Abstract
We have used genetic and microarray analysis to determine how ionizing radiation (IR) induces p53-dependent transcription and apoptosis in Drosophila melanogaster. IR induces MNK/Chk2-dependent phosphorylation of p53 without changing p53 protein levels, indicating that p53 activity can be regulated without an Mdm2-like activity. In a genome-wide analysis of IR-induced transcription in wild-type and mutant embryos, all IR-induced increases in transcript levels required both p53 and the Drosophila Chk2 homolog MNK. Proapoptotic targets of p53 include hid, reaper, sickle, and the tumor necrosis factor family member Eiger. Overexpression of Eiger is sufficient to induce apoptosis, but mutations in Eiger do not block IR-induced apoptosis. Animals heterozygous for deletions that span the reaper, sickle, and hid genes exhibited reduced IR-dependent apoptosis, indicating that this gene complex is haploinsufficient for induction of apoptosis. Among the genes in this region, hid plays a central, dosage-sensitive role in IR-induced apoptosis. p53 and MNK/Chk2 also regulate DNA repair genes, including two components of the nonhomologous end-joining repair pathway, Ku70 and Ku80. Our results indicate that MNK/Chk2-dependent modification of Drosophila p53 activates a global transcriptional response to DNA damage that induces error-prone DNA repair as well as intrinsic and extrinsic apoptosis pathways.
In organisms ranging from bacteria to humans, DNA damage induces transcription of genes that promote survival and limit mutations following genotoxic stress (22). In Escherichia coli, the SOS response to DNA damage induces the coordinated expression of multiple DNA repair systems, including an error-prone polymerase that leads to increased mutation rates (63). In yeast, the DNA damage checkpoint pathway activates both a general stress response as well as a DNA damage-specific response that upregulates genes involved in homologous recombination and subunits of ribonucleotide reductase (23). In mammalian cells, the p53 transcription factor regulates DNA damage-induced transcription (4, 19, 20, 68). p53 target genes include p21, which regulates cell cycle progression, RNR2, GADD45, which may enhance DNA repair, and Mdm2, which acts in a negative feedback pathway to downregulate p53 activity. At least 10 p53 target genes encode potential proapoptotic proteins, including regulators of the intrinsic apoptotic pathway, such as APAF1 and several BH3-containing proteins, as well as tumor necrosis factor receptor family members that regulate the extrinsic apoptotic pathway. Knock out of the mouse BAX or Bid genes decreases damage-induced apoptosis in some cell types (41, 55). Why so many proapoptotic genes are regulated by p53 and the relative contributions of these genes to damage-induced apoptosis are unclear.
Although p53 is not conserved in yeast, five mammalian proteins related to yeast DNA damage-responsive kinases may directly or indirectly regulate p53 activity. The ATM, ATR, and DNA-PKcs kinases are members of the phosphatidylinositol 3-kinase superfamily and have sequence similarity to budding and fission yeast checkpoint kinases (2, 32, 53). ATM mutant mice and cells are defective in p53 activation following ionizing radiation (IR) (5, 36, 76). Analysis of dominant-negative forms of ATR has led to conflicting reports of ATR-dependent activation of p53 (45, 65). Two downstream checkpoint kinases, Chk1 and Chk2, can phosphorylate p53 in vitro. Analysis of Chk2 mutant mice and dominant-negative constructs supports a central role for this kinase in p53 activation following DNA damage (13, 29, 30, 50).
These DNA damage-responsive kinases regulate p53 activity through multiple mechanisms (67, 69). In unstressed cells, the MDM2 protein mediates p53 ubiquitination and degradation. Following damage, phosphorylation of p53 and MDM2 blocks MDM2-mediated turnover of p53, leading to a rapid accumulation of nuclear p53. Overexpression of p53 can induce target gene expression, indicating that p53 accumulation is sufficient for at least some p53 functions. Direct phosphorylation and acetylation may also contribute to p53 activation. In vivo, p53 activation is generally accompanied by both modification and accumulation. The relative contributions of these activation mechanisms are likely to reflect factors including cell type and environmental signals.
Model organisms provide an opportunity to examine the most highly conserved aspects of a signaling pathway in the context of normal development. Drosophila melanogaster and Caenorhabditis elegans have a single p53 homolog required for DNA damage-induced apoptosis (11, 18, 48, 56). The proapoptotic gene reaper is a transcriptional target of Drosophila p53 and is part of a gene complex required for damage-induced apoptosis (11, 51, 61, 71, 72). Some damage-induced apoptosis can be induced in the absence of reaper, suggesting that Drosophila p53 activates additional proapoptotic genes. Regulation of other DNA damage responses by Drosophila p53 has not been described.
The mechanism of damage-induced activation of p53 is also unclear. The Drosophila genome contains homologs of the conserved checkpoint kinases, but it does not reveal an obvious MDM2 homolog (57); this observation indicates that either the homolog of MDM2 has too little sequence similarity to be identified by simple sequence searches or that Drosophila does not utilize protein turnover to regulate p53 activity.
In this study, we have characterized the regulation and function of Drosophila p53 following DNA damage. A null mutation of p53 (52) blocks damage-induced apoptosis but is not required for viability, fertility, or damage-induced cell cycle arrest. After IR, p53 protein exhibits a phosphatase-sensitive change in gel mobility, but p53 levels do not change. MNK, the Drosophila homolog of the Chk2 kinase (47, 75), is required for IR-induced modification of p53. These results suggest that posttranslational modification is sufficient to activate Drosophila p53. To identify cellular pathways regulated by p53, we have performed a genome-wide analysis of irradiation-induced gene expression in wild-type and mutant Drosophila embryos. IR-induced genes include regulators of apoptosis, cell-cell signaling, and DNA repair, but not cell cycle progression. Both mnk/Chk2 and p53 are required for all IR-induced increases in gene expression. Two targets of p53, Ku70 and Ku80, are involved in nonhomologous end joining, suggesting that error-prone DNA break repair may be an important feature of p53 signaling. Another target of p53, the Drosophila tumor necrosis factor (TNF) homolog Eiger (31, 43), can induce apoptosis when overexpressed but is not required for IR-induced apoptosis. We also demonstrate that three known regulators of apoptosis, rpr, skl (14, 62, 73), and hid (26), are targets of p53. We find that animals heterozygous for deficiencies spanning all three genes exhibit impaired IR induction of apoptosis and that hid in particular is haploinsufficient for this DNA damage response. Combined with previous observations that hid function is regulated by Ras activity (6, 7, 37) and micro-RNA expression (10), our results suggest that hid plays a central role in integrating signals from diverse signaling pathways to determine the apoptotic response to p53 activation.
MATERIALS AND METHODS
Genetics and transgenes.
All experiments were performed at 25°C unless otherwise indicated. The following alleles were used for analysis of damage-induced apoptosis and cell cycle arrest: mei-4129D (38), mei-41RT2 (27), mus304D1, mus304D2 (12), and grpsfs1 (60). Stocks were obtained from Hermann Steller, Kristin White, Scott Hawley, and the Bloomington Drosophila Stock Center.
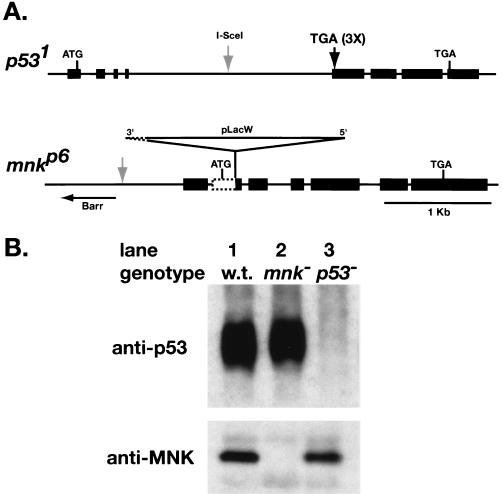
The mnkp6 allele was generated by transposase-mediated mobilization of a P[lacW] P-element insertion in the barren gene (8) followed by PCR to identify lines with insertions in the mnk coding region and not in barren. This line complemented the lethality associated with the original barren insertion. The insertion was in nucleotide position 465 of the long form of the mnk coding region, which corresponds to the second intron of the short form of mnk (47). A deletion associated with this insertion removed 218 nucleotides of mnk genomic sequence and 823 nucleotides of the 3′ end of the P[lacW] DNA. The sequence junction of this deletion was as follows: mnk genomic, GTGCTGGAGT /TCTTGAAGTG, P[lacW] DNA. A rescue construct for mnk was generated by PCR amplification. The oligonucleotide sequences used were as follows: 523 bases 5′ to the start of transcription, GGCCTCTAGAAACGACGCCGCAATTTAGGGC; 72 bases 3′ to the end of transcription, GGCCGCGGCCGCTGAGCAATTTGCCCGCCTCCG. The underlined sequences correspond to XbaI and NotI sites that were used to clone the genomic fragment into the pCaSpeR-2 transformation vector.
