Abstract
The androgen receptor (AR) may recruit multiple coregulators for proper or optimal transactivation. Here we report the identification and characterization of ARA67/PAT1 as an AR coregulator from a prostate cDNA library. ARA67/PAT1 was screened out as an AR N terminus interacting protein. Interaction mapping shows that the cooperation of multiple domains within ARA67/PAT1 may be required for the maximal interaction with AR. ARA67/PAT1 functions as a repressor with better suppressive effects on AR compared to glucocorticoid receptor and estrogen receptor. Further mechanism dissection reveals that the interrupted AR cytoplasmic-nuclear shuttling may play a major role in ARA67/PAT1 mediated suppression on AR. Together, these results suggest that ARA67/PAT1 may function as a novel repressor that can modulate AR function in prostate cancer.
Androgen receptor (AR) is a member of the nuclear receptor family and mediates androgen actions that are involved in a wide range of developmental and physiological responses, such as male sexual differentiation, virilization, and gonadotropin regulation (41). Besides its physiological roles, AR also contributes to pathological conditions, highlighted by its involvement in prostate carcinogenesis (41, 42). Like other members of the nuclear receptor family, the AR contains an amino-terminal transcription activation domain, a DNA-binding domain, and a carboxyl-terminal ligand-binding domain. Upon ligand binding, the AR dissociates from chaperone proteins, homodimerizes, translocates into the nucleus, and turns on the expression of its target genes by binding to the androgen-responsive element (41).
AR is classified with glucocorticoid receptor, mineralocorticoid receptor, and progesterone receptor as one group within the nuclear receptor superfamily, since they share high homology in the DNA-binding domain and recognize very similar hormone response elements (10, 27). However, the physiological responses mediated by these receptors upon cognate ligand activation are quite distinct and hormone specific. Apparently, these cannot be explained by a specific DNA binding through the DNA-binding domain, and factors located outside the DNA-binding domain may play a key role in determining the specific hormone responses. It is known that AR, as well as other nuclear receptors, recruits multiple coregulators for optimal transcriptional regulation (34, 35, 46). It has been suggested that regulation by coregulators is an efficient way to achieve cell- and promoter-specific activation (38). Although a large number of coregulators have been identified in recent years (reviewed in references 21 and 33), how these coregulators exert their influence on the specificity of nuclear receptor-mediated transcriptional responses remains largely unknown, since many coregulators have been shown to bind and influence multiple nuclear receptors in similar conditions.
For example, SRC-1 can serve as a coactivator to many nuclear receptors such as progesterone receptor, estrogen receptor, glucocorticoid receptor, thyroid hormone receptor, and retinoid X receptor (35). Although nuclear receptor corepressor and silencing mediator of retinoid and thyroid hormone receptor (SMRT) were initially identified as corepressors to mediate active suppression by unliganded thyroid hormone receptor and retinoid acid receptor (6, 22), later studies suggest that they also serve as corepressors of progesterone receptor (45), estrogen receptor (28), and AR (9, 30). It is assumed that the coregulators that can preferentially bind and influence an individual nuclear receptor at a specific subcellular environment may help to determine the specificity of nuclear receptor-mediated responses.
Compared to the highly conserved DNA-binding domain and ligand-binding domain, the N terminus is quite polymorphic in terms of sequence and length among nuclear receptors. Therefore, the N terminus is more likely to provide unique surfaces to recruit distinct factors that contribute to the specific action of a certain nuclear receptor. The AR N terminus (ARN) is large, and there are two distinct regions important for its transactivation function, amino acid residues 141 to 338, which are required for full ligand-inducible transactivation, and residues 360 to 494, where the ligand-independent activation function-1 (AF-1) region is located (21). Coactivators and corepressors have been identified to interact with the ARN (23, 24, 26, 29, 32, 40), however, whether and how these factors influence the specificity of androgen action remains unclear. Furthermore, although the ARN extends to more than one half of the full-length protein, its associated proteins are relatively fewer compared to those associated with AR DNA-binding domain and AR ligand-binding domain, presumably due to the existence of the AF-1 region. This limits the application of conventional yeast two-hybrid system with ARN as the bait. It is likely that additional ARN-associated proteins remain to be identified.
In order to identify proteins that are associated with ARN and possibly with AF-1, we employed a new yeast two-hybrid system, the CytoTrap Sos system (Stratagene), with full-length ARN as the bait, to screen a human prostate cDNA library. One of the clones identified was termed ARA67. A sequence alignment search revealed that ARA67 matched the sequence encoding protein interacting with amyloid precursor protein tail 1 (PAT1). ARA67/PAT1 contains 585 amino acids with a predicted molecular mass of 66.9 kDa. It shares homology with kinesin light chain (48), which is a molecular motor involved in the transportation of cargos along the microtubule. Studies have shown that ARA67/PAT1 can bind microtubules and is involved in amyloid precursor protein secretion (48).
In this report we demonstrate that ARA67/PAT1 is a novel AR-interacting protein that functions as a repressor of AR. Its influence on the subcellular distribution of AR in the presence of ligand may be a major contributor to the suppression effect.
MATERIALS AND METHODS
Plasmids.
The full-length open reading frame (ORF) cDNA of ARA67/PAT1 was generated by PCR with human testis cDNA library (Clontech) as the template, and subsequently constructed into pGEMT Easy vector. Plasmids pM-ARA67/PAT1, pSG5-ARA67/PAT1, and pcDNA4-ARA67/PAT1 (expressing His-tagged ARA67/PAT1) were constructed by releasing ARA67/PAT1 from pGEMT-ARA67/PAT1 with proper enzymes and inserted into the target vectors. Hemagglutinin (HA)-labeled ARA67/PAT1 constructs (in pKH3 vectors) and glutathione S-transferase (GST)-ARA67/PAT1 constructs (in pGEX vectors) were generated with pGEMT-ARA67/PAT1 as the PCR template and the PCR products were subsequently digested and ligated to their target vectors. The correct constructions and expression of these plasmid constructs were verified by sequencing, TNT in vitro expression, or Western blotting. pARE4-Luc contains four copies of synthetic tandem repeats of androgen-responsive element as has been described (31). The pGL3-PSA6.0Luc plasmid was kindly provided by A. Mizokami (Kanazawa University, Kanazawa, Japan).
Construction of DNA vector-based RNA interference plasmids.
Small interfering RNA (siRNA) target sites were selected by scanning the cDNA sequence for AA dinucleotides, recording the 19 nucleotides that start with G immediately downstream of the AA, and then analyzed by BLAST search to eliminate any sequences with significant homology to other genes. The siRNA inserts, containing selected 19-nucleotide coding sequences, followed by a 9-nucleotide spacer, an inverted repeat of the coding sequences, plus 5 T's, were generated as double-stranded DNAs with ApaI and EcoRI sites with primer extension, and then subcloned into plasmid BS/U6 (44) at the ApaI/EcoRI site. The corresponding oligonucleotides for generating the siRNA-H insert are 5′-AGT CGG GCC CGA AGG CAG AAC AGT TAA TTG TCA AGA GAA ATT AAC-3′ (forward) and 5′-CGG AAT TCT TCC AAA AAG AAG GCA GAA CAG TTA ATT TCT CTT GAC-3′ (reverse). A nonfunctional siRNA was constructed as a negative control, which contains nucleotide substitutions at the 19-nucleotide targeting sequence of siRNA-H. The corresponding oligonucleotides for generating this scrambled siRNA control are 5′-AGT CGG GCC CGA AGG TAG ATC AGC TAA TTG TCA AGA GAA ATT AGC-3′ (forward) and 5′-CGG AAT TCT TCC AAA AAG AAG GTA GAT CAG CTA ATT TCT CTT GAC-3′ (reverse).
Yeast two-hybrid screening.
The CytoTrap Sos system (Stratagene) was used for the yeast two-hybrid screening. pSos-ARN containing the human Sos gene fused with cDNA encoding ARN (amino acids 1 to 537) was generated as the bait to screen a human prostate cDNA library constructed in the pMyr vector (Stratagene), which expresses library proteins fused with a myristylation membrane localization signal. Expression of the myristylation sequence-tagged proteins is induced by galactose, but not glucose, and the expressed proteins are anchored to the cell membrane. The screening was carried out by cotransforming the pSos-ARN bait construct and library plasmids into a temperature-sensitive mutant Saccharomyces cerevisiae strain, cdc25H, which cannot grow at a stringent temperature of 37°C. Once the bait protein physically interacts with the target protein, the human Sos protein fused to the bait is recruited to the membrane, which subsequently activates the Ras signaling pathway, allowing the mutant S. cerevisiae strain to grow at 37°C.
Cell culture and transfection.
