseudoknot in stem-loop A that is required for BMV replication (Fig. 2A) (14, 15).
FIG. 2.
Effect of mutations in the template for minus-strand BMV RNA3 accumulation in barley protoplasts. (A) The relevant sequence and proposed structure of stem-loop A in the tRNA-like structure of BMV RNA3 (14). The initiation cytidylate (+1C) within the CCA sequence is in bold. Arrows denote the positions that were mutated and point to the changes that were made. (B) Northern blot analysis of mutant RNA3 demonstrates that there is repair of the CCA sequence. The identities of the wild-type (WT) and mutant RNA3s transfected into barley protoplasts are indicated above the autoradiograms. Total RNA was extracted from transfected barley protoplasts at 21 h postinoculation as described in Materials and Methods. Progeny minus-strand, plus-strand, and 18S rRNAs were detected with radiolabeled and strand-specific probes. The identity of the most relevant input transcripts is denoted at the top of the lane. The identities of the bands are shown on the sides of the autoradiograms. The asterisk denotes a minus-strand RNA that is likely transcribed from plus-strand RNA4. (C) Examination of the effects of mutations on the kinetics of minus-strand RNA accumulation. The identities of the wild-type and mutant RNA3s transfected into protoplasts are shown above the autoradiogram, and the identities of the bands are shown to the sides of the autoradiogram. The shaded shape above the autoradiogram denotes the relative time after transfection at which total RNAs were harvested. Each set of RNA was extracted at 0, 3, 10, 18, and 24 h after transfection.
To determine whether the repair of the CCA end will take place efficiently in barley protoplasts, we changed the CCA to GGA in a variant of RNA3 named M1,2CC/GG and also deleted the terminal two nucleotides in a mutant named M1,2ΔCA. The replication of these two RNAs was assayed in barley protoplasts cotransfected with wild-type RNA1 and RNA2 (Fig. 2B). The minus-strand Μ1,2ΔCΑ accumulated at 59% of wild-type RNA3 at 21 h posttransfection. This decrease likely contributed to an observed decrease in the accumulation of plus-strand RNA3 (33%) (Fig. 2B; Table 1). The replacement of CCA to GGA, a single nucleotide change at +1C to G, and a change of the +2C to A, U, or G all resulted RNA levels greater than 85% of that of the wild type (Fig. 2B; Table 1).
Since the CCA site directs the initiation of minus-strand RNA synthesis (47), these results suggest that the changes we made are rapidly repaired either by the ATP(CTP):tRNA nucleotidyl transferase, by RNA recombination, or a combination of the two processes. Similar rapid repair of the ends of BMV RNAs had been reported by Rao et al. (37). While repair is a mechanistically interesting process, it hampers our ability to discern the effect of mutations on RNA accumulation. In order to decrease the possibility of repair, we examined the effects of nucleotide substitutions that are 5′ of the CCA. Rapid repair of the sequence upstream of the CCA site by ATP(CTP):tRNA nucleotidyl transferase was not previously reported (37). The effects of these mutations on RNA recombination-mediated repair of the 3′ terminus will be addressed in the section below.
In a single-point replication assay at 21 h, mutations at the +3A, +5A, and +7A positions were tested because they likely would be less disruptive to the tRNA-like structure (Fig. 2A). All three nucleotide substitutions at the +3 position were tested. However, only an A to G change was tested at the +5 and +7 positions because these should need to retain the ability to form the RNA pseudoknot structure. We found that all changes at the +3A position and the ones at the +5 and +7 positions accumulated RNAs at more than 56% of the wild-type level (Table 1). These results suggest that the bases at positions +3, +5, and +7 are not critical for RNA accumulation.
It is unlikely that there is active recombination-mediated repair at these three positions by 21 h after transfection. However, to ensure that we were not examining the effects of recombinational repair, we performed a time course experiment to examine minus-strand RNA synthesis early in infection. Recombination is expected to be a rare event, and a longer time is needed for the repair and significant accumulation of the repaired RNA. RNAs M1C/G, M3A/C, M3A/G, M5A/G, and M7A/G were transfected into protoplasts along with RNA1 and RNA2, and total RNA was harvested at 0, 3, 10, 18, and 24 h after transfection. All these mutants had kinetics of minus-strand RNA accumulation similar to those of the wild type (Fig. 2C). These results indicate that changes in the template for minus-strand RNA3 synthesis had only minimal effects on all of the mutations tested.
