Abstract
The semidominant mutation Liguleless3-O (Lg3-O) causes a blade-to-sheath transformation at the midrib region of the maize (Zea mays L.) leaf. We isolated a full-length lg3 cDNA containing a knotted1-like family homeobox. Six Lg3-O partial revertant alleles caused by insertion of a Mutator (Mu) transposon and two deletion derivatives were isolated and used to verify that our knotted1-like cDNA corresponds to the LG3 message. In wild-type plants the LG3 mRNA is expressed in apical regions but is not expressed in leaves. In mutant plants harboring any of three dominant lg3 alleles (Lg3-O, -Mlg, and -347), LG3 mRNA is expressed in leaf sheath tissue, indicating that the Lg3 phenotype is due to ectopic expression of the gene. The Lg3-O revertant alleles represent two classes of Lg3 phenotypes that correlate well with the level of ectopic Lg3 expression. High levels of ectopic LG3 mRNA expression results in a severe Lg3 phenotype, whereas weak ectopic Lg3 expression results in a mild Lg3 phenotype. We propose that ectopic Lg3 expression early in leaf development causes the blade-to-sheath transformation, but the level of expression determines the extent of the transformation.
Maize (Zea mays L.) leaf development is thought to be divided into three distinct stages (Sylvester et al., 1990, 1996). In the first stage the vegetative meristem recruites an overlapping ring of founder cells that will form the next phytomer: a repeating segment of the maize plant composed of the leaf, internode, node, and bud (Galinat, 1959). In the second stage, a subset of the founder cells, the leaf founder cells divide equally into an undifferentiated primordium. During the third stage, growth and differentiation of the primordium occur to form the mature leaf. Maize leaves are divided into three parts: the sheath, the ligular region, and the blade, as shown in Figure 1. The proximal sheath wraps around the culm and provides support for the plant. The distal blade grows out from the main axis of the plant and is its major photosynthetic organ. The ligular region, composed of the ligule and two wedge-shaped auricles, separates the sheath from the blade. Many mutations affect maize leaf development and in particular disrupt or displace the blade-sheath boundary and the associated ligule and auricle (Freeling and Hake, 1985; Becraft et al., 1990; Becraft and Freeling, 1994; Fowler and Freeling, 1996; Harper and Freeling, 1996; Schichnes and Freeling, 1998).
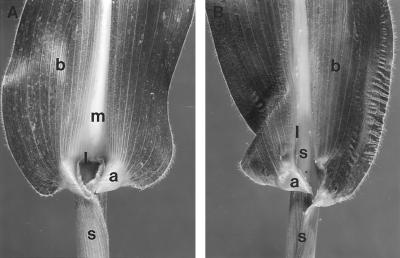
Figure 1.
Wild-type and Lg3 leaves. A, Wild-type maize leaf. B, Lg3-O maize leaf. a, Auricle; b, blade; l, ligule; m, midrib; s, sheath.
The semidominant Liguleless3-O (Lg3-O) mutation results in a blade-to-sheath transformation at the midrib region of the maize leaf (Fig. 1B; Fowler and Freeling, 1996). Blade, auricle, and ligule regions are replaced by sheath at the midrib region, and at the midrib the ligule is removed. The blade-to-sheath transformation in the Lg3-O mutant causes the ligule to develop at the new blade-sheath boundary, distal to the location of wild-type ligules. The displaced ligule gradually approaches the wild-type position at the leaf margin (Fig. 1B; Muehlbauer et al., 1997). Homozygous Lg3-O plants exhibit a more severe Lg3 phenotype than heterozygous Lg3-O plants, hence, the semidominant designation (Fowler and Freeling, 1996). Similar blade-to-sheath transformation phenotypes have been observed in the other dominant maize leaf mutants: Knotted1-O (Kn1-O; Freeling and Hake, 1985), Rough Sheath1-O (Rs1-O; Becraft and Freeling, 1994), and Liguleless4-O (Lg4-O; Fowler and Freeling, 1996). These mutations have been described as “ligule-polarity” mutations because the transformation occurs only in the blade-to-sheath (or distal-to-proximal) direction (Freeling, 1992). However, in each mutant the transformed region is positioned in different lateral regions of the leaf. For example, the Kn1-O mutation causes a transformation over the lateral veins, whereas the Lg3-O mutation causes a transformation at the midrib region (Freeling and Hake, 1985; Fowler and Freeling, 1996).
The genes for kn1 and rs1 have been isolated and shown to encode homeodomain proteins (Vollbrecht et al., 1991; Schneeberger et al., 1995). The homeodomain is a sequence-specific DNA-binding motif composed of a helix-turn-helix structure, suggesting that homeodomain proteins act as transcriptional regulators. The kn1 and rs1 genes are expressed in apical tissues, suggesting that they are involved in regulating the meristem, segmentation, and early organogenic events (Smith et al., 1992; Jackson et al., 1994; Schneeberger et al., 1995). It is interesting that neither kn1 nor rs1 expression is detectable in the wild-type leaf. However, the dominant mutant transformation phenotypes exhibited by Kn1-O and Rs1-O are attributed to the ectopic expression of their respective proteins in the transformed regions. Furthermore, two dominant mutations in tomato that affect leaf morphology, Curl (Cu) and Mouse-ear (Me), are likely to be due to ectopic expression of a KN1-like homeodomain protein (Chen et al., 1997; Parnis et al., 1997). In barley, the dominant Hooded mutation, which causes floral meristems to develop on a leaf-like awn structure, has been correlated with ectopic expression of a kn1-like gene (Müller et al., 1995). Thus, the morphogenetic programs operating during leaf development appear to be sensitive to the activity of homeodomain proteins encoded by the kn1-like family.