The p531 mutation was generated by homologous recombination (52). The p53 cDNA transgene (GUS-p53) has been described previously (11). This construct moderately overexpresses p53 in the developing eye at a level insufficient to generate a rough eye phenotype. Much higher levels of expression are generated by coexpression of GMR-Gal4, resulting in the rough eye phenotype seen below in Fig. 4. Rescue of p53-dependent apoptosis in the developing eye was accomplished using GUS-p53 in the absence of GMR-Gal4.
FIG. 4.
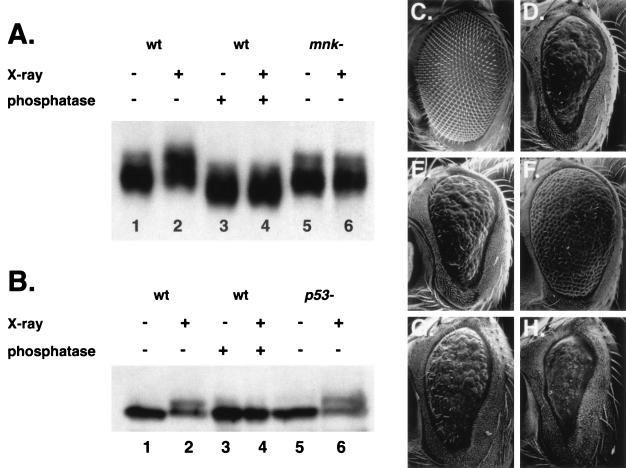
MNK/Chk2 is required for IR-induced modification of p53. (A) p53 protein was detected in lysates of untreated (lanes 1, 3, and 5) or irradiated (lanes 2, 4, and 6) 2- to 16-h-old embryos. p53 was immunoprecipitated with mouse anti-p53 monoclonal antibody, run on an SDS-PAGE gel, and detected with guinea pig anti-p53 polyclonal antibody. p53 mobility was reduced following irradiation of wild-type (lanes 1 and 2) but not mnk mutant (lanes 5 and 6) embryos. Phosphatase treatment (lanes 3 and 4) increased the mobility of p53 from both unirradiated (lane 3 versus lane 1) and irradiated (lane 4 versus lane 2) embryos. (B) MNK protein was detected in lysates of untreated (lanes 1, 3, and 5) or irradiated (lanes 2, 4, and 6) 2- to 16-h-old embryos. Lysates were run on an SDS-PAGE gel, and MNK was detected with rabbit anti-MNK polyclonal antibody (47). An anti-MNK signal with reduced mobility was detected in wild-type (lanes 1 and 2) and p53 mutant (lanes 5 and 6) embryos. Phosphatase treatment (lanes 3 and 4) eliminated the signal, with reduced mobility from irradiated embryos (lane 4 versus lane 2). (C to H) Adult eyes from transgenic female animals were dehydrated and viewed by scanning electron microscopy. All animals carried a transgene expressing the Gal4 transcription factor under the control of the eye-specific promoter GMR. Expression of Gal4 alone did not affect eye development and is shown here as the wild-type control. Animals were raised at either 25°C (C to F) or 18°C (G and H). (C) GMR-Gal4/+; wild-type eye morphology. (D) GMR-Gal4 GUS-p53/+; p53-dependent rough eye phenotype at 25°C. (E) GMR-Gal4 GUS-p53/GUS-GRPS; the p53-dependent rough eye phenotype was unaffected by GUS-GRPS. (F) GMR-Gal4 GUS-p53/GUS-MNK(kd); the p53-dependent rough eye phenotype was suppressed by the kinase-dead form of MNK. (G) GMR-Gal4 GUS-p53/+; p53-dependent rough eye phenotype at 18°C. (H) GMR-Gal4 GUS-p53/GUS-MNK; the p53-dependent rough eye phenotype was enhanced by wild-type MNK.
The Eigere1 and Eigere2 mutations were generated by transposase-mediated mobilization of the KG02299 P-element insertion immediately upstream of the Eiger transcript. CyO-Δ2-3 was used as a source of transposase. PCR was used to screen chromosomes that had lost the white marker on the KG02299 element.
Overexpression constructs for mnk and grps were generated by PCR amplification of the coding region of each gene flanked by gateway recombination sites (oligonucleotide sequences available on request). Amplification oligonucleotides introduced a translation start sequence immediately 5′ of the start codon and included the stop codon for each gene. Gateway recombination reactions transferred these sequences into pGUSgw, a gateway-modified pGUS plasmid (M. H. Brodsky, unpublished data). A predicted kinase-dead form of MNK was generated by using PCR to introduce an Asp (GAC) to Ala (GCC) mutation at amino acid position 303 in the mnk coding region. The mnk coding sequence with this mutation was introduced into pGUSgw.
Detection of p53 and MNK/Chk2.
p53 antibodies were generated using C-terminal and N-terminal fragments of p53 (11). Two mouse monoclonal lines raised against the C-terminal fragment of p53, C7A10 and B9A10, were capable of recognizing p53 overexpressed in Drosophila S2 tissue culture cells by immunofluorescence, immunoblotting, and immunoprecipitation. Guinea pig polyclonal antibodies were raised against glutathione S-transferase fusions to N-terminal and C-terminal fragments of p53. These antibodies were affinity purified using maltose binding protein fusions to the same fragment. These antibodies also detect overexpressed p53 in Drosophila tissue culture cells.
To detect damage-induced modification of p53, protein lysates were isolated from staged (3- to 15-h) embryos treated with irradiation or mock treated and incubated for 30 min at room temperature. Embryos were dechorionated and frozen in liquid nitrogen. Frozen embryos were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 300 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, and 0.5 μg each of aprotinin, pepstatin, leupeptin, chymotrypsin, and antipain/μl) and phosphatase inhibitors (10 mM sodium fluoride, 0.4 mM sodium meta- and ortho-vanadate [each], 0.1 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1 μg of phosvitin/ml, 0.5 μM microcystin-LR, and 0.5 μM okadaic acid) using a Wheaton glass homogenizer (A pestle). Homogenized lysates were centrifuged at 35,000 rpm in a Beckman Ti70 rotor at 4°C for 30 min. Total protein was determined by Bradford assay, and 50 mg was used in each immune precipitation (IP). IPs were performed for 20 h at 4°C, with 20 μg of an equal mixture of two p53 monoclonal antibodies covalently coupled to protein A/G-agarose (Oncogene Research Products). Precipitates were washed five times in RIPA buffer, resuspended in 50 μl of 2× Laemmli sample buffer, and heated to 100°C for 5 min. For phosphatase treatment, samples were resuspended to 50 μl in RIPA buffer, 2 mM MnCl2, and 400 U of Lambda phosphatase (New England Biolabs) and incubated for 30 min at room temperature. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with 9% gels, transferred to nitrocellulose, and detected with a 1:5,000 dilution of affinity-purified, p53 N-terminal-specific polyclonal guinea pig antiserum.
To detect MNK, clarified lysates from the p53 IPs were mixed with Laemmli sample buffer and heated to 100°C for 5 min. Protein samples (approximately 10 to 20 μg of total protein/sample) were separated by SDS-9% PAGE and probed with a 1:1,000 dilution of affinity-purified rabbit polyclonal anti-MNK antiserum (47).
Irradiation-induced phenotypes.
Irradiation-induced apoptosis and cell cycle arrest were performed as previously described (11, 12).