A minus-strand RNA approximately the length of RNA4 was apparent in most of our experiments (e.g., see Fig. 2B, upper panel, band denoted by an asterisk). This RNA is less abundant than the other minus-strand RNAs and is observed only when BMV RNA4 is produced. Probing of the Northern blots with the sequence coding for the capsid protein lit up this band and minus-strand RNA3, but not BMV RNA1 or RNA2, indicating that it is derived from RNA3 (or RNA4 [Hema, data not shown]). In time course experiments, however, this RNA appeared after the synthesis of RNA4. It is possible that this RNA is produced from subgenomic RNA4. While this RNA was observed throughout the results, we will not pursue its genesis further.
Genomic plus-strand RNA synthesis.
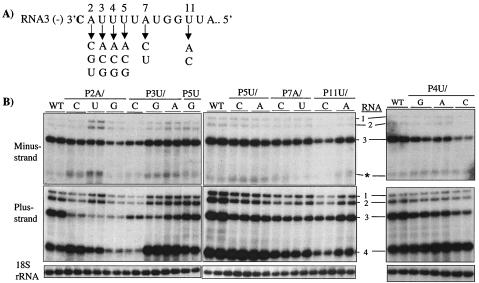
The sequence immediately 5′ of the initiation site for plus-strand synthesis is rich in A and U nucleotides. Marsh et al. (31) and Sivakumaran et al. (46) proposed that this sequence contributes to efficient RNA synthesis. Mutational analysis of a functional promoter-template that is capable of genomic plus-strand RNA initiation by the enriched BMV replicase in vitro showed that sequences from position +2 to +6 likely contribute to the initiation process (45, 46). We made a series of base substitutions starting at the 3′ end of the BMV minus-strand RNA3 from positions +2 to +11 and tested them in barley protoplasts for RNA replication (Fig. 3A). All of the mutations reduced minus-strand RNA3 accumulation to between 40 and 90% of the wild-type level (Fig. 3B, upper panel, and Table 1). However, several of these mutations, especially ones near the initiation cytidylate, had a more severe effect on plus-strand RNA3 synthesis (Fig. 3B, lower panel). For example, all changes at the +2 and +3 positions resulted in less than 10% and 25%, respectively, of plus-strand synthesis compared to that of the wild type (Fig. 3B; Table 1). Mutations from positions +4 to +11 had less severe effects, with RNA accumulation being between 32 and 62% of that of the wild type (Fig. 3B; Table 1). All of these results suggest that the presence of an A and a U at the +2 and +3 positions, respectively, is required for RNA synthesis, but that the requirement for specific bases during plus-strand RNA synthesis may be relaxed starting from the +4 position.
FIG. 3.
Accumulation of RNAs with mutations near the initiation site in BMV plus-strand RNA3 template. (A) Schematic of the 3′ 13 nucleotides of minus-strand BMV RNA3 that direct plus-strand RNA3 synthesis. The initiation cytidylate is in bold, and the numbers represent the position relative to the initiation site. Arrows point to the substitutions made at each nucleotide. (B) Northern blot analysis of progeny RNA accumulated in barley protoplasts at 21 h posttransfection. The mutated RNAs named above the lanes in the autoradiogram were transfected into protoplasts, and the progeny RNAs were processed for minus-strand and plus-strand BMV RNA accumulation. The 18S rRNA accumulation is shown to establish that similar amounts of RNAs were loaded in each lane.
Several mutations in the templates for genomic plus-strand RNA synthesis had an unexpected effect on subgenomic RNA synthesis. The relationship between the two RNAs is somewhat complex; mutants P2A/C, P3U/G, P3U/A, P4U/A, P4U/C, P5U/C, and P5U/A all increased subgenomic plus-strand RNA levels by up to threefold (Fig. 3B; Table 1), while mutants P2A/G and P3U/C actually decreased RNA4 accumulation to less than 18%. These effects were reproducible in two independent experiments and suggest that, under some conditions, an inability to produce RNA3 allows increased RNA4 accumulation.
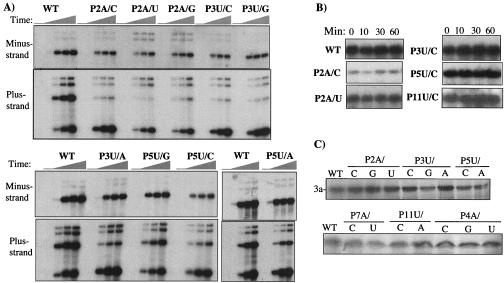
To further confirm the effects of the mutations and to reduce the possibility that the observed effects were not due to repair of the mutations during replication, we performed a time course experiment. Protoplasts were transfected and total RNA was extracted at 0, 3, 10, 18, and 24 h after transfection. While only the mutations up to position P5U are shown (Fig. 4A), the entire set of mutants in the template for plus-strand RNA synthesis were tested. Relative to transfections with wild-type RNA3, the effects of the mutations on minus-strand RNA accumulation was much less than those on plus-strand RNA3 (Fig. 4A). In addition, several of these RNAs, such as mutants P2A/C, P2A/U, P3U/C, P3U/G, and P3U/A, failed to increase over time at rates similar to that of wild-type RNA3 (e.g., compare the results in the uppermost panel in Fig. 4A). This is to be expected because plus-strand RNA3 synthesized de novo during infection also contributes to the accumulation of minus-strand RNA synthesis, and several of these mutants are defective for plus-strand RNA3 synthesis (Fig. 4A, top panel). These results confirm that the majority of the mutations affect plus-strand RNA synthesis (Fig. 4A). Lastly, we note that the increases in RNA4 accumulation by P2A/C, P3U/G, and P3U/A were observed throughout the time courses (Fig. 4A).