Homeobox genes have been studied from a variety of plant species. Loss-of-function mutations at the kn1 locus exhibit impenetrant, relatively subtle phenotypes including fewer branches and spikelets on the tassel, ears absent or small with few spikelets, extra carpels in female florets, abnormally proliferated ovule tissue, and extra leaves in the axils of vegetative leaves (Kerstetter et al., 1997). These phenotypes may indicate that the KN1 gene product is involved in keeping meristematic cells from differentiating prematurely. In maize, a family of knox (kn1 homeobox)-like genes encoding homeodomain proteins have been isolated (Kerstetter et al., 1994). These genes exhibit a high degree of sequence similarity in the homeobox region and, in general, are expressed in apical tissues. The Arabidopsis SHOOTMERISTEMLESS (STM) gene encodes a kn1-like homeodomain protein (Long et al., 1996). Loss-of-function mutations at the STM locus result in plants that can form cotyledons but are unable to form subsequent leaves, indicating that the role of the STM protein is in meristem maintenance (Barton and Poethig, 1993; Clark et al., 1996; Endrizzi et al., 1996). Several other kn1-like homeobox genes have been isolated from rice, soybean, and Arabidopsis (Matsuoka et al., 1993; Lincoln et al., 1994; Ma et al., 1994; Serikawa et al., 1997), and the available evidence suggests that the kn1-like genes act to regulate basic morphogenetic programs in the apex.
In this paper we describe the isolation and characterization of the maize lg3 gene. The lg3 gene encodes a homeodomain protein and is a member of the knox gene family. In wild-type plants lg3 is expressed in apical tissues. In Lg3 mutant plants ectopic Lg3 expression appears to cause the Lg3 phenotype. In addition, the level of ectopic Lg3 expression correlates well with the severity of the Lg3 phenotype.
MATERIALS AND METHODS
Genetic Stocks
Stocks containing the Lg3-O allele were provided by the Maize Genetics Stock Center (Urbana, IL). The Lg3-347 allele was a gift from Steve Briggs (Pioneer Hi-Bred International, Johnston, IA). The Lg3-Multiple ligule (-Mlg) allele was identified in an ethyl methanesulfonate mutant screen at the sequencing facility in Berkeley, CA. The genetic lesions in the Lg3-O, Lg3-347, and Lg3-Mlg alleles are not known. Partly isogenic lines were derived by introgressing Lg3-O, Lg3-347, and Lg3-Mulg into B73 and Mo17. Six partially revertant Lg3 alleles, Lg3-Or81, Lg3-Or211, Lg3-Or331, Lg3-Or422, Lg3-Or671, and Lg3-Or1021 were isolated in a genetic screen described below and introgressed three to four times into a laboratory-inbred line carrying bz-m4 sh1. Two lg3 deletion mutations, Lg3-Or553 and Lg3-Or1091, were also isolated. The recessive yellow stripe3 (ys3) mutation causes yellowing of tissue between the lateral veins, which is symptomatic of an iron deficiency; ys3 plants were treated with an iron supplement (Greenol, Ortho no. 3428, Monsanto, San Ramon, CA) to overcome the difficulties due to reduced vigor. The ys3 mutation, linked in cis 5 centimorgans to Lg3-O, was used to distinguish homozygous Lg3-O plants from heterozygous Lg3-O plants by the presence of the ys3 phenotype in homozygotes. The bz1-mum9 allele was used to monitor Mutator (Mu) activity; the bz1-mum9 allele in the homozygous condition or with bz1-m4 sh1 confers purple spotting on a bronze background in the aleurone of maize kernels in which Mu elements are active.
Revertant Screen
To screen for revertants of Lg3-O, a stock homozygous for the Lg3-O and ys3 alleles was constructed in a Mu transposon-active line. To provide a convenient assay for the Mu activity of each plant, the stocks were either homozygous for the bz1-mum9 allele or heterozygous for bz1-mum9/bz1-m4 sh1. Mu-active, Lg3-O homozygotes were crossed to wild-type plants. Approximately 3 × 104 Mu-active kernels were planted and screened for a wild-type or partial Lg3 phenotype. To control for pollen contamination, putative revertants were tested for the presence of bz1-mum9 and ys3 by crossing them to bz1-m4 sh1 homozygotes and ys3 heterozygotes, respectively. The ys3 mutation was used to distinguish homozygous Lg3 partial revertant alleles from heterozygous Lg3 partial revertant alleles.