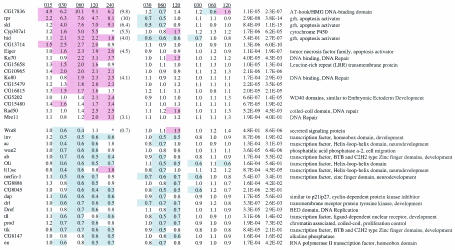
Gene expression analysis was performed using embryos collected 3 to 5 h after egg laying. Each embryo collection plate was cut in half; one half was X-irradiated for 15 min with 4,000 rads, and the other half was used as the untreated control. Embryos were incubated at room temperature and frozen in liquid nitrogen. Embryos treated at the 15-min time point were irradiated for 12 min with 3,200 rads. Time points indicate the time following the initiation of irradiation. Each experimental condition was performed in triplicate. Fifty to 100 μl of packed embryos was isolated for each sample. Total RNA was isolated by using RNAwiz (Ambion). Five micrograms of total RNA was used to generate biotin-labeled cRNA following a standard protocol provided by Affymetrix. Washes and scanning were performed according to the Affymetrix protocol. Image analysis was performed using Microarray Suite version 4. Expression values were determined using PM/MM models in dCHIP (39). Analysis of variance by least squares using the SAS proc glm program (SAS Institute, Cary, N.C.) was performed with the 30-, 60-, and 120-min time points from each genotype. P values for treatment effects and treatment by genotype effects were determined, and genes were selected with a false discovery rate of less than or equal to 5%. Only genes that increased or decreased at least 1.7-fold are included below in Table 2.
TABLE 2.
Gene expression changes following IRa
mRNA was prepared 15 to 240 min following treatment of wild-type, p53 mutant, or mnk mutant embryos. Each value is the average of three experiments determining the increase or decrease of expression in irradiated and untreated samples. The Q-PCR values indicate the increases following irradiation of wild-type embryos. Q-PCR was performed using samples collected 60 min following irradiation, except for Ku80 and Mre11, which were analyzed at 240 min following irradiation. P values are given for treatment (t) and treatment by genotype (t × g). Red shading indicates increases greater than or equal to 1.5-fold. Blue shading indicates decreases less than or equal to 0.7-fold. Gene names are from www.flybase.org. Asterisks indicate that negative expression values resulted in negative (Cyp307a1) or very high (Wnt8) expression ratios.
Real-time PCR was performed using the same total RNA samples used for microarray analysis. mRNA was isolated from 50 μg of total RNA using Oligotex purification resin (Qiagen). Predesigned Taqman oligonucleotides and probes for most genes were purchased from Applied Biosystems (ABI). Oligonucleotides and probes for ku80 and mre11 were designed using Primer Express software (sequences available on request). mRNA was isolated from 380 ng of total RNA using Oligotex beads (Qiagen). Real-time PCR was performed on an ABI 5700 apparatus using reverse transcription and PCR reagents from ABI. Relative levels in untreated and treated samples were determined using a standard curve for each probe. Glyceraldehyde-3-phosphate dehydrogenase probes were used to normalize for differences in total mRNA levels in each experiment.
mei-41 and ATM cDNAs.
A putative full-length clone for mei-41 was isolated from the LD embryonic cDNA library prepared by Ling Hong (Berkeley Drosophila Genome Project [www.fruitfly.org]). This clone is approximately 300 bp longer than the previously described cDNA sequence for mei-41 and contains an open reading frame with 100 additional amino acids. A putative full-length clone for ATM was isolated from the LP adult head cDNA library prepared by Ling Hong. This clone contains a 5-bp deletion when compared to a shorter head cDNA clone (GenBank accession number AY395749) or to the genome sequence (www.fruitfly.org). Addition of the missing 5 bp created a large open reading frame that extended across the entire cDNA and showed sequence similarity to ATM throughout the predicted amino acid sequence.
Nucleotide sequence accession numbers.
The full-length cDNA sequences obtained for mei-41 and CG6535/ATM have been deposited in GenBank with the accession numbers AY282465 and AY395748, respectively.
RESULTS
Regulation of DNA damage-induced apoptosis by p53 and mnk.
To identify genes required for IR-induced apoptosis in Drosophila, we examined several homologs of yeast and mammalian damage response genes (Table 1). BLAST searches and ClustalW alignments with these sequences confirmed earlier conclusions (57) that MEI-41 is most similar to mammalian ATR and CG6535 is most similar to ATM (S. Oikemus and M. H. Brodsky, unpublished data). Based on these observations, we refer to CG6535 as Drosophila ATM. No clear homolog of the related kinase DNA-PKcs was present in the Drosophila genome. Homologs of Chk1 (GRPS), Chk2 (MNK), and p53 have been previously described (11, 21, 47, 48). MUS304 is a DNA damage response gene with nearly identical mutant phenotypes to MEI-41 (12). ATR-IP appears to be the mammalian homolog of MUS304 and the yeast checkpoint proteins Rad 26 and Ddc2 (16). In support of this conclusion, we found that MUS304 interacted with MEI-41 in a yeast two-hybrid assay (M. H. Brodsky, G. Tsang, and G. M. Rubin, unpublished data).
TABLE 1.
DNA damage response homologs in mammals, flies, and yeast
| Mammalian proteina | Fly protein | Yeast proteins (budding, fission) |
|---|---|---|
| ATR (upstream kinase) | MEI-41 | Mec1, Rad3 |
| ATRIP (ATR binding protein) | MUS304 | Ddc2, Rad26 |
| ATM (upstream kinase) | ATM | Tel1, Tel1 |
| DNA-PKcs (upstream kinase) | ||
| Chk1 (downstream kinase) | GRPS | Chk1, Chk1 |
| Chk2 (downstream kinase) | MNK | Rad53, Cds1 |
| p53 (transcription factor) | p53 | |
| p63 (transcription factor) | ||
| p73 (transcription factor) |
A general description of the activity of each human protein is indicated in parentheses.
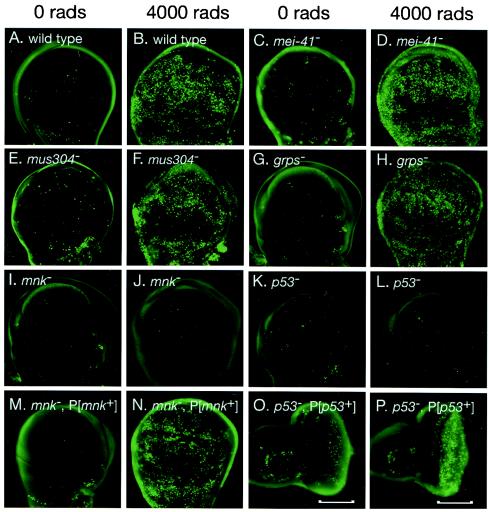
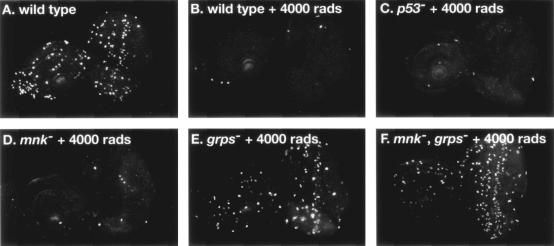
Previous genetic analysis demonstrated that mei-41, mus304, and grps are required for DNA damage and DNA replication checkpoints (12, 27, 59, 60). We examined X-ray-induced apoptosis in developing wings (wing disks) from larvae mutant for each of these genes (Fig. 1). Very low levels of apoptosis were observed in the absence of irradiation (Fig. 1A). Following irradiation, extensive induction apoptosis was induced in wild-type wing disks (Fig. 1B). Damage-induced apoptosis was still observed in mei-41, mus304, and grps disks (Fig. 1C to H). Thus, each of these genes is essential for IR-induced cell cycle arrest, but not for apoptosis.
FIG. 1.
Drosophila p53 and mnk/Chk2 are required for IR-induced apoptosis. Imaginal wing or eye disks were dissected from untreated (A, C, E, G, I, K, M, and O) or irradiated (B, D, F, H, J, L, N, and P) third instar larvae and stained with the vital dye acridine orange. IR-induced apoptosis was observed 4 h following irradiation in wild-type (B), mei-41 (D), mus304 (F), and grps (H) mutant larvae. No induction of apoptosis was observed in mnk (J) or p53 (L) mutant larvae. Damage-induced apoptosis was restored to mnk mutant disks by a transgene containing the mnk promoter and coding sequence (M and N). Damage-induced apoptosis was restored to the posterior (shown by brackets) of p53 mutant eye disks by a transgene driving expression of a p53 cDNA under control of a Glass-responsive promoter (described in Materials and Methods).