FIG. 4.
Time course experiments to examine the effects of mutations in the template for genomic plus-strand RNA3 synthesis. (A) Autoradiograms showing the accumulation of progeny minus- and plus-strand RNAs in protoplasts transfected and harvested between 0 and 24 h. The identity of each mutant is indicated above each lane of the autoradiogram. (B) Autoradiograms examining the stability of labeled mutant RNAs in transfected barley protoplasts. The time of RNA isolation (in minutes) is shown above the autoradiogram. (C) Results from rabbit reticulocyte translation extract programmed with wild-type and mutant RNA3 transcripts. Shown are the 3a proteins by SDS-12% PAGE. The identity of each mutant RNA is named above the autoradiograms.
The synthesis of minus-strand RNA3 and subgenomic RNA with mutations at the +2 and +3 positions suggests that there is no significant effect on RNA stability. Nonetheless, we examined this directly. Radiolabeled and capped RNAs were transfected into barley protoplasts along with unlabeled BMV RNA1 and RNA2 in a reaction mimicking a normal transfection experiment. The cells were harvested for total RNA at several time points up to 60 min. Although the labeling efficiency varied somewhat for each transcript, none was rapidly degraded within the 1-h period (Fig. 4B).
We also examined whether the mutations affected the translatability of the transcripts in rabbit reticulocyte extracts. While the amount of different translated products varied by approximately 20%, mutations that had an effect on RNA3 synthesis did not have an obvious effect on the amounts of protein produced (Fig. 4C). For example, mutations at the +2A and the +3U positions that reduced plus-strand RNA3 accumulation in protoplasts to ≈10% of the wild-type level directed translation of movement protein (3a) at levels similar to those from wild-type RNA3 (Fig. 4C). These experiments suggest that the effects of the mutations are on RNA accumulation rather than on RNA stability or translatability.
To examine whether the effects observed on BMV replication in protoplasts correlates to BMV infection in plants, we inoculated the mutant transcripts along with RNAs 1 and 2 into C. quinoa, a local lesion host for BMV. Wild-type transcripts gave rise to approximately 30 chlorotic lesions per inoculated leaf (Fig. 5A). In contrast, mutations in the plus-strand RNA3 template at the +2A and +3U positions resulted in no obvious virus-associated symptoms by 7 days after inoculation, consistent with observations of decreased replication in protoplasts (Fig. 5A). These experiments indicate that repair of the mutations at the 3′ end of the minus-strand RNA3 is at a sufficiently low level that the plants do not exhibit clear symptoms. Mutations at the +5U and +7A positions gave approximately 16 and 19 easily discernible lesions per inoculated leaf, respectively (Fig. 5A, and data not shown), suggesting that these mutations had only minor effects on BMV replication, as evidenced by a reduced number of lesions. Two weeks postinoculation, plants inoculated with wild-type transcripts and mutations at the +5U and the +7A positions of RNA3 developed systemic mottling symptoms, while plants inoculated with RNA3 containing mutations at the +2, +3, and +11 positions did not show obvious signs of systemic infection (Hema, unpublished results).
FIG. 5.
Plant plaque assay with wild-type and representative mutant RNAs. (A) Local lesions induced on C. quinoa after inoculation with wild-type RNA 1 and 2 and either wild-type RNA3 or RNA3 with mutations in the template for genomic plus-strand RNA3 synthesis. Representative leaves are shown, with the identity of the most relevant RNA denoted to the side of each leaf. Leaves inoculated with mutants P2A/U, P2A/C, 3PU/C, and P11U/C are all asymptomatic, and the leaf inoculated with P2A/U is shown to represent this group. All leaves were photographed 5 to 7 days postinoculation (dpi). (B) Representative examples of lesions induced on C. quinoa by wild-type RNA1, RNA2, and RNA3 variants with mutations in the subgenomic template. Leaves were photographed 7 days after inoculation.