Lg3 Cloning
Maize genomic DNA from an Lg3-O homozygote in a B73 line was digested to completion with BamHI and fractionated through a 10% to 40% Suc gradient. Fragments in the 5- to 15-kb range were recovered and ligated into the BamHI site of λ-DASH (Stratagene). A library of 7 × 105 recombinants was screened by nylon filter lifts (Sambrook et al., 1989) at low stringency with the kn1 homeobox probe. The kn1 clone containing the homeobox region was a generous gift from S. Hake (U.S. Department of Agriculture-Agricultural Research Station, Albany, CA). The entire genomic BamHI insert of the desired clone and a 1.6-kb BglII fragment spanning the homeobox region of the insert were subcloned into pBluescript KS− (Stratagene) for sequencing and further characterization.
A partially purified cDNA homeobox clone in the λ-ZAP vector (Stratagene) was provided by R. Kerstetter (Furman University, Greenville, SC), B. Veit, and S. Hake as a candidate for the lg3 cDNA. This cDNA proved to be a partial transcript of the gene carried on the 1.6-kb BglII insert and was used to screen a B73 vegetative meristem λ-ZAPII cDNA library provided by B. Veit.
Analysis Nucleic Analysis
Maize genomic DNA isolations, DNA gel blotting, and hybridizations were conducted according to the method of Lisch et al. (1994). Sequencing was conducted at the University of California, Berkeley, sequencing facility. Positions of the Mu insertions in the partial revertant Lg3 alleles were identified by sequencing PCR products. PCR was conducted with a Mu end primer (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′) and lg3 primers. Amplified products were isolated and directly sequenced at the Berkeley sequencing facility.
Lg3 Expression Analysis
RNA was isolated following the TRIzol reagent (GIBCO-BRL) RNA isolation procedure. RNA gel-blot analysis was conducted according to the work of Schneeberger et al. (1995). Tissues for RNA isolations of roots, vegetative meristems, shoots, immature ears, immature tassels, mature tassels, and developing embryos were from the inbred line B73. Vegetative meristem tissue is defined as 2-week-old seedling vegetative meristems with three to five leaves still on the meristem. Leaf samples for determining the Lg3 expression pattern in homozygous Lg3-O mutant and nonmutant siblings were generated by growing a segregating Lg3-O family for 6 weeks and harvesting leaf tissue from similarly staged leaves. Sheath tissue was sampled from plants carrying the Lg3-347 and Lg3-Mlg alleles and nonmutant siblings from similarly staged leaves from field-grown plants. To generate tissue samples to determine the relative RNA message levels in the Lg3 revertant alleles, plants were grown for 6 weeks and sheath tissue from similarly staged leaves from at least two plants were sampled from heterozygous mutant and nonmutant sibling leaves. RNA isolations were performed on pooled samples from at least two plants. RT-PCR was conducted according to the work of Bauer et al. (1994). cDNA synthesis was performed on the RNA samples using RT (GIBCO-BRL) and a poly(dT) primer. cDNA was PCR-amplified with two Lg3 primers (Fig. 3), 5′-GTGGAACACGCACTACCGCTG-3′ (Lg3/4) and 5′-AGTGGTGTATGATTCAGGGTCC-3′ (Lg3-3′), and two ubiquitin primers, 5′-TAAGCTGCCGATGTGCCTGCGTCG-3′ (ubi3) and 5′-CTGAAAGACAGAACATAATGAGCACAGGC-3′ (ubi4). The same cDNA reactions were used for the PCR amplification of Lg3 and ubiquitin. PCR conditions were 1-min denaturation at 94°C, 1-min primer annealing at 55°C, and 1-min elongation at 72°C for 20, 25, and 30 cycles. Equal loading of amplified products was achieved by first determining the amount of amplified product in the ubiquitin reactions. The ubiquitin reactions were equalized and the same volume for each corresponding lg3 reaction was analyzed. Equalizing the ubiquitin products and loading the same volume of lg3 product allows a semiquantitative assessment of the amount of LG3 message.
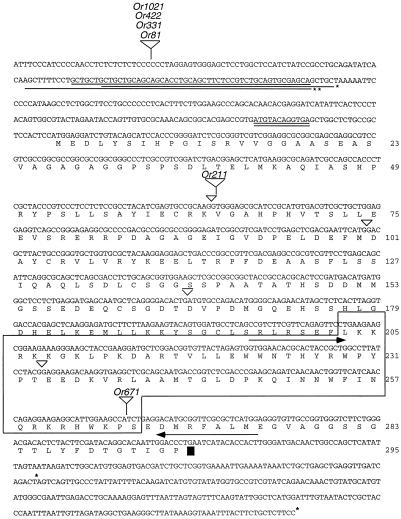
Figure 3.
Nucleotide and predicted amino acid sequence of Lg3. The numbers indicate amino acid residues. The single underline in the 5′ untranslated region indicates a deleted portion of the gene (between nucleotides 90 and 140) for the Lg3-Or81 allele. The single underline with the asterisk indicates a deleted portion of the gene (between nucleotides 96 and 145) for the Lg3-Or422 allele. The single underline with two asterisks indicates a deleted portion of the gene (between nucleotides 80 and 147) for the Lg3-Or211 allele. The double underline in the 5′ untranslated region indicates a short open reading frame. The small inverted triangles indicate the positions of the introns. The large inverted triangles indicate the positions of the Mu transposons for each allele. The outlined region shows the homeodomain. The arrows are the nucleotide sequences used for the lg3 RT-PCR; the 5′ arrow is primer Lg3/4 and the 3′ arrow is primer Lg3-3′. The asterisks in the 3′ untranslated region indicate the site of poly(A+) addition identified in the cDNAs. The accession number is AF100455.