To test if other damage response homologs are required for IR-induced apoptosis, we examined loss-of-function mutations in the remaining genes shown in Table 1. Initial characterization of ATM mutant animals indicated that this gene is required for viability (S. Oikemus, unpublished results), preventing a straightforward analysis of its role in IR-induced apoptosis. Mutations in mnk and p53 (52) were created by transposon-mediated mutagenesis and homologous recombination, respectively. p53 protein was detectable in wild-type and mnk mutant embryos, but not in p53 mutant embryos (Fig. 2B, upper panel). These antibodies recognize a C-terminal portion of p53, indicating that translation of p53 does not reinitiate following the stop codon. Anti-MNK antibodies detected a protein in wild-type and p53 mutant embryos, but not in mnk mutants (Fig. 2B, lower panel). These results indicate that we have isolated null mutations in mnk and p53.
FIG. 2.
Loss-of-function mutations in Drosophila MNK/Chk2 and p53. (A) Maps of p53 and mnk loci. Exons are depicted as rectangles, and introns are lines. Homologous recombination was used to introduce stop codons in three reading frames at the beginning of the second exon of p53, eliminating the DNA binding domain and the C-terminal tetramerization and regulatory domains. The I-SceI site used for double-strand break-mediated recombination (arrow) was present in the targeting construct but not the final mutant. The mnkp6 allele contains a P-element insertion at amino acid position 52 and an associated deletion that removes 218 nucleotides of genomic sequence, including the mnk start codon. (B) p53 and MNK protein in lysates of 2- to 16-h embryos. p53 was immunoprecipitated with mouse anti-p53 monoclonal antibody, run on an SDS-PAGE gel, and detected with guinea pig anti-p53 polyclonal antibody. Signal was detected in wild-type (lane 1) and mnk mutant (lane 2) embryos, but not in p53 mutant embryos (lane 3). These antibodies recognized a C-terminal portion of p53, indicating that translation of p53 does not reinitiate following the stop codon. MNK was detected with a polyclonal rabbit antibody (47). Signal was detected in wild-type (lane 1) and p53 mutant (lane 3) embryos, but not in mnk mutant embryos (lane 2).
We examined IR-induced apoptosis in p53 and mnk mutant larvae. In the absence of DNA damage, both mutants exhibited the low level of spontaneous apoptosis seen in wild-type animals (Fig. 1I and K). However, no increase in apoptosis was observed following irradiation of either mnk (Fig. 1J) or p53 (Fig. 1L) mutant larvae. IR-induced apoptosis could be rescued in mnk mutant disks by a transgene containing mnk (see Materials and Methods) (Fig. 1M and N). Similarly, IR-induced apoptosis could be restored in the third-instar eye imaginal disk of p53 mutant animals by a transgene expressing the p53 cDNA under the control of a Glass-dependent promoter (11). In these animals, IR-induced apoptosis occurred only in the posterior region of the disk, where the Glass-responsive promoter directed expression of the p53 transgene (Fig. 1O and P).
We also examined the ability of the mnk and p53 mutants to undergo IR-induced cell cycle arrest. Following irradiation, wild-type imaginal disks exhibited a G2 arrest (Fig. 3A and B) that could be assayed with a phospho-specific antibody against histone H3, specific for mitotic cells (12, 28). IR-induced cell cycle arrest was normal in p53 mutant larvae (Fig. 3C), while mnk mutant larvae showed a very mild defect in IR-induced cell cycle arrest (Fig. 3D). In contrast, grps mutant larvae demonstrated a reduction in IR-induced arrest (Fig. 3E) (12). Larvae with mutations in both mnk and grps were completely defective in IR-induced cell cycle arrest (Fig. 3F). Our results demonstrate that while mnk and p53 function are absolutely required for IR-induced apoptosis, these genes do not play a major role in cell cycle arrest.
FIG. 3.
Cell cycle regulation in p53 and mnk/Chk2 mutant animals. Imaginal eye disks were dissected from untreated (A) or irradiated (B to F) larvae, fixed, and stained with a phospho-specific antibody, anti-phosphorylated histone H3, which specifically recognizes mitotic cells in eukaryotes. Cell cycle arrest was assayed by the absence of mitotic cells. IR-induced arrest was observed in wild-type (A and B), p53 (I), and mnk (C and D) mutant larvae. Partial arrest was observed in grps mutant larvae (E). No arrest was observed in mnk grps double mutant larvae (F).
p53-deficient mice generally develop normally and die from early-onset cancers. Mice deficient for the p53 homologs p63 and p73 exhibit developmental defects resulting in embryonic or neonatal lethality (42, 77, 78). Drosophila p53 and mnk mutant flies are viable, fertile, and show no striking defects in external morphology (data not shown).
mnk/Chk2 is required for IR-induced modification of p53.
To examine whether DNA damage induced changes in Drosophila p53 modification or abundance, we used IP and Western blotting to analyze p53 from untreated and X-irradiated embryos (Fig. 4A). p53-dependent changes in gene expression could be seen within 15 to 30 min of IR (see below). Thirty minutes following IR, p53 exhibited reduced mobility, indicating that p53 underwent a posttranslational modification (Fig. 4A, lanes 1 and 2). Total levels of p53 protein were not substantially altered following IR. Similar results were seen at 15 and 60 min (data not shown). Following phosphatase treatment, there was no difference in the mobility of p53 from untreated or irradiated embryos (Fig. 4A, lanes 3 and 4), indicating that the IR-induced shift was due to phosphorylation. In mnk mutant embryos, p53 mobility was not altered following irradiation (Fig. 4A, lanes 5 and 6). The absence of an IR-induced shift in p53 mobility indicated that mnk is specifically required for IR-induced phosphorylation of p53.
Treatment of unirradiated samples with phosphatase increased the gel mobility of p53 (Fig. 4A, lanes 1 and 3), indicating that p53 is constitutively phosphorylated in the absence of exogenous DNA damage. This constitutive phosphorylation of p53 did not require mnk (lanes 1 and 5).
We also examined the gel mobility of MNK following IR. In unirradiated embryos, MNK migrated as a single band on SDS-PAGE (Fig. 4B, lane 1). Following IR, a second band with slower mobility was observed (Fig. 4B, lane 2). Phosphatase treatment of embryo lysates did not affect the mobility of MNK in untreated embryos (Fig. 4B, lane 3), but it eliminated the appearance of a second band in IR-treated embryos (Fig. 4B, lane 4). These results were consistent with IR-induced phosphorylation of MNK. The IR-induced shift in MNK mobility was present in both wild-type and p53 mutant embryos (lanes 5 and 6), suggesting that p53 acts downstream of mnk.
Although MNK was required for p53 phosphorylation, initial attempts to detect a stable association between MNK and p53 by coimmunoprecipitation from untreated embryos or interaction in a yeast two-hybrid assay were unsuccessful (B. T. Weinert and G. Tsang, unpublished results). For an in vivo assay, we examined genetic interactions between mnk and p53 transgenes expressed during fly eye development. Overexpression of Drosophila p53 during eye development produced a reduced, rough eye phenotype (11, 48) (Fig. 4C and D). We have tested the effect of coexpressing wild-type grps, mnk, and a kinase-dead form of mnk, mnk(kd). Each of these transgenes had little or no effect on eye development on their own (data not shown). Coexpression of grps did not alter the p53-dependent rough eye phenotype (Fig. 4E). Coexpression of mnk(kd) suppressed the p53-dependent rough eye phenotype (Fig. 4F). When raised at 25°C, animals coexpressing p53 and wild-type mnk died during pupation (data not shown), probably reflecting enhanced activity of p53 in tissues other than the eye. When raised at 18°C, animals expressing p53 alone or p53 and mnk could be recovered. Under these conditions, coexpression of mnk enhanced the p53-dependent rough eye phenotype (Fig. 4G and H). Similar genetic interactions have been reported previously (50). These results are consistent with a direct interaction between MNK and p53.
Genome-wide analysis of IR-induced transcription by p53 and MNK/Chk2.