Subgenomic template initiation sequence.
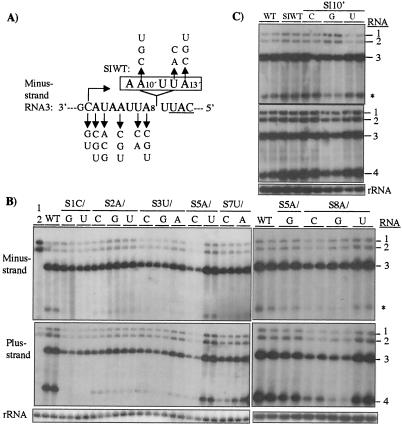
Subgenomic RNA4 is produced abundantly during BMV infection. The first nine positions of the template for RNA4 synthesis are devoid of C's and G's (Fig. 6A) with the translation initiation codon starting at nucleotide 10. Single nucleotide mutations were made up to the +8 position of the template for subgenomic RNA synthesis to examine their effects (Fig. 6A).
FIG. 6.
Effects of mutations in the subgenomic RNA template. (A) Schematic of the BMV subgenomic RNA template. The initiation cytidylate is in bold, and the start codon for coat protein translation is underlined. Arrows point to the single nucleotide mutations made at positions along the template for RNA4 synthesis. The open box encompasses a 5-nucleotide insertion (5′-AAUUA-3′) between +8 and +9 that increases the length of the nontranslated sequence to 14 nucleotides in an RNA named SIWT. The nucleotides inserted are distinguished from the normal order of the nucleotides in the subgenomic template by the prime after the nucleotide position. (B) Effects of mutations in the subgenomic RNA template on BMV RNA accumulation. Autoradiograms from Northern blot analyses are in the same format as was used in previous figures. The leftmost lane was transfected only with BMV RNA1 and RNA2. (C) RNA synthesis by SIWT and mutations at position 10′ made in the context of SIWT.
First, we determined whether the mutations within the subgenomic template affected other modes of BMV RNA synthesis. In general, mutations in the subgenomic template had minor effects on minus-strand and plus-strand RNA3 levels (Fig. 6B, Table 1). The most significant effect was with mutant S8A/C, which decreased plus-strand RNA3 to a third of the wild-type level. In contrast, all mutations in the first three template nucleotides for RNA4 synthesis decreased RNA4 levels to less than 14% of the wild-type level (Fig. 6B, bottom panel, and Table 1). Substitutions further away from +1C had generally more severe effects on RNA4 synthesis relative to the other RNAs (Table 1). Mutations at position +8 reduced RNA synthesis to between 12 and 66% of the wild-type level, with changes to C or G having a more severe effect than a change to a U.
The effects seen with substitutions at the +8 position suggest that we had not yet reached the 3′ boundary of the sequences that contribute to RNA4 accumulation. We did not change the U at the +9 position of the subgenomic RNA template because of its proximity to the initiation codon (present on the plus-strand RNA). To allow more extensive analysis of the length of the A/U-rich sequence required for RNA4 synthesis, we inserted a five-base sequence, 5′-AAUUA-3′, between positions +8 and +9 to result in an RNA named SIWT, increasing the length of the nontranslated sequence in the complementary strand to 14 nucleotides. We will refer to the nucleotides in the context of SIWT according to their positions relative to the initiation cytidylate, but with a prime next to the number denoting the nucleotide position (Fig. 6A).
Compared to wild-type BMV RNA, SIWT increased subgenomic RNA4 levels to 130% (Fig. 6C), and thus the insertion should have no negative effect on RNA stability. Mutation of the 10′A position to a G reduced RNA4 accumulation only slightly, to 90% of wild-type RNA3 and 63% of SIWT. Other mutations at the 12′ and 13′ positions had no obvious effects (Table 1). Thus, the subgenomic template requires an approximately 8-nucleotide A/U-rich sequence near the initiation site.
Several changes within positions 1 to 5 of the subgenomic template also resulted in decreased RNA3 accumulation, to between 40 and 60% of the level of wild-type RNA3 (Table 1; Fig. 6B), compared to the effects on RNA4 synthesis (reduced to between 1 and 14%). Contrary to the effects seen where several mutations affecting RNA3 synthesis unexpectedly increased RNA4 synthesis (Fig. 3; Table 1), we observed an increase in RNA3 accumulation only with the change to a U at the +5 position, a mutation that did not significantly affect RNA4 synthesis.