H1299 and COS-1 cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum. LNCaP and MCF-7 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. All media contained 100 units of penicillin, 100 μg of streptomycin per ml, and 2 mM l-glutamine. Cells were seeded to a density of 50 to 60% confluency for transfection.
In transfections where H1299 and COS-1 cells were used, the calcium phosphate precipitation method was used as described (37) unless otherwise noted. In mammalian two-hybrid assays, which did not require ligand treatment, luciferase activity of the reporter gene was assayed 20 to 24 h after transfection with the dual-luciferase reporter assay system (Promega). In other reporter gene assays, where ligand treatments were required, cell culture media were changed to Dulbecco's modified Eagle's medium containing 10% charcoal-dextran-stripped fetal bovine serum 2 h before transfection. After 16 to 18 h of transfection, cells were treated with medium containing either vehicle or ligands for another 20 to 24 h.
For assaying siRNA activity, cells were harvested 2 to 3 days after SuperFect transfections, which were performed according to the manufacturer's protocol (Qiagen). LNCaP cells were transfected by electroporation with 4 × 106 cells suspended in 0.4 ml of RPMI medium containing 2% fetal bovine serum and mixed with 10 μg of indicated plasmids. Electroporation was performed at 250 V and 950 μF with a Gene Pulser II (Bio-Rad). When the transfection efficiency was to be monitored, the control was vector pEGFP in an amount of 1/20 of the total DNA transfected.
GST pull-down assay.
GST-ARN, GST-ARA67/PAT1 fusion proteins, and GST control protein were expressed in Escherichia coli BL21(DE3)pLysS bacterial strain (Stratagene) and purified with glutathione-Sepharose beads as instructed by the manufacturer (Amersham Pharmacia). In vitro [35S]methionine-labeled AR, ARN, AR DNA-binding domain, AR ligand-binding domain, and ARA67/PAT1 proteins were generated with TNT-coupled reticulocyte lysate systems (Promega). For in vitro interactions, mixtures of glutathione bead-bound GST fusion proteins and 5 μl of [35S]methionine-labeled input proteins in 100 μl of interaction buffer (20 mM Tris, pH 8.0, 60 mM NaCl, 6 mM MgCl2, 1 mM EDTA, 0.05% Nonidet P-40, 1 μM dithiothreitol, 8% glycerol, and 1 mM phenylmethylsulfonyl fluoride) were incubated in the presence or absence of 10 μM dihydrotestosterone (DHT) on a rotating disk at 4°C for 2 h. After washing with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 6 mM MgCl2, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM dithiothreitol, 8% glycerol, and 1 mM phenylmethylsulfonyl fluoride) four times, the bound proteins were separated by sodium dodecyl sulfate-8 to 13% polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Northern blotting assay.
In multiple-tissue Northern blotting, the Human MTN Blot (Clontech, catalog 7760-1) was hybridized with a 32P-labeled cDNA probe covering amino acid residues 8 to 140 of ARA67/PAT1. The blot was subsequently probed with a β-actin cDNA probe. In cell line Northern blotting, total RNA was extracted from 13 cultured cell lines as indicated with Trizol reagent (Gibco) and 20 μg of each total RNA was transferred onto nylon membranes for Northern blotting. The RNA bound to the membrane was hybridized with the ARA67/PAT1 cDNA probe described above. 18S RNA was used as the RNA loading control.
Western blotting assay.
Protein samples collected from the cells were separated on SDS-8% PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk in TBST buffer (150 mM NaCl, 10 mM Tris, pH 8.0, and 0.5% Tween 20) at room temperature for 1 h. Then the membranes were immunoblotted with primary antibodies for 2 h at room temperature or overnight at 4°C, followed by incubation with secondary antibodies for 1 h at room temperature. Blots were developed with the AP color developing reagents (Bio-Rad) or the ECL-Plus reagent (Amersham).
Coimmunoprecipitation assay.
COS-1 cells seeded on 100-mm cell culture dishes were transiently transfected with AR and HA-ARA67/PAT1 expression plasmids in combinations as noted in Fig. 3C, with SuperFect transfection reagent (Qiagen) following company protocols. Other transfection and treatment procedures were the same as described above. Cells were harvested and dissolved in lysis buffer (1% Nonidet P-40, 10% glycerol, 135 mM NaCl, 40 mM Tris, pH 7.4, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 1x protease inhibitor cocktail [Roche]). Cell lysates containing 500 μg of proteins were precleared with 20 μl of protein A/G Plus-agarose and 1.0 μg of normal mouse IgG (Santa Cruz Biotechnology) for 0.5 h. The supernatant was then mixed with a 1:100 dilution of mouse monoclonal anti-Flag M2 antibody (Sigma) at 4°C for 2 h, followed by adding protein A/G Plus-agarose and mixing for another 2 h. Immunoprecipitates obtained by spinning down protein A/G Plus-agarose were washed with phosphate-buffered saline three times and separated on SDS-8% PAGE. The results were analyzed by Western blotting as described above.
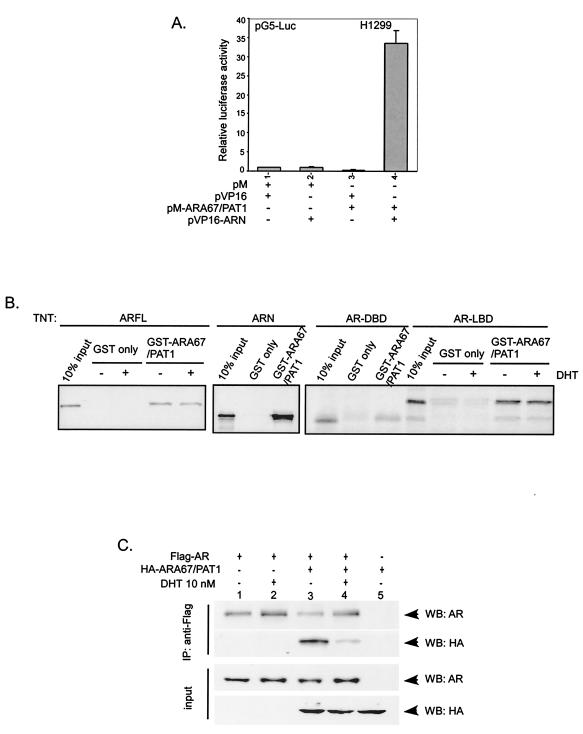
FIG. 3.
ARA67/PAT1 and AR interact in vitro and in vivo. (A) Mammalian two-hybrid assay; 0.5 μg each of pM, pVP16, pVP16-ARN, and pVP16-ARA67/PAT1 were cotransformed into H1299 cells in the combinations shown. The luciferase activity of the reporter, 0.5 μg of pG5-Luc, was normalized by the luciferase activity of 5 ng of internal control, pRL-TK, expressed as the increase over the control. The relative reporter gene activity was compared by setting the luciferase activity of the vector-alone group as 1. (B) Purified GST control protein and GST-ARA67/PAT1 fusion protein were incubated with 5 μl of [35S]methionine-labeled full-length AR (ARFL), ARN, AR DNA-binding domain (DBD), and AR ligand-binding domain (LBD) in the presence and absence of 1 μM DHT. Pulled-down proteins were separated on SDS-PAGE and visualized by autoradiography. (C) COS-1 cells were cotransfected with 1.5 μg of Flag-AR and 9.0 μg of HA-ARA67/PAT1 or vector. After transfection cells were treated with 10 nM DHT or vehicle for 24 h before harvesting; 500 μg of total cell lysate proteins from each samples was immunoprecipitated with anti-Flag antibody for Western blot analysis with anti-AR and anti-HA antibodies (Roche).
Immunofluorescence staining.
COS-1 cells were seeded on two-well Lab Tek Chamber slides (Nalge) in Dulbecco's modified Eagle's medium containing 10% charcoal-dextran-stripped fetal bovine serum 18 h before transfection. DNA was transfected with FuGENE 6 transfection reagent (Boehringer Mannheim). After transfection, cells were treated with either 10 nM DHT or vehicle for 12 h. Then cells were fixed with fixation solution (3% paraformaldehyde and 10% sucrose in phosphate-buffered saline) for 20 min on ice, followed by permeabilization with methanol for 10 min at −20°C. Slides were washed and blocked with 2% bovine serum albumin in phosphate-buffered saline for 15 min at room temperature. Then the cells were stained with 1 μg of rabbit polyclonal anti-AR antibody (NH27) per ml and 1 μg of mouse monoclonal anti-His antibody (Santa Cruz Biotechnology) per ml at room temperature for 1 h. After the first antibody incubation, cells were washed and incubated with Texas Red-conjugated goat anti-rabbit IgG and fluorescein isothiocyanate-conjugated goat anti-mouse IgG (ICN). Stained slides were mounted with coverslips and visualized with a fluorescence microscope.