Previously, we and others provided evidence that the proapoptotic genes reaper and sickle are targets of p53 following IR (11, 61). To determine which Drosophila mRNAs are induced by IR, we used Affymetrix oligonucleotide microarrays to probe expression of greater than 13,000 genes in both untreated and irradiated embryos (Table 2). We examined 3- to 5-h embryos, a stage when high levels of apoptosis can be induced by DNA damage but little developmentally regulated apoptosis is observed (3, 46). RNA was prepared from wild-type, p53 mutant, or mnk mutant embryos at five time points following irradiation. To limit gene expression variation due to minor age differences among different embryo collections, each collection was split and one half was irradiated while the other half was the unirradiated control. As a second assay for transcript levels, the relative expression level of a subset of genes was assayed at the 1- or 4-h time points using real-time quantitative PCR (Q-PCR) (Table 2). In each example tested, mRNA changes observed by microarray analysis were confirmed by Q-PCR.
Of over 13,000 genes represented on these arrays, we identified 17 induced and 18 repressed genes with at least a 1.7-fold change in expression levels (Table 2). The upregulation of all 17 induced genes was partially or entirely dependent on both p53 and mnk. The rapid (15- to 30-min) induction of many of these targets suggested that they may be direct targets of p53. In contrast, the decreased expression of only 1 of 18 repressed genes required p53, while all 18 required mnk for downregulation. These results suggest that mnk and p53 act in a linear pathway to activate gene expression following IR, but that a p53-independent mechanism can lead to decreased gene expression.
The annotations of the induced genes indicated that Drosophila p53 regulates both intrinsic and extrinsic apoptosis pathways following irradiation (Table 2). Three of the five most highly induced genes, rpr, skl, and hid, are part of a group of genetically linked, proapoptotic genes at cytological position 75C (14, 26, 62, 72, 73). Like the mammalian proapoptotic genes SMAC/Diablo and HTRA2/Omi, each of these genes can induce apoptosis by blocking the caspase-inhibiting activity of IAP proteins (25, 40, 70). Overexpression of one of these genes is sufficient to induce apoptosis, while deletions that remove these genes reduce or eliminate developmentally programmed and stress-induced apoptosis (51, 71). Previous experiments have demonstrated that the Drosophila Apaf1 homolog Ark is induced following UV irradiation (79, 80). Following IR, Ark and other known components of the core apoptosis pathway in Drosophila did not show altered expression. In mammals, members of the TNF and TNF receptor families act in the cell-cell signaling, or extrinsic, apoptosis pathway. Drosophila has one member each in the TNF ligand and TNF receptor families (31, 35, 43). The Drosophila TNF ligand homolog Eiger is induced following DNA damage (Table 2), while the TNF receptor homolog is not (data not shown). Eiger, rpr, skl, and hid RNAs were all induced within 30 min of IR treatment.
One hour following IR, several DNA repair genes are induced. Two genes, Mre11 and Rad50, encode proteins that are part of a heterotrimeric complex that also includes NBS1 (17). This repair complex has been implicated in multiple aspects of DNA repair, including homologous recombination, nonhomologous end joining, and delay of cell cycle progression. Two other induced genes, Ku70 and Ku80, have more specific functions in DNA break repair as part of the nonhomologous end-joining pathway. The coordinate upregulation of two components of each repair complex indicated that these changes are functionally important.
Other targets of mammalian p53 include cell cycle regulators, including the cyclin-dependent kinase inhibitor p21, which mediates p53-dependent G1-S arrest following DNA damage. In contrast, dacapo, the only member of the p21/p27 family in Drosophila, is repressed following IR (Table 2). RNA levels of other Drosophila cell cycle regulators are not altered following IR (data not shown). These results are consistent with our observation that Drosophila p53 is not required for IR-induced cell cycle arrest.
Besides proapoptotic genes and DNA repair genes, the remaining IR-induced targets do not resemble genes previously associated with mammalian p53 function. Two targets (CG17836 and CG5202) have motifs associated with known transcriptional regulators. Other targets include a putative transmembrane protein (CG15658) and a member of the cytochrome P450 family.
Among the IR-repressed genes identified in our experiments, many are known or likely regulators of embryonic patterning or cell fate determination. It is possible that the decreased expression of these genes was a secondary effect of an mnk-dependent checkpoint.
Genetic analysis of Eiger and Df(3L)Cat function during IR-induced apoptosis.
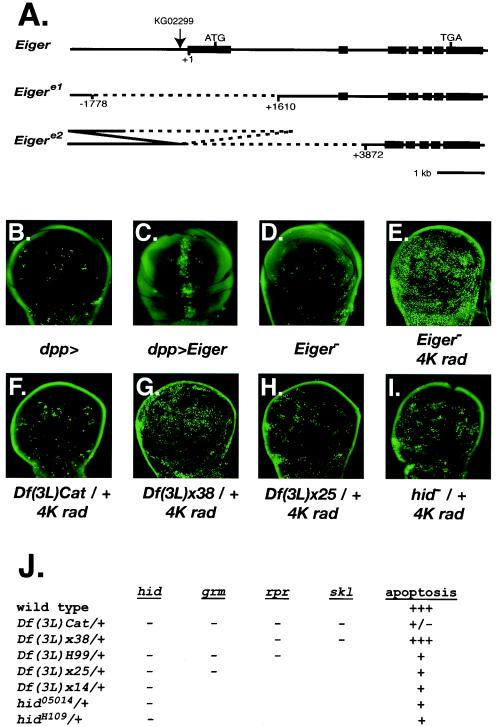
Although two TNF receptor family members, FAS and DR5/Killer, are targets of p53, neither is known to be required for damage-induced apoptosis. Previous analysis of the single Drosophila TNF ligand, Eiger, indicated that overexpression of Eiger can induce apoptosis by activating the Jnk pathway (31, 43). To examine the role of Eiger in IR-induced apoptosis, we generated gain- and loss-of-function alleles of Eiger (Fig. 5A). We confirmed that overexpression of Eiger in the developing wing was sufficient to induce apoptosis (Fig. 5B and C). Transposon-mediated mutagenesis was used to make deletions in the Eiger locus (Fig. 5A). Both deletions removed the predicted start codon. As previously reported (31), animals with these deletions were viable and fertile (data not shown). Eiger mutant animals had normal levels of IR-induced apoptosis at 4 and 8 h following irradiation with 4,000-rad X rays (Fig. 5D and E and data not shown). At lower X-ray doses, induction of apoptosis in wild-type animals was more variable. The levels observed in Eiger mutants overlapped with those in wild-type animals (data not shown). These results indicate that while Eiger overexpression is sufficient to induce apoptosis, it is not required for IR-induced apoptosis.
FIG. 5.
Genetic analysis of p53 targets and DNA damage-induced apoptosis. (A) Maps of Eiger mutations. Exons are depicted as rectangles, while introns are lines. The P-element insertion KG02299 (arrow) was used to generate two deletions in the Eiger locus. Eigere1 removes the first exon. Eigere2 removes the first two exons and leaves part of the KG02299 insertion. (B to F) Imaginal wing disks were dissected from third instar larvae and stained with the vital dye acridine orange to detect apoptotic cells. (B) dpp-Gal4 does not induce apoptosis. (C) dpp-Gal4 driving UAS-Eiger induces apoptosis at the anterior-posterior boundary. (D) Normal spontaneous apoptosis in homozygous Eigere1 disks. (E) Normal IR-induced apoptosis in homozygous Eigere1 disks 4 h following 4,000-rad X rays. (F to I) Imaginal wing disks were dissected from third instar larvae 4 h following 4,000-rad X rays and stained with the vital dye acridine orange. IR-induced apoptosis was reduced in Df(3L)Cat/+ (F), Df(3L)x25/+ (H), and hid05014/+ (I) and normal in Df(3L)x38/+ (G). (J) Summary of apoptotic response in animals heterozygous for deficiencies or point mutations in the 75C region. Genes disrupted in a given deficiency or mutant are indicated by a minus sign.