Time course experiments were performed for several of the most relevant mutants. All of the RNAs were electrophoresed on one gel along with the wild-type control, but the autoradiogram was cropped to allow the sets of RNAs to be displayed in a linear sequence (Fig. 7A). Consistent with the defects seen in mutations at the +1C, +2A, +3U, and +5A positions at 21 h postinoculation, changes from the wild-type nucleotide sequence decreased RNA4 accumulations even at 10 h posttransfection, when RNA4 is easily observed in protoplasts transfected with wild-type RNAs. Mutations at the +1C and +2A positions had little effect on plus-strand RNA3 accumulation at the early times. However, change of +5A to a C reduced the abundance of both minus-strand and plus-strand RNA3 accumulation (Fig. 7A). Quite interestingly, several mutants (e.g., S5A/U, S5A/G, S7U/C, S8A/C, and S8A/G) decreased plus-strand but not minus-strand RNA3 levels.
FIG. 7.
Time course analysis of progeny RNA accumulation by RNAs with mutations in the subgenomic template. (A) Northern analysis of total RNA extracted at 0, 3, 10, 18, and 24 h postinoculation and hybridized with strand-specific probes. The identities of the mutant RNAs are at the top of each lane of the autoradiogram. (B) Analysis of the effects of selected mutants on the stabilities of the RNAs in transfected protoplasts. The radiolabeled RNAs were transfected into protoplasts and extracted for analysis at the times indicated above each lane.
The stability of the RNA3 containing mutations in the subgenomic RNA template was examined. No significant decrease in several mutant RNAs was observed within 60 min after transfection into barley protoplasts in a manner that correlated with the decrease in RNA4 synthesis (Fig. 7B). When inoculated onto C. quinoa plants along with RNAs 1 and 2, wild-type RNA3 transcripts gave approximately 40 chlorotic lesions per inoculated leaf (Fig. 5B). In contrast, other mutants at the +2 to +8 positions resulted in approximately 40 to 80 necrotic lesions per inoculated leaf. The necrotic lesions coalesced over time, resulting in complete leaf necrosis (Fig. 5B).
All plants were held in the growth chamber for an additional 2- or 3-week period for observations. Plants inoculated with all three wild-type transcripts developed systemic mottling by 14 days postinoculation. None of the plants inoculated with transcripts of wild-type RNA1 and RNA2 and any of the other RNA3 variants showed obvious systemic symptoms on uninoculated leaves. We note with interest that deletions of some N-terminal amino acids in the capsid protein were previously reported to result in necrotic lesions (39). Our mutations do not affect the sequence of the capsid protein but may affect its level.
DISCUSSION
Analyses of the sequences of bromoviruses have led us to hypothesize that an A/U-rich template sequence regulates the level of RNA accumulation. First, all bromovirus plus-strand templates have a higher percentage of A/U nucleotides than templates for minus-strand RNA synthesis (Fig. 1B). Second, the relative abundance of the four BMV RNAs is correlated with the nucleotide position of the first C or G nucleotide in the template: RNA4 (10) > RNA3 (7) > RNA2 (4) > RNA1 (2) (Fig. 1B). Third, the second and third nucleotides of all bromoviral genomic plus-strand and subgenomic RNAs are A and U, respectively (2). This motif is seen even in Broad Bean Mottle Virus, which differs from other bromoviruses in that its RNA3 apparently initiates plus-strand RNA3 synthesis with an A rather than a G. These observations led us to examine how mutations in the template sequence for genomic plus-strand, genomic minus-strand, and subgenomic RNAs will affect RNA accumulation in transfected protoplasts. We believe that our analysis can lead to three main conclusions concerning BMV RNA synthesis: minus-strand RNA synthesis is not significantly affected by the template sequence; plus-strand RNA synthesis requires specific identities of +2A and +3U; and efficient subgenomic RNA synthesis requires an A/U-rich sequence. Each of these conclusions is expanded upon below.
Minus-strand RNA synthesis.
The effects of some of the mutations on minus-strand synthesis need to be interpreted cautiously because deletions of up to all three nucleotides of the CCA sequence in a tRNA can be repaired either by the ATP(CTP):tRNA nucleotidyl transferase (13) or by RNA recombination (37, 43). However, analysis of changes at the +3, +5, and +7 positions that should not be repaired by the ATP(CTP):tRNA nucleotidyl transferase revealed no significant change in RNA levels relative to wild-type RNAs. Also, the kinetics of minus-strand RNA accumulation by mutations at these positions suggest that RNA recombination is unlikely to be the cause for normal levels of RNA accumulation. The simplest explanation for the lack of obvious effects of these template changes is that the template nucleotides for minus-strand RNA synthesis do not affecting RNA synthesis as long as the template sequence fulfills other requirements for viral infection, such as forming the correct tRNA-like structure.
Initiation box.