Subcellular fractionation.
Cell fractionation was performed essentially as described (2). Cell monolayers were harvested with ice-cold phosphate-buffered saline and pelleted. Cold buffer A (10 mM HEPES-KOH, pH 7.9, at 4°C, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride) equal to 5 times the packed cell volume was used to resuspend the cells. After swelling on ice for 10 min, plasma membranes were disrupted by vortexing for 10 s. The nuclei were pelleted by centrifugation at 12,000 rpm for 20 s. Supernatants containing the cytoplasmic fraction of proteins were recovered. The remaining pellets were washed with cold buffer A once and resuspended in two times the pellet volume of cold buffer C (20 mM HEPES-KOH, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride) and incubated on ice for 20 min. Samples were then centrifuged at 12,000 rpm for 2 min at 4°C to remove the cellular debris. The supernatants containing the nuclear fraction were recovered.
RESULTS
Identification of ARA67/PAT1 as an AR N-terminus-interacting protein.
To identify ARN interacting proteins, we selected the CytoTrap Sos system (Stratagene) for the screening, since the ARN contains the AF-1 domain, which can be self-transactive, making it an unsuitable bait in the conventional yeast two-hybrid screening. The CytoTrap Sos system is based on generating fusion proteins whose interaction in the yeast cytoplasm induces cell growth by activating the Ras signaling pathway (Fig. 1A), which is advantageous over the conventional yeast two-hybrid system in screening interacting proteins for transcriptional activators. In this approach, cDNA encoding ARN (amino acids 1 to 537) was constructed into the pSos vector as the bait to screen a human prostate cDNA library. The bait and library constructs were cotransformed into S. cerevisiae strain cdc25H.
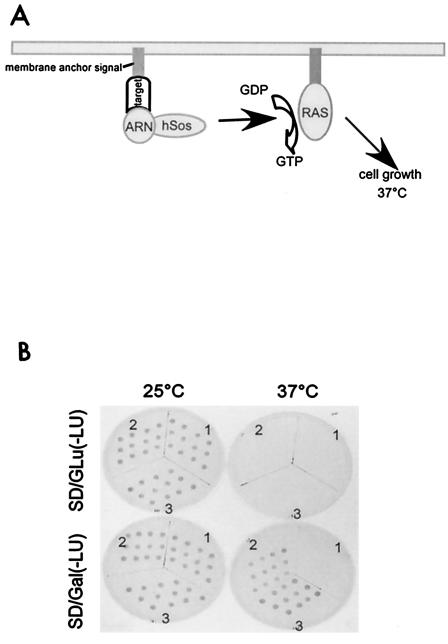
FIG. 1.
Identification of ARA67/PAT1 as ARN-interacting protein with the CytoTrap Sos system. (A) Model of CytoTrap Sos system screening strategy: the target protein is anchored to the cell membrane; human Sos fused with the bait protein (ARN) is recruited to the membrane through target-bait interaction, activating the Ras-signaling pathway by promoting GDP/GTP exchange; Ras activates the signaling cascade that permits mutant S. cerevisiae cdc25H to grow at the restrictive temperature of 37°C. (B) Interaction of ARN and ARA67/PAT1 in S. cerevisiae. S. cerevisiae cdc25H cells were cotransformed with different combinations of expression constructs and plated on different SD/Glu(-LU) plates. After the colonies appeared on plates incubated at the permissive temperature of 25°C, 12 colonies of each transformant were picked and spotted on SD/Gal(-LU) and SD/Glu(-LU) plates for interaction tests. S. cerevisiae cells were cotransformed with: section 1, the pSos vector and pMyr-ARA67/PAT1 (control to eliminate false-positive clones); section 2, pSos-ARN and pMyr-ARA67/PAT1; section 3, pSos-MAFB and pMyr-MAFB (positive control).
Of 8 × 105 clones screened, two positive clones were identified. One of the clones, named ARA67, matched a DNA sequence encoding amino acids 20 to 585 of the protein PAT1 (48) (GenBank accession no. AF017782) at an identity of 99.6%. 5′ rapid amplification of cDNA ends was used to obtain the full-length ARA67, which contains an open reading frame encoding 585 amino acids that matches the reported PAT1 sequence (48). The interaction in S. cerevisiae was confirmed by retransforming the pMyr-ARA67/PAT1 into S. cerevisiae cells pre-transformed with pSos-ARN and allowing the transformants to grow on synthetic drop-out (SD) glucose agar lacking leucine and uracil [SD/Glu(-LU)] and SD galactose agar lacking leucine and uracil [SD/Gal(-LU)] plates at the stringent temperature of 37°C. The clones that grow out on SD/Gal(-LU) plates but not on SD/Glu(-LU) plates at 37°C are interaction-positive clones. As shown in Fig. 1B, when pSos-ARN and pMyr-ARA67/PAT1 were both present, the clones showed positive growth.
ARA67/PAT1 selectively binds to ARN.
To test if the interaction between ARA67/PAT1 and ARN was specific, we cotransformed pMyr-ARA67/PAT1 with several other pSos constructs, including testicular receptor 2 (TR2) (4), testicular receptor 4 (TR4) (5), ARA55 (12), ARA70 (47), and two control plasmids (pSos-MAFB and pSos-Coll), into the temperature-sensitive mutant S. cerevisiae strain. After selection, only those S. cerevisiae cells that contained both ARN and ARA67/PAT1 expression plasmids showed positive growth, while S. cerevisiae cells containing all other pairs of plasmids could not grow on SD/Gal(-LU) plate at 37°C (summarized in Table 1). These data show that ARA67/PAT1 interacts with ARN and the interaction is rather selective.
TABLE 1.
ARA67/PAT1 selectively binds to ARN in S. cerevisiaea
| Cotransfection | Growth
|
|||
|---|---|---|---|---|
| SD/Glu(-LU)
|
SD/Gal(-LU)
|
|||
| 25°C | 37°C | 25°C | 37°C | |
| pSos + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-ARN + pMyr-ARA67/PAT1 | + | − | + | + |
| pSos-TR2 + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-TR4 + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-ARA55 + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-ARA70 + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-MAFB + pMyr-ARA67/PAT1 | + | − | + | − |
| pSos-Coll + pMyr-ARA67/PAT1 | + | − | + | − |
pMyr-ARA67/PAT1 was cotransformed with several other pSos fusion protein constructs. As shown, only ARN interacted with ARA67/PAT1, allowing the yeast host to grow at the stringent temperature of 37°C, while the other proteins tested could not.
Distribution of ARA67/PAT1 in human tissues and multiple cell lines.
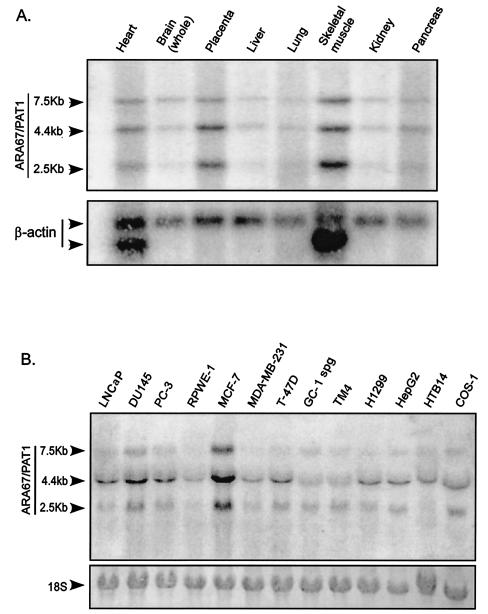
To detect the expression of ARA67/PAT1 in human tissues and cell lines, Northern blot analysis was performed with a probe covering amino acid residues 8 to 140. A BLAST search against GenBank database was performed to ensure that the probe selected did not share significant homology with other genes. Three transcripts with the sizes of 2.5 kb, 4.4 kb, and 7.5 kb were detected (Fig. 2). ARA67/PAT1 was widely expressed in multiple human tissues at variable levels. Strong expressions of all three transcripts were seen in skeletal muscle, placenta, and heart, while in other tissues, moderate to low expression levels were detected (Fig. 2A). The three ARA67/PAT1 transcripts were also seen in all the cell lines tested with the 4.4-kb transcript having the highest expression level (Fig. 2B). Among these cell lines, LNCaP, DU145, PC-3, and RPWE-1 originated from prostate, MCF-7, MDA-MB-231, and T-47D originated from breast, GC-1 spg and TM4 originated from mouse testis, H1299 originated from lung, HepG2 originated from liver, HTB14 originated from brain, and COS-1 originated from monkey kidney. An overexpression was seen in MCF-7. The different sizes of the transcripts may result from alternative mRNA splicing. Whether the three transcripts represent three different protein products remains to be answered. However, the variable levels of the three transcripts detected in different tissues and cell lines indicate that regulation of the gene products of ARA67/PAT1 may be required for maintaining different functions in various cells.