Previous studies have demonstrated that deletions that span the region containing the hid, grm, rpr, and skl genes reduce or eliminate apoptosis in Drosophila (51, 71). In embryos homozygous for Df(3L)H99, developmentally regulated and DNA damage-induced apoptosis was absent. However, the proapoptotic gene skl is not included in this deficiency but is highly induced following DNA damage. Thus, IR-induced expression of skl is not sufficient to induce apoptosis in the absence of the other proapoptotic genes in this region (14, 61). One possible explanation for this result is that following DNA damage, induction of multiple proapoptotic genes, hid, rpr, and skl, is required to produce a proapoptotic signal that exceeds a threshold level required to irreversibly commit cells to the apoptotic program. In this model, reducing the expression of all three genes by 50% might prevent the proapoptotic signal from reaching the threshold. To test this possibility, we examined IR-induced apoptosis in animals heterozygous for chromosome deficiencies that remove all of the damage-induced transcripts in the 75C region. In Df(3L)Cat/+ or Df(3L)WR10/+ animals, induction of apoptosis 4 h following IR was greatly reduced (Fig. 5F and data not shown). In contrast, animals heterozygous for other deficiencies that do not remove these genes showed normal induction of apoptosis (data not shown). Although the deficiencies showing reduced apoptosis also removed one copy of additional genes, rpr, skl, and hid are the only IR-induced genes within the affected regions. These results demonstrate a dose-sensitive requirement for genes in the 75C region for IR-induced apoptosis and are consistent with a threshold model for IR-induced apoptosis.
To further delineate which genes contribute to haploinsufficiency, we examined IR-induced apoptosis in animals heterozygous for deficiencies or point mutations removing a subset of genes in this region (Fig. 5G to K). Surprisingly, animals heterozygous for a deficiency that removes rpr and skl still exhibited a robust induction of apoptosis (Fig. 5G). In contrast, animals heterozygous for deficiencies or point mutations that remove hid all showed a significant decrease in IR-induced apoptosis.
DISCUSSION
Roles of MNK/Chk2 and p53 in damage response pathways.
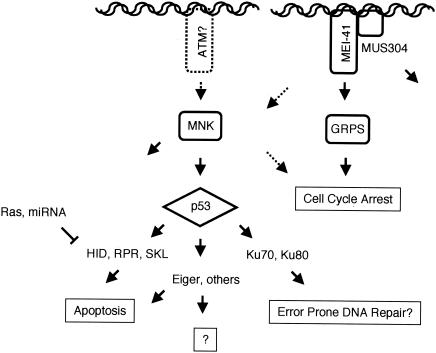
Previous studies have established that Drosophila p53 mediates X-irradiation-induced apoptosis and expression of rpr and skl (11, 48, 61). Here, we characterized the pathway that transduces the DNA damage signal to the apoptosis and cell cycle machineries. Our results indicated that a number of genes in this pathway are largely specific to the cell cycle or apoptotic response. Both cellular assays and transcriptional profiling suggest that Drosophila p53 is required for IR-induced regulation of apoptosis but is not required for G2 arrest. In contrast, mei-41, mus304, and grps were required for cell cycle arrest, but not induction of apoptosis. Our biochemical experiments suggested that mnk, which encodes a conserved damage-activated kinase, is required for phosphorylation of p53 following IR. All IR-induced transcription required both mnk and p53. The absence of genes that only required mnk or p53 was consistent with a linear signaling pathway of MNK activating p53, which acts as a global regulator of IR-induced transcription (Fig. 6).
FIG. 6.
A working model for the organization of DNA damage response pathways in Drosophila. See the text for a discussion.
Although mnk and p53 mutant animals have similar defects in IR-induced transcription, mnk also acts in p53-independent pathways. In animals with mutations in double-strand break repair enzymes, unrepaired breaks formed during meiotic recombination activate an mnk-dependent checkpoint signal that disrupts oocyte patterning and nuclear morphology (1, 24). Induction of the meiotic checkpoint differs from IR-induced transcription in at least two respects. First, activation of mnk during meiosis requires mei-41, the Drosophila homolog of ATR. Second, p53 is not required for this damage response pathway. In a different damage response pathway, mnk, but not p53, is required for damage-induced inactivation of centrosomes (64). In this study, we found that IR induced a p53-independent decrease in RNA levels of at least 17 genes, including many developmental regulators. Although this observation could indicate a transcriptional repressor that is regulated by mnk, we favor a model in which an mnk-dependent cell cycle delay following IR has a secondary effect on the developmental induction of these genes. Together, our results and previous studies indicate that mnk regulates multiple signaling pathways in addition to p53-dependent induction of gene expression.
In mammals, Chk2 and other checkpoint kinases block Mdm2-mediated turnover and inhibition of p53. Several lines of evidence suggest that this regulatory mechanism is not conserved in Drosophila. First, simple sequence searches have not revealed an obvious Mdm2 homolog in the Drosophila genome. Second, the Drosophila p53 protein sequence does not contain a conserved binding site for Mdm2. Finally, we found that p53 protein levels were not dramatically altered following IR. p53 did exhibit an IR-induced change in gel mobility due to mnk-dependent phosphorylation. Thus, our results provide a clear example of damage-induced activation of p53 without changes in p53 protein levels.
Phosphorylation of p53 by Chk2 may represent an important step in the evolution of DNA damage responses in multicellular animals. Checkpoint pathways regulating cell cycle control and DNA repair have been highly conserved in eukaryotes, including unicellular organisms such as yeast. In contrast, induction of apoptosis during development or in response to cellular stress is confined to multicellular organisms. We found that p53 phosphorylation by Chk2/MNK is a conserved molecular link between DNA damage detection and the core apoptotic machinery in metazoans. Mdm2 adds an additional layer of complexity to the regulation of mammalian p53 compared to Drosophila p53. Regulation of p53 turnover by Mdm2 may provide mammalian cells with greater control of the levels or timing of p53-dependent transcription.
DNA damage-induced targets of p53.
We have used microarray analysis to perform a comprehensive analysis of p53 targets following exposure to IR. The number of genes identified in these experiments was substantially smaller than the number of p53 targets identified in mammals. In part, this observation may reflect underlying differences in the damage response pathway in flies and mammals. For example, induction of p21 by mammalian p53 mediates G1 arrest following damage. To the best of our knowledge, IR-induced G1 arrest has not been described in Drosophila, consistent with our observation that the Drosophila p21/p27 homolog dacapo is not induced by IR. However, the smaller number of targets identified in Drosophila also reflects experimental differences. We have examined expression changes induced by IR and during a defined window of embryonic development. In contrast, targets of mammalian p53 have been identified in many different cell types following different types of DNA damage or simply overexpression of p53. It is likely that additional targets of Drosophila p53 will be identified using other types of cellular stresses in different cell types or developmental stages. For example, UV irradiation of Drosophila embryos has been shown to induce Apaf1 through either E2F or mei-41, depending on the developmental stage (80).
The most prominent group of p53 targets identified in our study regulates two apoptotic pathways that are also targeted by mammalian p53. hid, rpr, and skl are part of a group of genes that induce apoptosis by blocking the caspase-inhibiting activity of IAP proteins (25, 40, 70). Recent experiments have confirmed that HTRA2, a functional homolog of these genes, is a target of mammalian p53 (34). The Drosophila p53 target Eiger is a member of the TNF ligand family and can induce apoptosis when overexpressed (31, 43). In mammals, FAS and DR5/Killer are p53 targets that can regulate apoptosis by acting as receptors for TNF ligand family members (19, 68). Thus, we have identified two examples of mammalian and Drosophila p53 regulating common signaling pathways. Combined with the many other proapoptotic targets of mammalian p53, these results support the general hypothesis that multiple components of proapoptotic signaling pathways can be targets for transcriptional regulation following stresses such as DNA damage.
Although FAS and DR5/Killer are targets of mammalian p53 and act in the extrinsic apoptosis pathway, it is unclear what role they play in DNA damage-induced apoptosis (49, 58, 74). Our analysis of deletion mutations in the Drosophila p53 target Eiger indicated that this gene is not required to initiate IR-induced apoptosis. This negative result was not due to redundancy with a related molecule, since Eiger is the only TNF-related gene in the Drosophila genome sequence. It is possible that the conserved activation of the TNF pathway by p53 is required for the induction of apoptosis under specific conditions not tested in our experiments. Alternatively, induction of Eiger may activate other cellular responses to DNA damage. Further characterization of Eiger function should reveal how cell-cell signaling contributes to survival or genomic stability following DNA damage in multicellular organisms.