The BMV genomic plus-strand and subgenomic plus-strand RNA accumulations required highly specific nucleotides at three positions: +1C, +2A, and +3U (all in the template sense). Mutations of these three nucleotides caused more severe reduction in RNA levels in comparison to nucleotides further down the template. It is possible that these three nucleotides represent a site that specifically interacts with the viral replicase or is required to contact a cellular protein. The fact that genomic and subgenomic RNA share this motif suggests overlap in the mechanism of their initiation. Recombinant bovine diarrhea virus (BVDV) RNA polymerase has been demonstrated to specifically interact with nucleotides +1 through +3 at the 3′ end of the BVDV minus-strand RNA (25).
While +2A and +3U are important for plus-strand RNA3 accumulation, mutations at positions +4, +5, and +7 do not show a more severe effect when the A/U-rich sequence is replaced with C's or G's (Table 2). Thus, the primary template requirement for RNA3 accumulation is an initiation box, which may function in concert with sequences and structures downstream in the template for specific RNA synthesis (36, 46).
TABLE 2.
Correlation between the identity of the base in the template and RNA levelsa
| RNA and change | RNA level vs. wild type (fold) with indicated change at position:
|
||||||
|---|---|---|---|---|---|---|---|
| +2 | +3 | +4 | +5 | +6 | +7 | +8 | |
| Subgenomic | |||||||
| G/C | 1 | 11 | NT | 32 | NT | 25 | 19 |
| A/U | 8 | 13 | NT | 85 | NT | 72 | 66 |
| Genomic | |||||||
| G/C | 8 | 21 | 49 | 54 | NT | 50 | |
| A/U | 9 | 24 | 64 | 64 | NT | 50 | |
NT, not tested.
Subgenomic RNA and the A/U-rich sequence.
The substitution of any of the A or U nucleotides within the first eight nucleotides of the subgenomic RNA resulted in decreased RNA4 levels, with changes to G or C resulting in lower RNA accumulation than changes to A or U (Table 2). Consistent with the need for an A/U-rich sequence during RNA4 synthesis, increasing the length of the A/U-rich sequence from 9 to 14 nucleotides increased RNA accumulation to 130% (Fig. 6C; Table 1). These results indicate that an A/U-rich sequence is preferred for BMV subgenomic RNA accumulation.
What is the molecular basis for the preference of an A/U-rich sequence? One possibility is that A/U-rich sequences could affect translation. Some GC-rich sequences may prevent efficient ribosome scanning (27). However, we do not believe that the requirements for translation provide an adequate explanation of our observations. First, in vitro translations of the RNA3 transcripts containing single nucleotide changes that affected RNA synthesis did not have obvious defects in their translatability in reticulocyte extracts (Fig. 4C). Second, monocot plants have the preferred initiation context of 5′-C-(A/C)-(A/G)-(A/C)-C-AUG-3′ (initiation AUG is in italic) (21), similar to the comparable sequence from mammals (26). Therefore, the consensus translational initiation context does not preclude the presence of G's or C's, while several substitutions in the BMV subgenomic RNA that would mimic the consensus translational initiation context (e.g., positions 5 to 9 in RNA4) actually decrease RNA4 accumulation. In addition, the −3 position relative to the initiation AUG is equivalent to the +8 position for subgenomic RNA synthesis, and its change to a G (generally preferred for translation initiation) (27) decreased RNA accumulation to about a third of the wild-type level. The lack of correlation between the requirements for translation and effects on RNA accumulation lends more credence to the hypothesis that the A/U-rich sequence contributes to subgenomic RNA synthesis.
We propose that the initiation site and adjacent sequence regulate the level of RNA accumulation by modulating the interaction between the template and nascent RNA during the polymerization process. The interaction between the BMV replicase, the template, and the nascent RNA is known to affect RNA synthesis during the steps of initiation and termination (47-50) and regulates RNA recombination (24). The template sequence also can determine the frequency with which a recombinant RdRp bypasses base analogs (25). Perhaps the initiation sequence affects the progression of the ternary complex: a more AU-rich initiation sequence could allow faster transition of the replicase to elongative synthesis, thereby increasing the number of rounds of RNA synthesis. For minus-strand RNA synthesis, the transition from initiation to elongation may be kinetically slower, thus decreasing RNA accumulation and contributing to the asymmetric accumulation of plus-strand and minus-strand RNAs.
Pleiotropic effects on BMV RNA accumulation.
Several mutations had interesting effects on the synthesis of other classes of RNAs. For example, protoplasts transfected with RNA1 and RNA2 but not RNA3 had greater amounts of minus-strand but not plus-strand RNAs in comparison to cells transfected with all three BMV RNAs (Fig. 6B). The increase in plus-strand RNA1 and RNA2 synthesis in the presence of RNA3 suggests that BMV RNA3 or its gene products has a more active role in regulating RNA1 and RNA2 accumulation.