FIG. 2.
Distribution of ARA67/PAT1 mRNA in human tissues and multiple cell lines. (A) The human MTN blot (Clontech) was hybridized with a 32P-labeled cDNA probe covering amino acid residues 8 to 140 of ARA67/PAT1 and subsequently probed with β-actin. Three transcripts were detected, corresponding to the sizes of 2.5 kb, 4.4 kb, and 7.5 kb. (B) Total RNA from 13 cell lines (as indicated) was used to prepare the membrane. 18S RNA was used as the RNA loading control. The membrane was hybridized with the 32P-labeled probe as above.
Interaction of ARA67/PAT1 and AR in vitro and in vivo.
To confirm the results from yeast two-hybrid screening, the interaction between ARA67/PAT1 and AR was examined with multiple approaches. In mammalian two-hybrid assays, the reporter luciferase activity was highly induced in cells cotransfected with pM-ARA67/PAT1 and pVP16-ARN (Fig. 3A, lane 4) compared to negative controls (Fig. 3A, lanes 1, 2, and 3), indicating a strong interaction between ARA67/PAT1 and ARN. To prove that ARA67/PAT1 and AR directly interact, in vitro GST pull-down assay was carried out. In the presence and absence of DHT, ARA67/PAT1 showed interaction with full-length AR (Fig. 3B). Further dissection revealed that ARA67/PAT1 could interact with multiple regions of the AR but with different strengths; the interaction was strong in ARN, moderate in AR ligand-binding domain, and weak in AR DNA-binding domain (Fig. 3B).
The interaction between ARA67/PAT1 and AR was also verified by coimmunoprecipitation assay. Cell lysates from COS-1 cells transfected with either Flag-AR or Flag-AR with HA-ARA67/PAT1 were immunoprecipitated with anti-Flag antibody and were analyzed by Western blotting. NH27 anti-AR antibody was used to detect AR, and anti-HA antibody was used to detect HA-tagged ARA67/PAT1. As shown in Fig. 3C, ARA67/PAT1 was detected in the AR containing complex in the presence and absence of DHT (lane 3 and 4), while ARA67/PAT1 was not detected in negative control lanes (lanes 1, 2, and 5). Interestingly, a decreased interaction was observed in the presence of DHT, suggesting a potential competition between ARA67/PAT1 and DHT in binding to the AR. Together, results from yeast two-hybrid (Fig. 1B), mammalian two-hybrid (Fig. 3A), GST pull-down (Fig. 3B), and coimmunoprecipitation (Fig. 3C) assays demonstrate that ARA67/PAT1 can interact with AR in vitro and in vivo.
ARA67/PAT1 suppresses AR transactivation activity.
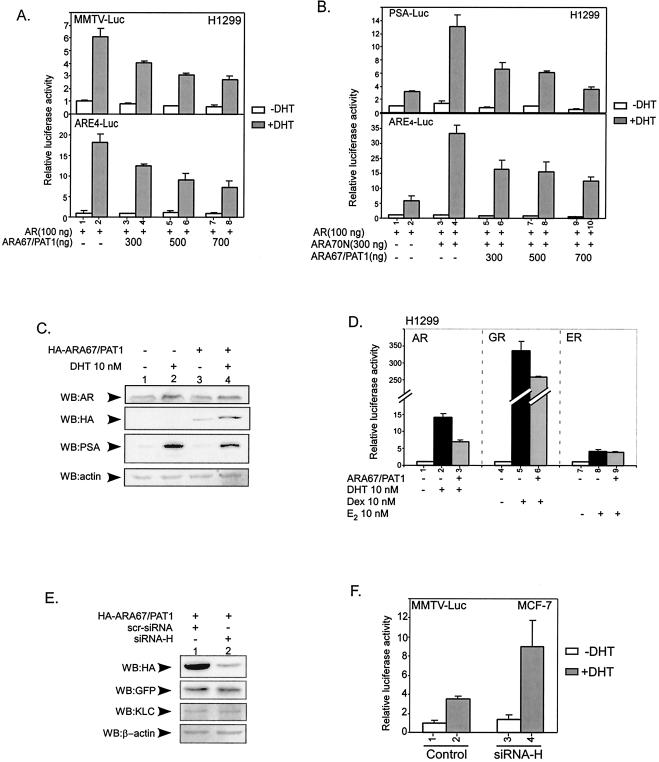
To test the consequences of the interaction between ARA67/PAT1 and AR, the influence of ARA67/PAT1 on AR transactivation was examined with reporter gene assays. Cotransfection of AR with different amounts of ARA67/PAT1 and reporter mouse mammary tumor virus-luciferase (MMTV-Luc)/ARE4-Luc in H1299 cells showed that DHT-induced AR transactivation was suppressed by ARA67/PAT1 in a dose dependent manner (Fig. 4A). Furthermore, ARA67/PAT1 could also counteract coactivator-enhanced AR transactivation. As shown in Fig. 4B, ARA70 N terminus (ARA70N) (47), a potent AR coactivator, significantly enhanced DHT-induced AR transactivation and addition of ARA67/PAT1 resulted in the suppression of AR transactivation in a dose-dependent manner with either prostate-specific antigen-luciferase (PSA-Luc) or ARE4-Luc reporters. In these experiments the internal control Renilla luciferase activity was stable. To eliminate the potential artificial effects linked to the reporter assay, we examined the influence of ARA67/PAT1 on the expression of AR's target gene at the protein level. As shown in Fig. 4C, 10 nM DHT induced prostate-specific antigen protein expression in LNCaP cells and addition of ARA67/PAT1 could then suppress this DHT-induced prostate-specific antigen expression.
FIG. 4.
ARA67/PAT1 suppresses AR transactivation. (A) We transfected 100 ng of pSG5-AR in combination with different doses of pSG5-ARA67/PAT1 (as shown in the figure) and/or pSG5 vector into H1299 cells together with 400 ng of MMTV-Luc or ARE4-Luc as the reporter and 2 ng of pRL-SV40 as the internal control. After transfection, cells were treated with 10 nM DHT or vehicle for 20 to 24 h. (B) pSG5-AR, pSG5-ARA70, and pSG5-ARA67/PAT1 were cotransfected in different combinations as shown into H1299 cells. After transfection, cells were treated with 10 nM DHT or vehicle for 20 to 24 h, and then assayed for luciferase activity; 400 ng of PSA-Luc or ARE4-Luc was used as the reporter, and the internal control was the same as above. (C) LNCaP cells were transfected with HA-ARA67/PAT1 or not (as shown) and cultured in a 60-mm cell culture dish with or without 10 nM DHT for 20 to 24 h; 50 μg of total cell lysate proteins from each sample was loaded onto the gel and Western blotted for AR, ARA67/PAT1 (HA tagged), PSA, and β-actin. (D) We cotransfected 100 ng of pSG5-AR, pSG5-glucocorticoid receptor (GR), and pSG5-estrogen receptor (ER) with 500 ng of pSG5-ARA67/PAT1 or pSG5 vector, respectively (as shown); 400 ngof MMTV-Luc was used as the reporter for both AR and glucocorticoid receptor, and 400 ng of estrogen-responsive element-Luc was used as the reporter for estrogen receptor. Each receptor group was treated with the cognate ligand or vehicle as shown for 24 h, and then assayed for luciferase activity; 2 ng of pRL-SV40 was used as the internal control. (E) COS-1 cells were transfected with HA-ARA67/PAT1 and scrambled control siRNA (scr-siRNA) or siRNA against HA-ARA67/PAT1 (siRNA-H), and harvested 2 days after transfection. pEGFP at an amount of 1/20 of total DNA was transfected as the transfection control; 50 μg of total cell lysate proteins from each sample was used for the Western blot of ARA67/PAT1 (HA tagged), GFP, kinesin light chain (KLC), and β-actin. (F) MCF-7 cells were transfected with MMTV-Luc and scrambled control siRNA or siRNA-H, cultured for 40 to 48 h, followed by 10 nM DHT or vehicle treatments for 20 to 24 h, and then assayed for luciferase activity. pRL-SV40 was used as the internal control.
A comparison of AR, glucocorticoid receptor, and estrogen receptor transactivation in the presence of ARA67/PAT1 was also carried out. At the same dose, ARA67/PAT1 showed the most significant suppression on AR transactivation (50%), compared to the moderate suppression of glucocorticoid receptor (20%), and little effect on estrogen receptor (Fig. 4D), suggesting the suppression on AR is relatively selective. Together, our data in Fig. 4A to D show that ARA67/PAT1 may function as a suppressor with better suppressive effects on AR compared to glucocorticoid receptor or estrogen receptor.