RESULTS
A kn1-Like Homeobox Sequence Is Tightly Linked to the Lg3-O Mutation
The similarity of the mutant phenotypes caused by Kn1-O and Lg3-O suggested to us that they might be caused by mutations in related genes (Fowler and Freeling, 1996). To address whether the lg3 gene is a member of the knox gene family, we first looked for cosegregation of a homeobox-containing RFLP with Lg3-O in families segregating 1:1 for Lg3-O mutant and nonmutant siblings. DNA from heterozygous mutant and nonmutant siblings was digested with several restriction enzymes, gel blotted, and hybridized at low stringency with a small fragment encompassing the kn1 homeobox. A 10-kb BamHI fragment was present in all mutant progeny analyzed and was absent in all nonmutant progeny (data not shown). In addition, a 1.6-kb BglII fragment cosegregated with the mutant phenotype (data not shown). We examined 81 potential recombination events by this method and found no recombination between the Lg3-O mutant phenotype and the 10-kb BamHI or 1.6-kb BglII fragments. Thus, the identified homeobox sequence is genetically linked to the lg3 locus, within 5 map units at the 95% confidence level.
We isolated the putative lg3-containing genomic BamHI fragment from an Lg3-O homozygote using the kn1 homeobox fragment as a probe. The 10-kb BamHI fragment contained an internal 1.6-kb BglII insert that hybridized to the kn1 homeobox probe, suggesting that the genomic fragment corresponded to the two knox RFLPs linked to lg3. When used as a probe at high stringency on the same DNA gel blots that identified the Lg3-O cosegregating homeobox fragment, the 1.6-kb BglII fragment recognized the cosegregating 10-kb BamHI fragment identified in mutant plants (Fig. 2). It also hybridized to an 11-kb fragment, corresponding to the nonmutant B73 allele, in all progeny. At this stringency the probe did not hybridize to other fragments on the gel blot, indicating that the clone represented a single-copy genomic sequence. Sequence analysis of the 1.6-kb BglII fragment revealed a knox family homeobox region. The tight linkage of this knox sequence to lg3, along with the genetic similarities of the dominant Lg3-O mutation to several dominant Kn1 mutant alleles, strongly suggested that the identified homeobox was within the lg3 gene itself.
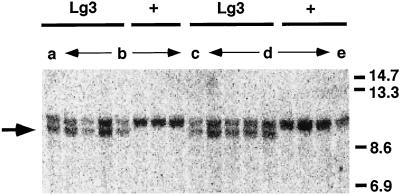
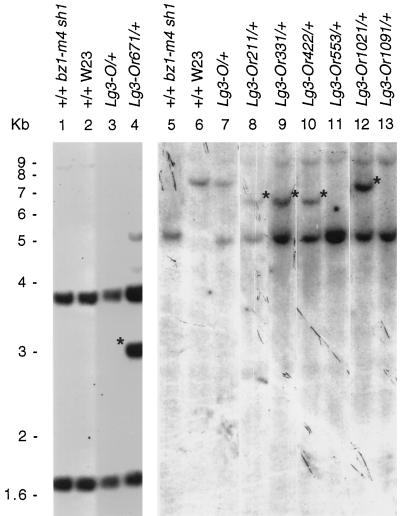
Figure 2.
DNA gel-blot analysis of two families segregating for the Lg3-O mutation in a B73 inbred line. The genomic DNA samples were digested with BamHI. Lane a, Lg3 plant 1678-7; lanes b (designated by arrows), seven progeny (four Lg3, three wild type) of the cross B73 wild type × plant 1678-7; lane c, plant 1678-8; lanes d (designated by arrows), seven progeny (four Lg3, three wild type) of the cross B73 wild type × plant 1678-8; lane e, B73 inbred tester. The DNA gel blot was probed with the BglII 1.6-kb fragment from the putative lg3 clone. The arrow marks a 10-kb fragment always present in Lg3-O heterozygotes but never present in wild-type siblings. The number scale on the right is in kilobases.
Isolation and Sequence Analysis of lg3 cDNAs
To analyze the transcript sequence corresponding to the lg3 gene, we plaque-purified a knox-containing partial cDNA given to us by R. Kerstetter and S. Hake. Sequence analysis indicated that this partial clone corresponded to the 3′ end of the sequence derived from the 1.6-kb genomic BglII fragment (data not shown). We isolated three more lg3 cDNAs using the partial cDNA as a probe on a B73 vegetative meristem library generously provided by B. Veit and S. Hake. The longest lg3 cDNA was 1536 nucleotides and was the clone used in all further characterization. RNA gel-blot analysis of immature ear poly(A)+ RNA probed with the lg3 cDNA revealed an approximately 1.5-kb transcript (data not shown), indicating that a cDNA containing the entire coding sequence for the lg3 gene was isolated. The lg3 cDNA contains an 885-nucleotide-coding sequence, a 316-nucleotide 5′ untranslated sequence, and a 334-nucleotide 3′ untranslated sequence (Fig. 3). It is interesting that a short open reading frame of unknown function is present in the 5′ untranslated region. Based on the 885-nucleotide-coding region, the lg3 gene encodes a 295-amino acid protein containing a 64-amino acid homeodomain region, with an overall predicted Mr of 32,000. The homeodomain region is a putative helix-turn-helix DNA-binding motif, indicating that the LG3 protein probably functions as a transcriptional regulator.