Our analysis of the remaining proapoptotic targets of p53 indicated that they are part of a dosage-sensitive mechanism that regulates IR-induced apoptosis. In contrast to Eiger, the proapoptotic genes in the genetic region containing hid, rpr, and skl are both sufficient and necessary for apoptosis. Animals with deletions that include genes in this region are defective in IR-induced apoptosis (51, 71). Because these proapoptotic genes act, at least in part, by inhibiting a common target (IAP1/Thread), it has been proposed that they contribute to a rheostat-like mechanism in which the added activity of all proapoptotic proteins present must pass a threshold before a cell undergoes the irreversible decision to undergo programmed cell death. Following our observation that three of these genes are induced following DNA damage, we tested the effect of lowering the dose of all proapoptotic genes in this region by half. We found that deletions in this region were haploinsufficient for IR-induced apoptosis. Dose sensitivity may represent an important feature of damage-induced apoptosis. Animals heterozygous for these deletions exhibit apparently normal morphology and fertility, suggesting that they are not haploinsufficient for developmentally regulated apoptosis. One possible interpretation of these results is that the apoptotic signal in many developmental contexts is well past the threshold required to commit to apoptosis, while the apoptotic signal following DNA damage is closer to that threshold. A lower apoptotic signal following DNA damage may allow cells to monitor DNA repair and block apoptosis if repair is successful. Haploinsufficiency of some tumor suppressor genes, including p53, has been proposed to contribute to cancer development (15, 66). If stress-induced apoptosis in mammals is sensitive to the dose of p53 target genes, haploinsufficiency of these genes may also contribute to suppression of apoptosis, particularly in cells with extensive aneuploidy.
Analysis of animals heterozygous for deletions that removed a subset of genes revealed that loss of one copy of hid was sufficient to reduce IR-induced apoptosis. A greater reduction was observed in larger deletions, indicating that additional genes in this region, likely rpr and skl, also contribute to IR-induced apoptosis. Previous analysis of animals heterozygous for two overlapping deletions [Df(3L)H99 and Df(3L)xr38] that remove both copies of rpr demonstrated reduced levels of IR-induced apoptosis (51). Our results indicate that part of that reduction is due to haploinsufficiency of hid and other genes in this region. Although the induction of hid RNA was lower than that observed for rpr and skl, hid may exhibit a greater absolute difference in RNA and protein levels following IR. Because null mutations in hid are embryonic lethal, we have not examined the effects of completely removing hid function. The dose-sensitive effects of hid suggest that total loss of hid would completely block IR-induced apoptosis. However, even in animals with normal levels of hid, increased levels of rpr and skl may be required to pass the proapoptotic signaling threshold required for a full DNA damage response. The Ras pathway (6, 7, 37, 54) and a micro-RNA in the bantam locus (10) regulate hid expression. These and other pathways regulating hid may help determine which cells in the developing wing are most sensitive to DNA damage.
The other class of p53 targets identified in our experiments includes components of the Ku and Mre11 DNA repair complexes. Both of these complexes participate in repair of double-strand DNA breaks by nonhomologous end joining (NHEJ) (33). Compared with homologous recombination, NHEJ is a potentially error-prone mechanism for DNA repair. Mutagenic DNA repair mechanisms are a prominent feature of the SOS response in bacteria that apparently promotes cell survival following DNA damage at the expense of genomic integrity (63). The ability of multicellular animals to eliminate damaged cells by apoptosis might suggest that low-fidelity mechanisms of DNA repair would not be favored following damage. However, the induction of NHEJ components by p53 suggests that mechanisms such as apoptosis or cell cycle arrest that are presumed to prevent mutations following DNA damage may compete with mechanisms that promote cell survival and prevent aneuploidy by error-prone DNA repair. The previous demonstration that an isoform of Ku86 is also a target of mammalian p53 (9, 44) suggests that this is an evolutionarily conserved response to DNA damage in metazoans that may modulate mutagenesis following DNA damage.
Addendum in proof
Jassim et al. have shown that hid is also induced by UV irradiation but that p53 protects cells from UV-induced apoptosis, apparently through enhanced DNA repair (O. W. Jassim, J. L. Fink, and R. L. Cagan, EMBO J. 22:5622-5632, 2003). Although the targets of p53 that mediate enhanced repair of UV damage are unknown, these results are consistent with our conclusion that Drosophila p53 regulates DNA repair.
Acknowledgments
M.H.B. was supported by fellowships from the American Cancer Society and the Alameda Cancer League and by a Worcester Foundation for Biomedical Research Annual Research Fund Innovation Grant.
We thank Isao Oishi for the generous gift of anti-MNK antibody, Ling Hong for providing cDNA libraries, and Anne Laurencon and Scott Hawley for providing the mei-4129D allele prior to publication. We thank Kristin White, Hermann Steller, and the Bloomington Stock Center for providing Drosophila stocks. We thank Elaine Kwan for isolation of anti-p53 monoclonal antibodies and Stephen Davis and Paul Spellman for assistance with microarray data analysis.
REFERENCES
- 1.Abdu, U., M. Brodsky, and T. Schupbach. 2002. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12:1645. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 3.Abrams, J. M., K. White, L. I. Fessler, and H. Steller. 1993. Programmed cell death during Drosophila embryogenesis. Development 117:29-43. [DOI] [PubMed] [Google Scholar]
- 4.Amundson, S. A., M. Bittner, P. Meltzer, J. Trent, and A. J. Fornace, Jr. 2001. Physiological function as regulation of large transcriptional programs: the cellular response to genotoxic stress. Comp. Biochem. Physiol. B 129:703-710. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, C., K. D. Brown, C. X. Deng, D. A. Tagle, and A. Wynshaw-Boris. 1997. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 17:453-456. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann, A., J. Agapite, K. McCall, and H. Steller. 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95:331-341. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann, A., M. Tugentman, B. Z. Shilo, and H. Steller. 2002. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev. Cell 2:159-170. [DOI] [PubMed] [Google Scholar]
- 8.Bhat, M. A., A. V. Philp, D. M. Glover, and H. J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87:1103-1114. [DOI] [PubMed] [Google Scholar]
- 9.Braastad, C. D., M. Leguia, and E. A. Hendrickson. 2002. Ku86 autoantigen related protein-1 transcription initiates from a CpG island and is induced by p53 through a nearby p53 response element. Nucleic Acids Res. 30:1713-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell, and S. M. Cohen. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25-36. [DOI] [PubMed] [Google Scholar]
- 11.Brodsky, M. H., W. Nordstrom, G. Tsang, E. Kwan, G. M. Rubin, and J. M. Abrams. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103-113. [DOI] [PubMed] [Google Scholar]
- 12.Brodsky, M. H., J. J. Sekelsky, G. Tsang, R. S. Hawley, and G. M. Rubin. 2000. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 14:666-678. [PMC free article] [PubMed] [Google Scholar]
- 13.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 14.Christich, A., S. Kauppila, P. Chen, N. Sogame, S. I. Ho, and J. M. Abrams. 2002. The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim, and hid. Curr. Biol. 12:137-140. [DOI] [PubMed] [Google Scholar]
- 15.Cook, W. D., and B. J. McCaw. 2000. Accommodating haploinsufficient tumor suppressor genes in Knudson's model. Oncogene 19:3434-3438. [DOI] [PubMed] [Google Scholar]
- 16.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 17.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 18.Derry, W. B., A. P. Putzke, and J. H. Rothman. 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294:591-595. [DOI] [PubMed] [Google Scholar]
- 19.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 20.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 21.Fogarty, P., S. D. Campbell, R. Abu-Shumays, B. S. Phalle, K. R. Yu, G. L. Uy, M. L. Goldberg, and W. Sullivan. 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7:418-426. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 23.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghabrial, A., and T. Schupbach. 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1:354-357. [DOI] [PubMed] [Google Scholar]