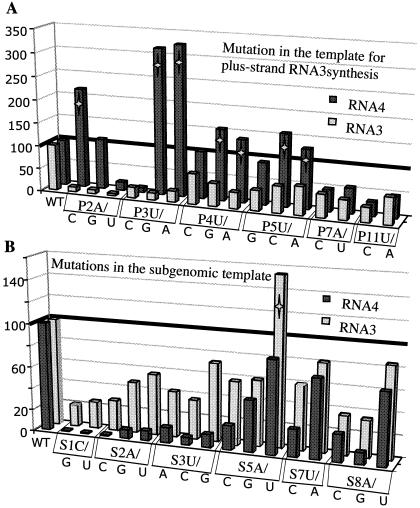
Some but not all mutations that decreased RNA3 levels increased subgenomic RNA4 (Fig. 8). These results suggest that the BMV replicase can perform subgenomic RNA synthesis when it is unable to make RNA3. We do not have a good explanation for why mutations that decreased RNA4 levels did not conversely increase RNA3 levels. It may be that the replicase(s) responsible for subgenomic RNA synthesis remains more stably bound to the subgenomic promoter even in the absence of productive RNA4 synthesis, while the interaction between the replicase and the template for RNA3 initiation is more labile.
FIG. 8.
Graphic summary illustrating the relationship between plus-strand RNA3 and RNA4 accumulation. The graphs contain a visual depiction of some of the results presented in Table 1. (A) Mutations in the template for RNA3 synthesis can affect subgenomic RNA4 accumulation. The identities of the RNAs are on the x axis. The dark line represents the level of RNA synthesis from protoplasts transfected with wild-type BMV RNAs. A star identifies all cases where RNA4 was present at higher than wild-type levels. (B) Mutations in the template for RNA4 synthesis generally do not increase RNA3 accumulation. The format of this graph is identical to that of the one used in panel A.
Mutations in the templates for plus-strand RNA3 and subgenomic RNA4 synthesis had only modest effects on minus-strand RNA3 synthesis (Fig. 3 and 6). Even changes in the template at the +2 or +3 position that significantly affected plus-strand RNA3 levels had only modest effects on minus-strand RNA3 levels (see mutations at the +2A and +3U positions, Fig. 3B). A current model for viral replication involves the circularization of the two ends of the RNA by protein-protein and protein-RNA interactions (17, 19). Should circularization take place for the BMV RNA, it is either not affected by the mutations at the 5′ end of RNA3 or not required for minus-strand RNA3 synthesis.
Comparison of results from protoplast and previous biochemical results.
Our interest in the sequence in template RNAs was first piqued by the observation that short single-stranded templates based on the BMV subgenomic template could direct RNA synthesis, but were less able to do so efficiently when a G was within five nucleotides of the initiation cytidylate (45). An examination of the effects of nucleotide substitutions to C/G and A/U revealed that lower subgenomic RNA levels are associated with C/G's present near the initiation cytidylate (Table 2). Evidence of how single nucleotide substitutions affect RNA3 initiation is not yet available, but the in vivo results do not suggest a correlation between the amount of genomic RNA3 synthesized and the specific effects of C/G bases in the template beyond position 3.
Accumulation of both subgenomic and genomic plus-strand RNAs in protoplasts requires an adenylate and a uridylate at the +2 and +3 positions. In the biochemical assay, we previously observed that a change of the +2A in the template for subgenomic RNA synthesis in vitro to C or G reduced RNA synthesis to 37 and 4%, respectively, of the wild-type level, while a change to a U retained 66% of the RNA synthesis (2). For genomic plus-strand RNA synthesis, a direct comparison is not possible because the mutations were tested in vitro in the context of RNA2 (46). However, there are some indications that the results also do not completely agree with those from protoplasts. A change of +2A to G reduced synthesis to 11%, but a change to a U retained 77% of the synthesis of wild-type RNA3. At the +3 position, changes to C or G resulted in RNA synthesis at less than 18% of the wild-type level, but a change to an A retained 88% of the wild-type level of synthesis (46). These results indicate that there are requirements for RNA synthesis in protoplasts that are not revealed by the enriched BMV replicase. We do note that mutations that negatively affected RNA synthesis in vitro are more revealing about requirements in vivo.
In summary, a combination of mechanisms likely regulates the timing and levels of viral RNA synthesis. In this work, we demonstrate that the BMV template sequence modulates RNA accumulation and that accumulation of plus-strand, minus-strand, and subgenomic RNAs responds differently to the template sequence.