Figure 4A depicts sequence comparisons of the LG3 protein with the maize KN1 and RS1 proteins that exhibit 76% and 75% identity in the homeodomain region, respectively, and 100% identity in the third recognition helix region. However, outside the homeodomain region, the LG3 amino acid identity to RS1 and KN1 is greatly reduced. Comparisons of the LG3 protein sequence to other plant homeodomain-containing proteins shows that the LG3-coding sequence is most closely related to a protein encoded by a rice gene (accession no. AF050180) isolated from an embryogenesis-specific cDNA library (A.D. Postma-Haarsma, I.I.G.S. Verwoert, O.P. Stronk, J. Koster, G.E.M. Lamers, J.H.C. Hoge, and A.H. Meijer, unpublished data). The maize LG3 protein and its putative rice ortholog exhibit 78% identity and are notably shorter than other characterized genes of the kn1 family (Fig. 4A), and thus belong to a separate subgroup (Fig. 4B). Comparisons with other homeodomain proteins from tomato, Arabidopsis, soybean, and barley all show a similar trend: high identity within the homeodomain and low identity outside. These sequence comparisons show that lg3 is a member of the knox gene family.
Figure 4.
Comparisons of the LG3 amino acid sequence with other KNOX amino acid sequences. A, Comparisons of the predicted maize LG3, RS1, and KN1 and putative rice LG3 amino acid sequences. B, A dendrogram comparison of the plant KNOX-like protein sequences. Entire protein sequences were compared. The maize and rice LG3 proteins (LG3ZM and LG3OS, respectively) form a separate subgroup. The figure was compiled using the software program PileUp (Genetics Computer Group, Madison, WI). Protein sequences in B are from Arabidopsis (KNAT1-5, STM1), Glycine max (SBH1), Hordeum vulgare (HOODED), Lycopersicon esculentum (LET6, TKN1), Oryza sativa (LG3OS, OSH1), Solanum tuberosum (POTH1), and Zea mays (LG3ZM, KN1, RS1).
Lg3-O Revertant Screen and Characterization of Alleles
To genetically verify that we had identified authentic lg3 sequences, a Mu transposon insertional mutagenesis of the Lg3-O allele was conducted (see Methods for details). We looked for a correlation between a heritable change in the Lg3-O mutant phenotype and an alteration in the genomic DNA structure at the putative lg3 locus corresponding to our clone. A Mu-active stock homozygous for the Lg3-O mutation was outcrossed to a wild-type tester. All progeny generated from this cross were heterozygous for Lg3-O and should display the mutant phenotype. Insertions of Mu elements into the Lg3-O mutant allele that resulted in lowering or eliminating the mutant activity were recovered by selecting for reversion to the wild-type phenotype. Figure 5 shows six partial revertant alleles, Lg3-Or81, Lg3-Or211, Lg3-Or331, Lg3-Or422, Lg3-Or671, and Lg3-Or1021, which were among the alleles isolated and will be discussed in detail below. Two complete revertant alleles, Lg3-Or553 and Lg3-Or1091, were also isolated.
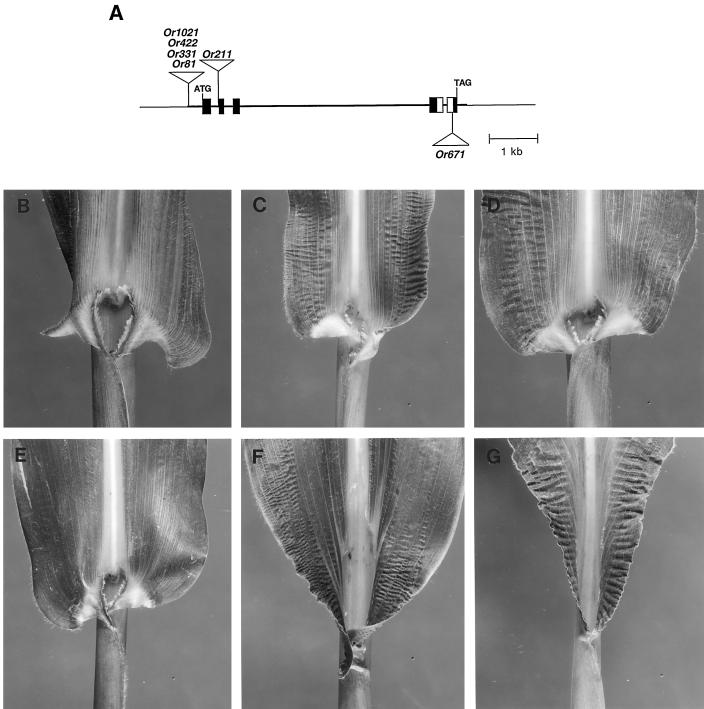
Figure 5.