To further reduce the potential artificial effects linked to exogenous transfection of ARA67/PAT1 and prove that endogenous ARA67/PAT1 can also function as a suppressor for AR activity, we applied siRNA techniques to modulate endogenous ARA67/PAT1 expression and its potential influence on AR transactivation. As shown in Fig. 4E, siRNA-H, with the 19-nucleotide targeting sequence of 5′-GAAGGCAGAACAGTTAATT-3′, at about 1:1 transfection ratio with ARA67/PAT1, suppressed ARA67/PAT1 expression over 65%. In this experiment vectors containing scrambled sequences served as negative controls. The silencing effect of siRNA-H on ARA67/PAT1 is specific as it has little influence on the expression of kinesin light chain (Fig. 4E), which shares homology with ARA67/PAT1 (48). We also used enhanced green fluorescent protein as a transfection control and β-actin as a sample loading control. The expression of these two controls was not obviously affected by siRNA-H.
MCF-7 cells were then chosen for assaying siRNA-H's effects on AR-mediated DHT induction of genes, because MCF-7 expresses functional AR (1) and relatively higher amounts of ARA67/PAT1 (Fig. 2B). We found that the inhibition of ARA67/PAT1 expression correlated with a better DHT induction. As shown in Fig. 4F, DHT induced MMTV-Luc activity about threefold (lane 2 versus 1), while in the presence of siRNA-H, a further induction of two- to threefold (lane 4 versus 2) was observed, suggesting that endogenous ARA67/PAT1 has the ability to suppress AR transactivation.
Interaction domains between ARA67/PAT1 and AR and their influence on AR transactivation.
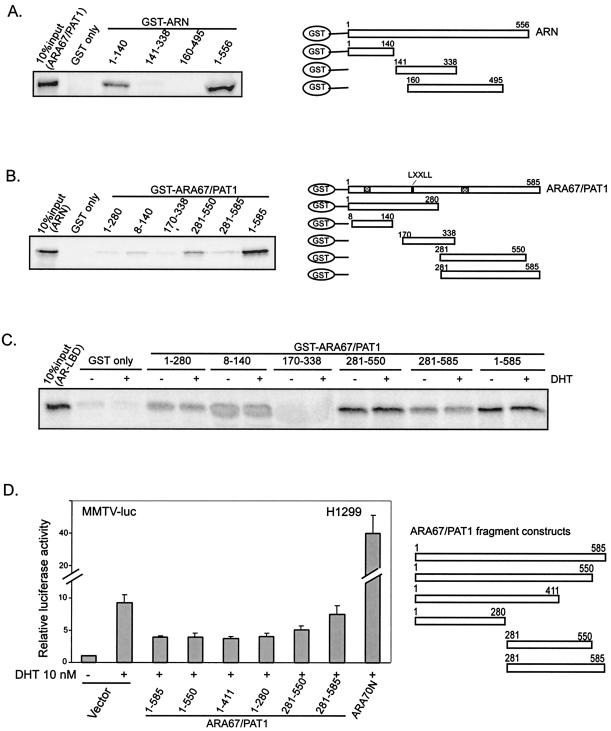
Our data in Fig. 3A and B demonstrated that ARN interacts with ARA67/PAT1 strongly. To further map the interaction domains within AR and ARA67/PAT1, several ARN-fragment constructs were generated for testing their ability to interact with ARA67/PAT1 in GST pull-down assay. As shown in Fig. 5A, after incubation with in vitro [35S]methionine-labeled ARA67/PAT1, only ARN1-140 showed positive interaction although not as strong as that seen in full-length ARN (ARN1-556). These data indicate that residues 1 to 140 within ARN are critical for the interaction with ARA67/PAT1. Since important regions for AR transactivation within ARN are in residues 141 to 338, which are required for full ligand inducible transcription, and residues 360 to 494, which contain the AF-1 region that is also required for full AR function (21), the evidence that AR residues 1 to 140 interact with ARA67/PAT1 suggests that another new region within ARN may play an important role in mediating the AR transactivation.
FIG. 5.
Interaction domains between ARA67/PAT1 and AR and their influence on AR transactivation. (A) GST only and GST-fused ARN fragments (as shown) were incubated with [35S]methionine-labeled ARA67/PAT1. [35S]methionine-labeled ARN (B) and AR ligand-binding domain (C) were incubated with GST only and GST-fused ARA67/PAT1 fragments (as shown). GST pull-down assays were performed as described. (D) H1299 cells were transfected with 100 ng of pSG5-AR in combination with 600 ng of other plasmid constructs (as shown); 300 ng MMTV-Luc was used as the reporter and 3 ng of pRL-TK as the internal control.
Motif scans suggested several protein-protein interaction motifs/domains, including the signature motif of transcriptional coactivators, LXXLL (20), exist in ARA67/PAT1 (15). Several truncated ARA67/PAT1 fragments were constructed (Fig. 5B) for mapping the regions that may play key roles in mediating the interaction with AR. GST pull-down assays were performed. Our results showed that both the N-terminal (ARA67/PAT11-280) and C-terminal (ARA67/PAT1281-585) regions of ARA67/PAT1 interacted with ARN but the interaction was relatively weak. ARA67/PAT18-140 and ARA67/PAT1281-550 showed slightly stronger interaction with ARN than their bigger counterparts ARA67/PAT11-280 and ARA67/PAT1281-585, respectively, while ARA67/PAT1281-550 was stronger than ARA67/PAT18-140. Although no fragment construct of ARA67/PAT1 strongly interacted with ARN, full-length ARA67/PAT1 showed strong interaction with ARN, suggesting participation of different parts of ARA67/PAT1 may be required for the interaction (Fig. 5B).
The interaction pattern between ARA67/PAT1 fragments and AR ligand-binding domain was similar to that between ARA67/PAT1 fragments and ARN, but ARA67/PAT1 C-terminal fragment showed an interaction strength similar to full-length ARA67/PAT1 (Fig. 5C), which indicates that the interaction between ARA67/PAT1 and AR ligand-binding domain may not need the cooperation between the N- and C-termini of ARA67/PAT1. Amino acid sequences located within 8 to 140 and 281 to 550 of ARA67/PAT1 may contribute more to its interaction with AR (Fig. 5B and 5C). ARA67/PAT1 contains an LXXLL motif, which is a signature motif in many nuclear receptor coactivators that is important for their binding to nuclear receptors (20). In our data, GST-ARA67/PAT1170-338, which contains the LXXLL motif, showed very weak interaction with ARN and no interaction with AR ligand-binding domain, suggesting that the LXXLL motif in ARA67/PAT1 is not critical for the interaction with AR.
We further asked if there is any domain or sequence that is essential for ARA67/PAT1 to suppress AR. Several truncated ARA67/PAT1 fragments were constructed and their influences on AR transactivation were tested with reporter gene assays. ARA67/PAT1 contains a PEST sequence at its C-terminal end (15), which is often seen in regulatory proteins with high turnover rate. We expected that the ARA67/PAT1 deletion construct lacking the PEST sequence (ARA67/PAT11-550) may be more stable and thus more potent as an AR repressor, but as seen in Fig. 5D, ARA67/PAT11-550 did not show a stronger suppression effect than full-length ARA67/PAT1. We also did Western blot to test the expression of ARA67/PAT1 fragment constructs and found the protein level of ARA67/PAT11-550 was similar to that of ARA67/PAT11-585 24 h after transfection (data not shown).
ARA67/PAT1 also contains nuclear localization signals at its C terminus and ARA67/PAT11-411, a truncated form lacking the nuclear localization signals, remains in the cytoplasm, and cannot enter the nucleus (15). We asked whether the nuclear localization of ARA67/PAT1 is required for its suppression on AR. As shown in Fig. 5D, ARA67/PAT11-411 had a suppression effect similar to full-length ARA67/PAT1, indicating that the nuclear localization of ARA67/PAT1 may not be critical for its effect on AR. The N-terminal (ARA67/PAT11-280) and C-terminal (ARA67/PAT1281-550, ARA67/PAT1281-585) regions of ARA67/PAT1 could also suppress AR transactivation, however ARA67/PAT11-280 was a better suppressor than ARA67/PAT1281-550 and ARA67/PAT1281-585. Together, Fig. 5B to D show that both the N- and C-terminal regions of ARA67/PAT1 are involved in the interaction with and suppression of AR, and the interaction strength is not the sole determinant of suppression potency.
Influence of ARA67/PAT1 on AR N- and C-terminal (N/C) interaction.