Acknowledgments
We thank R. Kumar, K. Sivakumaran, and L. Kao for edits during the preparation of the manuscript and K. Sivakumaran for establishing BMV replication in barley protoplasts for our lab.
Funding was provided by the National Science Foundation.
REFERENCES
- 1.Adkins, S., R. W. Siegel, J. H. Sun, and C. C. Kao. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634-647. [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. Kao. 1998. Mechanistic analysis of RNA synthesis by an RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Ahlquist, P., V. Luckow, and P. Kaesberg. 1981. Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 153:23-38. [DOI] [PubMed] [Google Scholar]
- 5.Allison, R., C. Thompson, and P. Ahlquist. 1990. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat protein genes for systemic infection. Proc. Natl. Acad. Sci. USA 87:1820-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y.
- 7.Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification in brome mosaic virus replicase protein 1a that mediates association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1996. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, M. R., and C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, M., A. L. N. Rao, and C. Kao. 1998. The 5′ ends of BMV RNAs promote initiation of (−)-strand RNA synthesis in vitro and in vivo. Virology 252:458-467. [DOI] [PubMed] [Google Scholar]
- 11.Chapman, M., R. Tayon, and C. Kao. 1999. Cis-acting sequences for minus-strand RNA synthesis in plant-infecting RNA viruses. Curr. Top. Virol. 1:175-189. [Google Scholar]
- 12.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by the helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher, M. P. 1982. tRNA nucleotidyltransferase. Enzymes 15:183-215. [Google Scholar]
- 14.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus-strand promoter activity. J. Mol. Biol. 201:31-40. [DOI] [PubMed] [Google Scholar]
- 15.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA J. Mol. Biol. 201:41-55. [DOI] [PubMed] [Google Scholar]
- 16.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie, D. R. 2002. Protein-protein interactions required during translation. Plant Mol. Biol. 50:949-970. [DOI] [PubMed] [Google Scholar]
- 18.Goldbach, R., O. LeGall, and J. Wellink. 1991. Alpha-like viruses in plants. Semin. Virol. 2:19-25. [Google Scholar]
- 19.Herold, J., and Andino, R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 21.Joshi, C. P., H. Zhou, X. Huang, and V. L. Chiang. 1997. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 35:993-1001. [DOI] [PubMed] [Google Scholar]
- 22.Kao, C. C. 2002. Lessons learned from the core RNA promoters of Brome Mosaic Virus and Cucumber Mosaic Virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 23.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 24.Kim, C. H., and C. C. Kao. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA 7:1476-1485. [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, M. J., W. Zhang, Z. Hong, and C. Kao. 2000. The bovine viral diarrhea virus RNA-dependent RNA polymerase recognizes different nucleotide moieties during de novo initiation and elongation of RNA synthesis. J. Virol. 74:10312-10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroner, P. A., B. M. Young, and P. Ahlquist. 1990. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, with linker insertion mutagenesis. J. Virol. 64:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 31.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh, L. E., C. H. Huntley., G. P. Pogue., J. P. Connell, and T. C. Hall. 1991. Regulation of plus/(−) strand asymmetry in replication of brome mosaic virus RNA. Virology 182:76-83. [DOI] [PubMed] [Google Scholar]
- 33.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 34.Mise, K., R. F. Allison, M. Janda, and P. Ahlquist. 1993. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 67:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogue, G. P., and T. C. Hall. 1992. The requirement for a 5′ stem-loop structure on brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol. 66:674-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogue, G. P., L. E. Marsh, J. P. Connell, and T. C. Hall. 1992. Requirement for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology 188:742-753. [DOI] [PubMed] [Google Scholar]
- 37.Rao A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the transfer RNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, A. L. N., and G. L. Grantham. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 211:42-52. [DOI] [PubMed] [Google Scholar]
- 39.Rao, A. L. N., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif, and its role in encapsidation, movement, and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 40.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize and 1a independently localizes reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shi, P. Y., N. Maizels, and A. M. Weiner. 1998. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J. 17:3197-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivakumaran, K., and C. C. Kao. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 73:6415-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sivakumaran, K., C. H. Kim, R. Tayon Jr., and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, J. H., S. Adkins, G. Faurote, and C. C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: Synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 48.Sun, J. H., and C. C. Kao. 1997. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology 233:63-73. [DOI] [PubMed] [Google Scholar]
- 49.Sun, J. H., and C. C. Kao. 1997. Characterization of RNA products associated with or aborted by a viral RNA-dependent RNA polymerase. Virology 236:348-353. [DOI] [PubMed] [Google Scholar]
- 50.Tayon, R., Jr., M.-J. Kim, and C. Kao. 2001. Nucleotides near the 5′-terminus of the template contributes to the completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 29:3583-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]