Lg3 partial revertant alleles. A, Genomic map of the Lg3 gene showing the sites of Mu insertions. B, Lg3-Or81 leaf. C, Lg3-Or671 leaf. D, Lg3-Or331 leaf from a Mu-inactive plant. E, Lg3-Or422 leaf from a Mu-inactive plant. F, Lg3-Or1021 leaf from a Mu-inactive plant. G, Lg3-Or211 leaf from a Mu-inactive plant. The leaves are from plants carrying the alleles in the heterozygous condition. Note the degree of ligule displacement in the alleles. For example, in the Lg3-Or211 allele the ligule is removed and in the Lg3-Or331 allele the ligule is only slightly displaced.
Each of these eight alleles contain an alteration in the progenitor Lg3-O phenotype that is linked to the lg3 locus, based on the closely linked ys3 marker. These data indicate that the phenotypic changes are due to mutations within the lg3 gene and not to dominant extragenic suppressors of Lg3-O. Heterozygotes for both the Lg3-Or553 and Lg3-Or1091 alleles exhibit a wild-type, lg3+ phenotype. In the heterozygous condition, the Lg3-Or81 and Lg3-Or671 alleles cause a mild Lg3 mutant phenotype (Fig. 5). Four alleles, Lg3-Or211, Lg3-Or331, Lg3-Or422 and Lg3-Or1021, are Mu suppressible; i.e. their phenotypic effect is modified by the presence or absence of Mu activity (Martienssen et al., 1989). Phenotypes of plants harboring the Mu-suppressible Lg3 alleles were observed in both Mu-active and Mu-inactive plants, as assayed by the bz-mum9 reporter allele (see Methods). In Mu-active plants each of the four Mu-suppressible alleles confers a wild-type phenotype, whereas in Mu-inactive plants these alleles cause an Lg3 mutant phenotype (Fig. 5; Fowler et al., 1996; Muehlbauer et al., 1997). These data strongly suggest that the Lg3-Or211, Lg3-Or331, Lg3-Or422, and Lg3-Or1021 alleles contain Mu insertions.
To determine whether the alteration in phenotype corresponded to a genomic change within the putative lg3 sequences, we characterized the genomic structure of the revertant alleles. In plants heterozygous for the revertant alleles Lg3-Or211, Lg3-Or331, Lg3-Or422, Lg3-Or671, and Lg3-Or1021, changes in the restriction fragment patterns of genomic DNA were revealed by hybridization with lg3 cDNA probes (Fig. 6). Further characterization of the Lg3-Or81, Lg3-Or211, Lg3-Or331, Lg3-Or422, Lg3-Or671, and Lg3-Or1021 alleles at the nucleotide level showed that Mu elements were inserted throughout the lg3 gene (Figs. 3 and 5). We sequenced PCR products amplified from each allele using a Mu end primer and lg3 primers (see Methods). Lg3-Or81, Lg3-Or331, Lg3-Or422, and Lg3-Or1021 carry Mu elements at identical nucleotide positions in the 5′ untranslated region. The Lg3-Or211 allele carries a Mu1 element at the last nucleotide of the first intron. The Lg3-Or671 allele carries a Mu element in the homeobox region in the gene's fifth exon. These data show that Mu element insertion in the progenitor Lg3-O allele resulted in a loss (or partial loss) of mutant activity in each revertant and proves that the isolated homeobox clone corresponds to the lg3 gene.
Figure 6.
DNA gel blots showing alterations in lg3 genomic sequence associated with a change in Lg3 phenotype. Lanes 1 to 4 were digested with BglII and probed with the entire lg3 cDNA. Lanes 5 to 13 were digested with XbaI and probed with the 5′ untranslated region of the lg3 cDNA. Asterisks indicate polymorphic fragments due to DNA alterations at the lg3 locus due to Mu element insertions into the lg3 gene.
Although the Mu insertions in the Lg3-Or81, Lg3-Or331, Lg3-Or422, and Lg3-Or1021 alleles are in the same nucleotide, several observations show that they are independent alleles. The Lg3-Or81 allele does not respond to Mu activity, whereas the Lg3-Or331, Lg3-Or422, and Lg3-Or1021 alleles respond to Mu activity and are Mu suppressible. In addition, the Lg3-Or1021 RFLP is larger than the Lg3-Or331 and Lg3-Or422 RFLP (Fig. 5), indicating that Lg3-Or1021 is caused by insertion of a different Mu element than in the Lg3-Or331 and Lg3-Or422 alleles. Also, our sequence analysis showed that small deletions in the 5′ untranslated region of the Lg3-Or331 and Lg3-Or422 alleles span different nucleotides (Fig. 3). These data indicate that the Lg3-Or81, Lg3-Or331, Lg3-Or422, and Lg3-Or1021 alleles are independently derived.