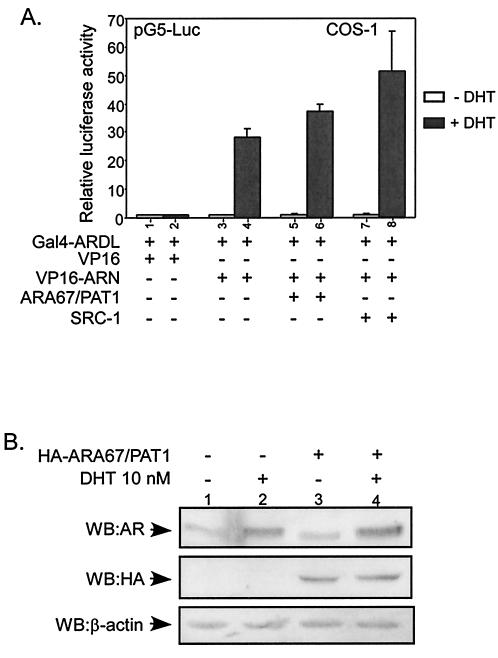
Early reports suggest that AR N/C interaction may influence the stability of AR (49) and AR N terminus is required for the full ligand-induced AR transactivation (43). Since ARA67/PAT1 can interact with both AR N terminus and AR C terminus (Fig. 3B), it is possible that ARA67/PAT1 may influence AR transactivation by blocking AR N/C interaction. With mammalian two-hybrid assay, we showed that DHT promoted AR N/C interaction (Fig. 6A). When ARA67/PAT1 was present, DHT-promoted AR N/C interaction was slightly enhanced rather than suppressed. This is in contrast to a coactivator of AR, SRC-1, which has been reported to be able to enhance AR N/C interaction (25).
FIG. 6.
ARA67/PAT1 influences AR N/C interaction and AR protein level. (A) COS-1 cells were transfected with different plasmid constructs in the combination shown in the figure and treated with 10 nM DHT or vehicle. DNAs of pSG5-ARA67/PAT1 and pSG5-SRC-1 were cotransfected with the AR N/C interaction pair (VP16-ARN and Gal4-ARDL) in a ratio of 4 to 3. The assays were carried out as described for the mammalian two-hybrid assay. (B) H1299 cells were cotransfected with pSG5-AR and HA-ARA67/PAT1 or vector in a ratio of 1 to 6. Cells were treated with 10 nM DHT or vehicle for 24 h before harvesting; 40 μg of proteins from total cell lysate from each sample was loaded onto the gel and Western blotted for AR, ARA67/PAT1 (anti-HA), and β-actin.
We further tested the influence of ARA67/PAT1 on AR protein level. Consistent with the AR N/C interaction data, the AR protein level was slightly increased rather than decreased in the presence of ARA67/PAT1 (Fig. 6B). Therefore, the suppression effect of ARA67/PAT1 on AR cannot be explained by its influence on AR N/C interaction or protein stability and other mechanisms may apply.
Histone deacetylase activity is not involved in ARA67/PAT1-mediated suppression effect on AR.
It has been suggested that coactivator and corepressor complexes, which exhibit histone transferase and histone deacetylase activities, respectively, play an important role in regulating nuclear receptor transactivation activity (46). AR is one of the nonhistone proteins that can be acetylated, and a point mutation at the acetylation site abrogates DHT-induced AR transactivation in cultured cells (11). In the same report, trichostatin A, a specific histone deacetylase inhibitor, is shown to enhance ligand-induced AR transactivation.
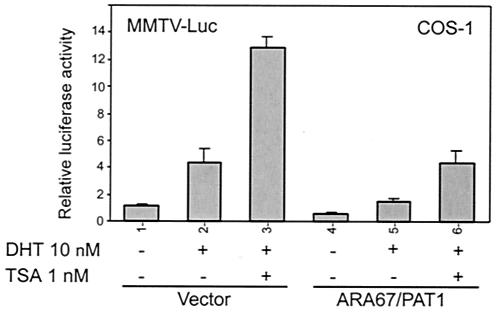
ARA67/PAT1 contains several putative protein-protein interaction domains, and our data also show that ARA67/PAT1 can interact with AR through multiple sites (Fig. 5B and C). It is possible that it behaves as an adapter between AR and regulatory multiprotein complexes that contain histone deacetylase activity. To test this hypothesis, we examined the effect of trichostatin A on ARA67/PAT1 function. We first tested several different trichostatin A concentrations to determine the best working conditions. In our system, trichostatin A at 10 nM and above caused significant cell death in COS-1 and H1299, while 1 nM trichostatin A showed no obvious toxic effect and gave the best activation of AR (data not shown). As shown in Fig. 7, 1 nM trichostatin A enhanced DHT-induced AR transactivation. In the presence of ARA67/PAT1, trichostatin A-enhanced AR transactivation was repressed to a similar extent as DHT-induced AR transactivation was repressed by ARA67/PAT1, suggesting that trichostatin A has little influence on the relative ability of ARA67/PAT1 to repress AR transactivation, which indicates that histone deacetylase activity may not be involved in ARA67/PAT1-mediated suppression on AR transactivation.
FIG. 7.
Histone deacetylase activity is not involved in ARA67/PAT1-mediated suppression effect on AR. COS-1 cells were transfected with pSG5-AR in combination with 6× pSG5-ARA67/PAT1 or pSG5 vector. Cells were then treated with DHT and trichostatin A (TSA) as indicated in the figure. Luciferase activities of reporter MMTV-Luc were assayed to monitor AR transactivation activity.
ARA67/PAT1 influences the subcellular distribution of AR.
It is known that upon ligand binding, AR translocates from the cytoplasm into the nucleus where it binds to the androgen-responsive element of its target gene and turns on the expression of its target gene. Decrease of AR nuclear translocation has been reported to lead to suppression of AR transactivation and androgen induced cell growth (16). ARA67/PAT1 is present in both cytoplasm and nucleus (15), shares homology with kinesin light chain (48) that is involved in protein trafficking, and can interact with the microtubule (48), a cytoskeleton structure. Thus, it is possible that ARA67/PAT1 may suppress AR transactivation through interrupting AR nuclear translocation.
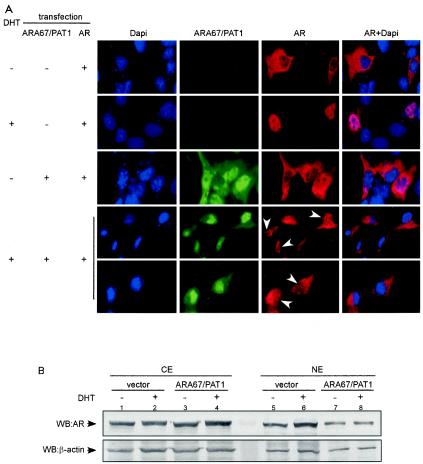
To test this possibility, we performed immunofluorescence staining analyses. COS-1 cells were cotransfected with AR and His-tagged ARA67/PAT1 expression plasmids or vector and then treated with either vehicle or DHT. The subcellular localization of AR was examined as red fluorescence signal under the microscope. In the absence of ARA67/PAT1, the unliganded AR was mainly cytoplasmic, and the liganded AR was predominantly nuclear (Fig. 8A). In contrast, the addition of ARA67/PAT1 did not obviously alter the cytoplasmic localization of unliganded AR, while the DHT induced nuclear localization of AR was interrupted as the AR signal remained mostly in the cytoplasm and the signal in nucleus was very weak (Fig. 8A). Therefore, ARA67/PAT1 may promote a cytoplasmic retention of AR in the presence of its ligand DHT.
FIG. 8.
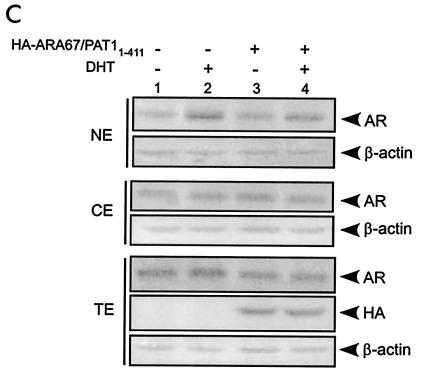
ARA67/PAT1 influences the subcellular distribution of AR. (A) COS-1 cells were transfected with pCMV-AR in combination with sixfold pcDNA4-ARA67/PAT1 or pcDNA4 vector. Immunofluorescence staining was performed as described. Arrowheads point out the cells where the AR was prevented from entering the nuclei. (B) COS-1 cells were transfected with pSG5-AR in combination with sixfold pSG5-ARA67/PAT1 or pSG5 vector, and then treated with 10 nM DHT or vehicle for 16 to 20 h. Subcellular fractionation of cells were performed as described, followed by Western blotting for AR and β-actin. (C) COS-1 cells were transfected with pSG5-AR in combination with sixfold HA-ARA67/PAT11-411 or vector. Treatment was the same as described in panel B. NE, nuclear extract; CE, cytoplasmic extract; TE, total cell extract.