- 25.Goyal, L., K. McCall, J. Agapite, E. Hartwieg, and H. Steller. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grether, M. E., J. M. Abrams, J. Agapite, K. White, and H. Steller. 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9:1694-1708. [DOI] [PubMed] [Google Scholar]
- 27.Hari, K. L., A. Santerre, J. J. Sekelsky, K. S. McKim, J. B. Boyd, and R. S. Hawley. 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82:815-821. [DOI] [PubMed] [Google Scholar]
- 28.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 29.Hirao, A., A. Cheung, G. Duncan, P. M. Girard, A. J. Elia, A. Wakeham, H. Okada, T. Sarkissian, J. A. Wong, T. Sakai, E. De Stanchina, R. G. Bristow, T. Suda, S. W. Lowe, P. A. Jeggo, S. J. Elledge, and T. W. Mak. 2002. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22:6521-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 31.Igaki, T., H. Kanda, Y. Yamamoto-Goto, H. Kanuka, E. Kuranaga, T. Aigaki, and M. Miura. 2002. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 21:3009-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson, S. P. 2001. Detecting, signalling and repairing DNA double-strand breaks. Biochem. Soc. Trans. 29:655-661. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 34.Jin, S., M. Kalkum, M. Overholtzer, A. Stoffel, B. T. Chait, and A. J. Levine. 2003. CIAP1 and the serine protease HTRA2 are involved in a novel p53-dependent apoptosis pathway in mammals. Genes Dev. 17:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanda, H., T. Igaki, H. Kanuka, T. Yagi, and M. Miura. 2002. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J. Biol. Chem. 277:28372-28375. [DOI] [PubMed] [Google Scholar]
- 36.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 37.Kurada, P., and K. White. 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95:319-329. [DOI] [PubMed] [Google Scholar]
- 38.Laurencon, A., A. Purdy, J. Sekelsky, R. S. Hawley, and T. T. Su. 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164:589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, S. J. 2002. Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell 109:793-796. [DOI] [PubMed] [Google Scholar]
- 41.McCurrach, M. E., T. M. Connor, C. M. Knudson, S. J. Korsmeyer, and S. W. Lowe. 1997. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc. Natl. Acad. Sci. USA 94:2345-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 43.Moreno, E., M. Yan, and K. Basler. 2002. Evolution of TNF signaling mechanisms. JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 12:1263. [DOI] [PubMed] [Google Scholar]
- 44.Myung, K., C. Braastad, D. M. He, and E. A. Hendrickson. 1998. KARP-1 is induced by DNA damage in a p53- and ataxia telangiectasia mutated-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7664-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nghiem, P., P. K. Park, Y. S. Kim, B. N. Desai, and S. L. Schreiber. 2002. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277:4428-4434. [DOI] [PubMed] [Google Scholar]
- 46.Nordstrom, W., P. Chen, H. Steller, and J. M. Abrams. 1996. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180:213-226. [DOI] [PubMed] [Google Scholar]
- 47.Oishi, I., S. Sugiyama, H. Otani, H. Yamamura, Y. Nishida, and Y. Minami. 1998. A novel Drosophila nuclear protein serine/threonine kinase expressed in the germline during its establishment. Mech. Dev. 71:49-63. [DOI] [PubMed] [Google Scholar]
- 48.Ollmann, M., L. M. Young, C. J. Di Como, F. Karim, M. Belvin, S. Robertson, K. Whittaker, M. Demsky, W. W. Fisher, A. Buchman, G. Duyk, L. Friedman, C. Prives, and C. Kopczynski. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91-101. [DOI] [PubMed] [Google Scholar]
- 49.Owen-Schaub, L. B., W. Zhang, J. C. Cusack, L. S. Angelo, S. M. Santee, T. Fujiwara, J. A. Roth, A. B. Deisseroth, W. W. Zhang, E. Kruzel, et al. 1995. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol. Cell. Biol. 15:3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters, M., C. DeLuca, A. Hirao, V. Stambolic, J. Potter, L. Zhou, J. Liepa, B. Snow, S. Arya, J. Wong, D. Bouchard, R. Binari, A. S. Manoukian, and T. W. Mak. 2002. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc. Natl. Acad. Sci. USA 99:11305-11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson, C., G. E. Carney, B. J. Taylor, and K. White. 2002. reaper is required for neuroblast apoptosis during Drosophila development. Development 129:1467-1476. [DOI] [PubMed] [Google Scholar]
- 52.Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani, P. Bandyopadhyay, B. M. Olivera, M. Brodsky, G. M. Rubin, and K. G. Golic. 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16:1568-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 54.Sawamoto, K., A. Taguchi, Y. Hirota, C. Yamada, M. H. Jin, and H. Okano. 1998. Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ. 5:262-270. [DOI] [PubMed] [Google Scholar]
- 55.Sax, J. K., P. Fei, M. E. Murphy, E. Bernhard, S. J. Korsmeyer, and W. S. El-Deiry. 2002. BID regulation by p53 contributes to chemosensitivity. Nat. Cell Biol. 4:842-849. [DOI] [PubMed] [Google Scholar]
- 56.Schumacher, B., K. Hofmann, S. Boulton, and A. Gartner. 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11:1722-1727. [DOI] [PubMed] [Google Scholar]
- 57.Sekelsky, J. J., M. H. Brodsky, and K. C. Burtis. 2000. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 150:F31-F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheikh, M. S., and A. J. Fornace, Jr. 2000. Death and decoy receptors and p53-mediated apoptosis. Leukemia 14:1509-1513. [DOI] [PubMed] [Google Scholar]
- 59.Sibon, O. C., A. Laurencon, R. Hawley, and W. E. Theurkauf. 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9:302-312. [DOI] [PubMed] [Google Scholar]
- 60.Sibon, O. C., V. A. Stevenson, and W. E. Theurkauf. 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388:93-97. [DOI] [PubMed] [Google Scholar]
- 61.Sogame, N., M. Kim, and J. M. Abrams. 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA 100:4696-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srinivasula, S. M., P. Datta, M. Kobayashi, J. W. Wu, M. Fujioka, R. Hegde, Z. Zhang, R. Mukattash, T. Fernandes-Alnemri, Y. Shi, J. B. Jaynes, and E. S. Alnemri. 2002. sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr. Biol. 12:125-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 64.Takada, S., A. Kelkar, and W. E. Theurkauf. 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113:87-99. [DOI] [PubMed] [Google Scholar]
- 65.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venkatachalam, S., S. D. Tyner, C. R. Pickering, S. Boley, L. Recio, J. E. French, and L. A. Donehower. 2001. Is p53 haploinsufficient for tumor suppression? Implications for the p53+/− mouse model in carcinogenicity testing. Toxicol. Pathol. 29(Suppl.):147-154. [DOI] [PubMed] [Google Scholar]
- 67.Vousden, K. H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602:47-59. [DOI] [PubMed] [Google Scholar]
- 68.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 69.Wahl, G. M., and A. M. Carr. 2001. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat. Cell Biol. 3:E277-E286. [DOI] [PubMed] [Google Scholar]
- 70.Wang, S. L., C. J. Hawkins, S. J. Yoo, H. A. Muller, and B. A. Hay. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98:453-463. [DOI] [PubMed] [Google Scholar]
- 71.White, K., M. E. Grether, J. M. Abrams, L. Young, K. Farrell, and H. Steller. 1994. Genetic control of programmed cell death in Drosophila. Science 264:677-683. [DOI] [PubMed] [Google Scholar]
- 72.White, K., E. Tahaoglu, and H. Steller. 1996. Cell killing by the Drosophila gene reaper. Science 271:805-807. [DOI] [PubMed] [Google Scholar]
- 73.Wing, J. P., J. S. Karres, J. L. Ogdahl, L. Zhou, L. M. Schwartz, and J. R. Nambu. 2002. Drosophila sickle is a novel grim-reaper cell death activator. Curr. Biol. 12:131-135. [DOI] [PubMed] [Google Scholar]
- 74.Wu, G. S., K. Kim, and W. S. el-Deiry. 2000. KILLER/DR5, a novel DNA-damage inducible death receptor gene, links the p53-tumor suppressor to caspase activation and apoptotic death. Adv. Exp. Med. Biol. 465:143-151. [DOI] [PubMed] [Google Scholar]
- 75.Xu, J., S. Xin, and W. Du. 2001. Drosophila Chk2 is required for DNA damage-mediated cell cycle arrest and apoptosis. FEBS Lett. 508:394-398. [DOI] [PubMed] [Google Scholar]
- 76.Xu, Y., E. M. Yang, J. Brugarolas, T. Jacks, and D. Baltimore. 1998. Involvement of p53 and p21 in cellular defects and tumorigenesis in Atm−/− mice. Mol. Cell. Biol. 18:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 78.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 79.Zhou, L., Z. Song, J. Tittel, and H. Steller. 1999. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell 4:745-755. [DOI] [PubMed] [Google Scholar]
- 80.Zhou, L., and H. Steller. 2003. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev. Cell 4:599-605. [DOI] [PubMed] [Google Scholar]