Genetic and molecular analyses indicate that the Lg3-Or553 and Lg3-Or1091 alleles represent deletions of the lg3 locus. Lg3-Or553 and Lg3-Or1091 heterozygotes exhibited no RFLP that corresponded to the progenitor Lg3-O allele (Fig. 6). We monitored populations derived from plants carrying these alleles by following the revertant allele-linked ys3 mutation. Homozygous ys3 plants exhibit a yellow-stripe phenotype on the leaves. We observed no or very low transmission through the male and only slightly higher transmission through the female gametophyte (data not shown), suggesting the presence of a genetic lesion that reduces haploid viability. Reduced transmission through the gametophyte is characteristic of chromosomal deficiencies in plants (Birchler, 1994), but the extent of the deletions is not known. Therefore, elimination of the Lg3-O allele (and its corresponding RFLP) by deletion removes the Lg3 phenotype from the plant. These deletion alleles provide additional evidence to verify the lg3 cDNA.
Lg3 Expression in Wild-Type and Lg3 Plants
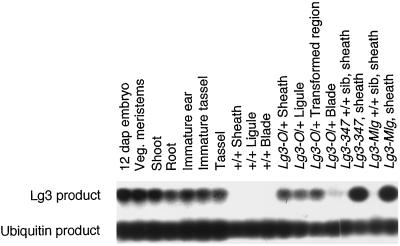
To determine the expression pattern of lg3 in wild-type and Lg3-O mutant plants, we conducted RT-PCR on a variety of tissues. We used the amount of ubiquitin PCR-amplified product to equalize our Lg3 PCR reactions to obtain a semiquantitative estimate of the amount of LG3 product (see Methods). In wild-type plants we found LG3 mRNA in vegetative meristems, shoots, roots, immature ears and tassels, mature tassels, and embryos (Fig. 7). We did not observe LG3 mRNA in sheath, ligule, or blade tissue of Lg3-O wild-type sibling plants. These data indicate that the wild-type expression pattern of lg3 is predominantly confined to apical regions. However, in homozygous Lg3-O mutant plants, we observed LG3 mRNA in sheath tissue, the ligular region, and the transformed region of developing leaves (Fig. 7). LG3 mRNA was observed at a very low level in the blade tissue of Lg3-O mutant plants. The expression pattern in Lg3-O leaves suggests that ectopic expression of LG3 mRNA is the cause of the Lg3 mutant transformation phenotype.
Figure 7.
RT-PCR analysis of Lg3 expression in wild-type and mutant tissues. cDNA from different tissues of wild-type and mutant plants was used for amplification of LG3 and ubiquitin sequences. Thirty cycles of PCR were used to amplify the LG3 and ubiquitin sequences. The PCR products were hybridized with the lg3 cDNA and a ubiquitin probe. dap, Days after pollination; Veg., vegetative.
To provide additional evidence that ectopic LG3 mRNA expression causes the dominant Lg3 phenotype, we investigated expression in plants carrying either of the two independently isolated Lg3 alleles, Lg3-Mlg or Lg3-347. These alleles were previously mapped to the centromeric region of chromosome 3 (Lg3-O is positioned on the short arm of chromosome 3 close to the centromere) using the waxy-marked translocation stocks (Harper et al., 1995; Fowler and Freeling, 1996). Each of these mutants cosegregated with an lg3 RFLP in families segregating 1:1 for mutant and nonmutant plants, indicating that they are lg3 alleles. Ectopic expression of LG3 mRNA in the sheath was observed in both Lg3-Mlg and Lg3-347, whereas leaf-sheath tissue of wild-type siblings showed no detectable LG3 mRNA expression (Fig. 7). These data provide further evidence that ectopic Lg3 expression in the leaf causes the dominant Lg3 phenotype.
The Partially Revertant Lg3-O Alleles Represent Two Classes of Lg3 Phenotypes
Based on the area of the leaf affected by the dominant Lg3 phenotype, the partially revertant Lg3-O alleles represent two classes of Lg3 phenotypes (Fig. 5). When we examined these alleles in the heterozygous condition we observed very little ligule present in the Mu-inactive Lg3-Or211 allele, sections of remnant ligule in the Mu-inactive Lg3-Or1021 allele, and slight ligule displacement in the Mu-inactive Lg3-Or331, Mu-inactive Lg3-Or422, and Lg3-Or671 alleles. Therefore, the phenotypes caused by these alleles can be categorized in two classes represented by the Mu-inactive Lg3-Or211 and Lg3-Or1021 alleles that confer the strongest Lg3 phenotypes, and by the Mu-inactive Lg3-Or331, Lg3-Or422, and Lg3-Or671 alleles that confer weaker Lg3 phenotypes (Fig. 5). In addition, Lg3-O homozygotes exhibit a stronger Lg3 phenotype than Lg3-O heterozygotes (Fowler and Freeling, 1996).