To further support the immunofluorescence staining result, we performed Western blot analyses with separate cytoplasmic and nuclear fractions of proteins. As shown in Fig. 8B, after DHT treatment, the AR level in the nuclear fraction decreased with ARA67/PAT1 transfection (lane 8) compared to nontransfection (lane 6). Together, results from both immunofluorescence staining and Western blotting demonstrate that ARA67/PAT1 can influence subcellular distribution of AR in the presence of DHT, which may contribute to its suppression effect on AR transactivation.
ARA67/PAT11-411, lacking the nuclear localization signal, has been shown to be present exclusively in cytoplasm (15), and we found ARA67/PAT11-411 can suppress AR transactivation to a similar extent as the full-length ARA67/PAT1 (Fig. 5D). Examining the ability of ARA67/PAT11-411 to influence the subcellular localization of the AR may provide clues to answer whether the decreased nuclear localization of liganded AR in the presence of ARA67/PAT1 is due to a decreased nuclear import or an increased nuclear export, or if other potential mechanisms exist leading to the suppression. Subcellular fractionation with addition of ARA67/PAT11-411 was performed. Before being subjected to fractionation, a portion of the cells were separated for preparing total cell extract. We found that ARA67/PAT11-411 decreased the nuclear AR level in the presence of ligand (Fig. 8C, nuclear extract panel, lane 4 versus 2). However, a reduction of the total AR protein level was also observed in the presence of ARA67/PAT11-411 (Fig. 8C, total cell extract panel, lane 4 versus 2). Therefore, the decreased nuclear AR signal could be due to the decrease in total AR level. These findings are in contrast to what has been found for the full-length ARA67/PAT1, which has been shown to increase AR protein level, suggesting that the fragment constructs of ARA67/PAT1 may mediate the suppression of AR transactivation through mechanisms different from that for their full-length partner.
DISCUSSION
In this report, we present the identification of a novel AR-associated protein, ARA67/PAT1, which functions as a repressor of AR. Many coregulators of AR have been identified and characterized. Compared to coactivators, the identified corepressors of AR are relatively fewer and less well characterized. Calreticulin can bind to AR DNA-binding domain and suppress AR transactivation by blocking AR binding to target DNA sequences (3, 7). Cyclin D1 has been reported to suppress AR function presumably through influencing androgen-dependent transactivation function in ARN (40). Since androgen action involves dissociation of AR from heat shock protein complex, homodimerization, nuclear translocation, and binding to target genes, all these processes can be influenced by coregulators.
Nuclear localization of androgen bound AR is a prerequisite for its transactivation function. However, relatively little is known about the mechanism of its nucleocytoplasmic trafficking and its potential interacting proteins that may contribute to this process. Our data show that ARA67/PAT1 is able to alter the subcellular distribution pattern of AR in the presence of DHT, indicating that it may play a role in AR trafficking. Early studies showed that ARA67/PAT1 shares homology with kinesin light chain, a molecular motor driving the trafficking of cargos along the microtubule, directly interacts with the microtubule, and is functionally related to amyloid precursor protein trafficking/processing (48). Our data correlate with these findings, supporting the role of ARA67/PAT1 in protein trafficking. However, the detailed mechanisms need further investigation.
Studies with glucocorticoid receptor suggest that an intact cytoskeleton network is required for the shuttling of glucocorticoid receptor between the cytoplasm and nucleus in physiological conditions (13, 14). It is not clear whether this represents a common feature in nucleocytoplasmic shuttling among all steroid hormone receptors, since the ligand-dependent translocation of progesterone receptor has been suggested as independent of cytoskeleton integrity (39). ARA67/PAT1 can bind microtubules and the binding can be enhanced 5- to 10-fold in the presence of Mg-ATP (48), suggesting the possibility that the microtubule network may be an important component for ARA67/PAT1 to promote cytoplasmic retention of AR. Whether the decreased nuclear AR level in the presence of DHT is due to a decreased nuclear import or a increased nuclear export remains unclear. We observed a decreased interaction between ARA67/PAT1 and AR in the presence of DHT (Fig. 3C), and also a reverse of ARA67/PAT1's inhibitory effect by a high dose (1,000 nM) of DHT (unpublished data), suggesting a potential competition between ARA67/PAT1 and the ligand in binding to the AR. Since ligands promote the dissociation of AR from heat shock protein in the cytoplasm, and the subsequent nuclear translocation of AR, ARA67/PAT1 may interfere at these steps which may lead to the trapping of AR in the cytoplasm. Besides heat shock proteins, filamin has been reported to be involved in the nuclear translocation of AR (36). It is also known that nuclear import of steroid receptors is mediated by importins, although the nuclear export mechanism remains largely unknown (8). Whether ARA67/PAT1 can be associated with these factors and influence their function requires further study.
Although we demonstrate that ARA67/PAT1 can influence the subcellular distribution of liganded AR, which may contribute to its suppression effect on AR transactivation, other mechanisms may still exist. The possession of inherent repression activity has also been examined for ARA67/PAT1 by testing its ability to influence Gal4-DNA-binding domain-ARN transcriptional activity. Results show ARA67/PAT1 can enhance Gal4 DNA-binding domain-ARN transcriptional activity by two- to threefold rather than suppressed it (data not shown), indicating the lack of an inherent repression activity of ARA67/PAT1. Nevertheless, this one-hybrid assay result still provides extra evidence showing that ARA67/PAT1 can interact and influence AR function. However, since ARA67/PAT1 presents in both cytoplasm and nucleus (15), a direct influence on the nuclear AR transactivation is still possible. Several deletion constructs of ARA67/PAT1 can also sufficiently inhibit AR transactivation (Fig. 5D), even without effective interaction with AR compared to the full-length ARA67/PAT1 (Fig. 5B and 5C), which may further support this hypothesis. However, we would also like to point out that small fragment constructs may act independently, and may not represent the behavior of its full-length protein. ARA67/PAT11-411, a deletion construct that remains in the cytoplasm (15), can mediate a repression on AR transactivation (Fig. 5D) and decrease both nuclear and total AR protein level (Fig. 8C). This is distinct from the full-length ARA67/PAT1, which can increase total AR protein level (Fig. 6B) while suppressing the nuclear AR signal (Fig. 8A and 8B).
It is known that AR N and C termini can directly interact through the LXXLL-like motif present in the AR N terminus and the AF-2 domain in AR C terminus (18, 19). The N/C interaction may help to stabilize AR protein and modulate the transactivation activity of AR (49, 18). Coregulators that influence the AR N/C interaction could affect AR transactivation. One of the mechanisms by which coactivators enhance AR transactivation is through facilitating AR N/C interactions as seen in SRC-1 and CBP-mediated coactivation (25). Since ARA67/PAT1 can interact with both AR N and C termini, it is reasonable to hypothesize that ARA67/PAT1 may influence the AR N/C interaction. Our results show ARA67/PAT1 enhances the interaction between AR N and C termini, and accordingly we also observed a mild increase in AR protein level that may result from an increased AR stability. These seem to be contradictory to the role of ARA67/PAT1 as a repressor. However, we show that ARA67/PAT1 can decrease nuclear localization of AR upon AR ligand binding. This may prevent elevated AR protein from enhancing AR transactivation, leading to the repression of AR transactivation as a net result, since only nuclear AR can exert its influence on its target genes.
The suppression effect of ARA67/PAT1 may differ among various cells, since subcellular environments may vary. It is possible that some unknown modifications on ARA67/PAT1 may weaken its ability to block AR nuclear translocation, while the increased AR protein level may be dominant, in which case ARA67/PAT1 may function as either a weaker repressor or even as a coactivator.
In summary, we demonstrate that ARA67/PAT1 can interact with AR and suppress AR transactivation. The interrupted nuclear localization of AR in the presence of its ligand may contribute to ARA67/PAT1-mediated suppression. In addition, ARA67/PAT1 has the potential to enhance AR transactivation through enhancing the AR N/C interaction and AR stability. Since AR is one of the key players in prostate carcinogenesis, it is possible that subcellular environments in some prostate cancer cells may modify the ability of ARA67/PAT1 to repress AR and favor cancer growth. Furthermore, ARA67/PAT1 can also bind specifically to amyloid precursor protein (48), a protein that is involved in the pathogenesis of Alzheimer's disease. Testosterone has been reported to protect neurons from neurotoxic damage through the AR-mediated pathway (17). Whether AR may play a role in neuron-related diseases through the linkage of ARA67/PAT1 remains as an interesting area for further investigation.
Acknowledgments
We thank K. Wolf for help in editing the manuscript and our lab members for helpful discussion.
This work was supported by NIH grants DK60912, DK60948, and DK60905 and the George Whipple Professorship Endowment.
REFERENCES
- 1.Andò, S., De Amicis, F., V. Rago, A. Carpino, M. Maggiolini, M. L. Panno, and M. Lanzino. 2002. Breast cancer: from estrogen to androgen receptor. Mol. Cell. Endocrinol. 193:121-128. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burn, K., B. Duggan, E. A. Atkinson, K. S. Famulski, M. Nemer, R. C. Bleackley, and M. Michalak. 1994. Modulation of gene expression by calreticulin binding to the glucocorticoid receptor. Nature 367:476-480. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C., and J. Kokontis. 1988. Identification of a new member of the steroid receptor super-family by cloning and sequence analysis. Biochem. Biophys. Res. Commun. 155:971-977. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C., da Silva, S. L., R. Ideta, Y. Lee, S. Yeh, and J. P. H. Burbach. 1994. Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proc. Natl. Acad. Sci. 91:6040-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. D., and R. M. Evans. 1995. A transcriptional corepressor that interacts with nuclear hormone receptors. Nature 377:4-7. [DOI] [PubMed] [Google Scholar]
- 7.Dedhar, S., M. Shago, C. L. Hagesteijn, H. Yang, J. Filmus, R. G. Hawley, N. Bruchovsky, H. Cheng, R. J. Matusik, and V. Giguere. 1994. Inhibition of nuclear hormone receptor activity by calreticulin. Nature 367:480-483. [DOI] [PubMed] [Google Scholar]
- 8.DeFranco, D. B. 1999. Regulation of steroid receptor subcellular trafficking. Cell Biochem. Biophys. 30:1-24. [DOI] [PubMed] [Google Scholar]
- 9.Dotzlaw, H., U. Moehren, S. Mink, A. C. Cato, Iniguez Lluhi, J. A., and A. Baniahmad. 2002. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol. Endocrinol. 16:661-673. [DOI] [PubMed] [Google Scholar]
- 10.Forman, B. M., and H. H. Samuels. 1990. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol. Endocrinol. 4:1293-1301. [DOI] [PubMed] [Google Scholar]
- 11.Fu, M., C. Wang, A. T. Reutens, J. Wang, R. H. Angeletti, Siconolfi- L. Baez, V. Ogryzko, M. L. Avantaggiati, and R. G. Pestell. 2000. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 275:20853-20860. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto, N., S. Yeh, H. Kang, S. Inui, H. Chang, A. Mizokami, and C. Chang. 1999. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J. Biol. Chem. 274:8316-8321. [DOI] [PubMed] [Google Scholar]
- 13.Galigniana, M. D., J. L. Scruggs, J. Herrington, M. J. Welsh, C. Carter-Su, P. R. Housley, and W. B. Pratt. 1998. Heat shock protein 90-dependent (geldanamycin-inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Mol. Endocrinol. 12:1903-1913. [DOI] [PubMed] [Google Scholar]
- 14.Galigniana, M. D., P. R. Hoousley, D. B. DeFranco, and W. B. Pratt. 1999. Inhibition of glucocorticoid receptor nucleocytoplasmic shuttling by okadaic acid requires intact cytoskeleton. J. Biol. Chem. 275:16222-16227. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Y., and S. W. Pimplikar. 2001. The γ-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc. Natl. Acad. Sci. 98:14979-14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes, M. J., T. D. Dang, M. Larsen, and D. R. Rowley. 1998. Transforming growth factor-β1 induces nuclear to cytoplasmic distribution of androgen receptor and inhibits androgen response in prostate smooth muscle cells. Endocrinology 139:3569-3577. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, J., Q. Le, C. Goodyer, M. Gelfand, M. Trifiro, and A. LeBlanc. 2001. Testosterone-mediated neuroprotection through the androgen receptor in human primary neurons. J. Neruochem. 77:1319-1326. [DOI] [PubMed] [Google Scholar]
- 18.He, B., J. A. Kemppainen, J. J. Voegel, H. Gronemeyer, and E. M. Wilson. 1999. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 274:37219-37225. [DOI] [PubMed] [Google Scholar]
- 19.He, B., J. A. Kemppainen, and E. M. Wilson. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275:22986-22994. [DOI] [PubMed] [Google Scholar]
- 20.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 21.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 22.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, and C. K. Glass. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 23.Hsiao, P. W., and C. Chang. 1999. Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J. Biol. Chem. 274:22373-22379. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao, P. W., D. Lin, R. Nakao, and C. Chang. 1999. The linkage of Kennedy's neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. J. Biol. Chem. 274:20229-20234. [DOI] [PubMed] [Google Scholar]
- 25.Ikonen, T., J. J. Palvimo, and O. A. Janne. 1997. Interaction between the amino and carboxyl terminal regions of rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivator. J. Biol. Chem. 272:29821-29828. [DOI] [PubMed] [Google Scholar]
- 26.Knudsen, K. E., W. K. Cavenee, and K. C. Arden. 1999. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 59:2297-2301. [PubMed] [Google Scholar]
- 27.Laudet, V., C. Hänni, J. Coll, F. Catzeflis, and D. Stéhelin. 1992. Evolution of the nuclear receptor gene superfamily. EMBO J. 11:1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavinsky, R. M., K. Jeppsen, T. Heinzel, et al. 1998. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complex. Proc. Natl. Acad. Sci. 95:2920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, D. K., H. O. Duan, and C. Chang. 2000. From androgen receptor to the general transcription factor TFIIH: indication of cdk activating kinase (CAK) as an androgen receptor NH2-terminal associated coactivator. J. Biol. Chem. 275:9308-9313. [DOI] [PubMed] [Google Scholar]
- 30.Liao, G., L. Y. Chen, A. Zhang, A. Godavarthy, F. Xia, J. C. Ghosh, H. Li, and J. D. Chen. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278:5052-5061. [DOI] [PubMed] [Google Scholar]
- 31.Lu, M. L., M. C. Schneider, Y. Zheng, X. Zhang, and J. P. Richie. 2001. Caveolin-1 interacts with androgen receptor. J. Biol. Chem. 276:13442-13451. [DOI] [PubMed] [Google Scholar]
- 32.Markus, S. M., S. S. Taneja, S. K. Logan, W. Li, S. Ha, A. B. Hittelman, I. Rogatsky, and M. J. Garabedian. 2002. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol. Biol. Cell. 13:670-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear Receptor Coregulators: Cellular and Molecular Biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 34.Meyer, M. E., H. Gronemeyer, B. Turcotte, M. T. Bocquel, D. Tasset, and P. Chambon. 1989. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 57:433-442. [DOI] [PubMed] [Google Scholar]
- 35.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 36.Ozanne, D. M., M. E. Brady, S. Cook, L. Gaughan, D. E. neal, and C. N. Robson. 2000. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol. Endocrinol. 14:1618-1626. [DOI] [PubMed] [Google Scholar]
- 37.Pan, H. J., H. Uno, S. Inui, N. O. Fulmer, and C. Chang. 1999. Roles of testosterone in the growth of keratinocytes through bald frontal dermal papilla cells. Endocrine. 11:321-327. [DOI] [PubMed] [Google Scholar]
- 38.Pearce, D., and K. R. Yamamoto, 1993. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 259:1161-1164. [DOI] [PubMed] [Google Scholar]
- 39.Perrot-Applanat, M., P. Lescop, and E. Milgrom. 1992. The cytoskeleton and the cellular traffic of the progesterone receptor. J. Cell Biol. 119:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petre, C. E., Y. B. Wetherill, M. Danielsen, and K. E. Knudsen. 2002. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 277:2207-2215. [DOI] [PubMed] [Google Scholar]
- 41.Quigley, C. A., De Bellis, A., K. B. Marschke, M. K. el-Awady, E. M. Wilson, and F. S. French. 1995. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr. Rev. 16:271-321. [DOI] [PubMed] [Google Scholar]
- 42.Santen, R. J. 1992. Clinical review 37: Endocrine treatment of prostate cancer. J. Clin. Endocrinol. Metab. 75:685-689. [DOI] [PubMed] [Google Scholar]
- 43.Simental, J. A., M. Sar, M. V. Lane, F. S. French, and E. M. Wilson. 1991. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 266:510-518. [PubMed] [Google Scholar]
- 44.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, B. L., J. D. Norris, T. A. Knotts, N. L. Weigel, and D. P. McDonnell. 1998. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol. Cell. Biol. 18:1369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, L., Glass, C. K., and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 47.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, P. J. Eastman, S. V. Pol, and S. W. Pimplikar. 1998. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc. Natl. Acad. Sci. 95:14745-14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, Z. X., M. V. Lane, J. A. Kemppainen, F. S. French, and E. M. Wilson. 1995. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol. Endocrinol. 9:208-218. [DOI] [PubMed] [Google Scholar]