We reasoned that these phenotypic differences could be due to the level of ectopic expression of lg3. Therefore, we conducted RNA expression studies on plants heterozygous for each of these alleles to determine whether the level of ectopic LG3 mRNA expression correlated with the severity of the Lg3 phenotype. RNA was isolated from pooled samples of sheath tissue from similarly staged leaves from at least two plants from heterozygous mutant and nonmutant sibling leaves. RT-PCR analysis was conducted on RNA isolated from sheath tissue of heterozygous Lg3-Or671 plants and heterozygous Mu-inactive Lg3-Or211, Lg3-Or331, Lg3-Or422, and Lg3-Or1021 plants. We used the amount of ubiquitin PCR-amplified product to equalize our lg3 PCR reactions to obtain a semiquantitative estimate of the amount of LG3 product (see Methods). We observed the highest levels of LG3 mRNA in sheath tissue from Mu-inactive plants carrying the Lg3-Or211 and Lg3-Or1021 alleles, and much lower levels in the Lg3-Or671 allele and in Mu-inactive plants carrying the Lg3-Or331 and Lg3-Or422 alleles (Fig. 8). Compared with the 30 cycles of PCR shown in Figure 8, the relative levels of the amplified products observed in 20 cycles of PCR were the same, indicating that our amplified products were within the linear range (data not shown). Furthermore, RNA-blot analysis of sheath tissue from Lg3-O homozygotes and heterozygotes shows that there is a higher level of LG3 mRNA in the Lg3-O homozygotes (data not shown). These data show a trend indicating that the level of ectopic Lg3 expression correlates with the extent of the blade-to-sheath transformation and likely determines the phenotypic differences. High ectopic expression (e.g. Mu-inactive Lg3-Or211) results in a severe phenotype, whereas lower ectopic expression (e.g. Mu-inactive Lg3-Or311) results in a milder phenotype.
Figure 8.
RT-PCR analysis of Lg3 expression in sheath tissue from partially Lg3 revertant alleles. cDNA from sheath tissue from several Lg3 alleles and nonmutant siblings was used for amplification of Lg3 and ubiquitin sequences. Thirty cycles of PCR were used to amplify the Lg3 and ubiquitin sequences. The relative levels of the amplified products were similar at 20 cycles of PCR (data not shown). The PCR products were hybridized with the lg3 cDNA and a ubiquitin probe.
DISCUSSION
The lg3 Gene Encodes a KNOX-Family Homeodomain Protein
In this paper we report the isolation and molecular characterization of the lg3 gene, which encodes a protein containing the homeodomain DNA-binding motif. We verified that our cDNA clone corresponds to the lg3 gene by showing that changes in the Lg3 dominant phenotype are accompanied by genomic alterations at the locus identified by our clone. The LG3 protein is highly homologous to the maize KN1 and RS1 proteins, showing a high level of amino acid identity in the homeodomain region and 100% identity within the homeodomain's third helix. The third helix has been identified in other homeodomain proteins as the putative recognition helix and provides much of the specificity for the DNA-binding activity (Scott et al., 1989). Outside the homeodomain, the identity between these maize proteins is much lower. A putative rice lg3 ortholog has been identified and exhibits high identity throughout the protein to the maize LG3 protein. Sequence comparisons to KNOX proteins from other plant species shows a similar trend: high identity in the homeodomain region and lower identity outside the homeodomain. The presence of the homeobox indicates that knox genes (including lg3) are likely to encode DNA-binding transcriptional regulators. In fact, the barley HOODED homeodomain protein possesses DNA-binding activity (Krusell et al., 1997), supporting this hypothesis. These sequencing data, the genetic analysis, and the similar blade-to-sheath transformation phenotypes caused by dominant mutations in the gene (Fowler and Freeling, 1996) demonstrate that lg3 is a member of the kn1-like family of homeobox genes.
In addition to the knox genes, other distinct classes of plant homeobox genes have been isolated. A unique class, referred to as HD-Zip proteins (Schena and Davis, 1992), contains a homeodomain and a carboxy-terminal Leu zipper region. HD-Zip homeobox genes have putative functions in several processes, including photomorphogenesis, vascular development, root hair development, and defense gene regulation (Schindler et al., 1993; Korfhage et al., 1994; Quaedvlieg et al., 1995; Carabelli et al., 1996; Tornero et al., 1996). Two maize genes in this class, ZmHox1a and ZmHox1b, have been isolated (Bellman and Werr, 1992; Uberlacker et al., 1996). The ZMHOX1a homeodomain protein binds to the SHRUNKEN feedback-control element, further supporting the notion that plant homeodomain proteins function as DNA-binding regulators (Bellman and Werr, 1992). Finally, the Arabidopsis glabra2 (gl2) gene represents a third class of homeobox genes (Rerie et al., 1994). The gl2 gene is thought to regulate cell fate decisions, because recessive mutations at the gl2 locus result in defective trichome development (Rerie et al., 1994). These studies demonstrate that homeobox genes are found throughout plant species and are involved in regulating a wide range of functions.
Possible Developmental Roles for LG3
Our expression analysis shows that in wild-type plants LG3 mRNA is found in all apical tissues examined, including roots, shoot tips, immature ears, immature tassels, mature tassels, and embryos. However, the LG3 transcript is not present in wild-type leaves. Thus, although the leaf-transformation phenotype caused by dominant mutant alleles of lg3 indicates that LG3 can influence cell fate decisions, it is not likely to play a role in wild-type leaf differentiation. Our data indicate that lg3 may be expressed in a pattern similar to the kn1 and rs1 genes (Smith et al., 1992; Jackson et al., 1994; Schneeberger et al., 1995). KN1 mRNA is found throughout the L2 layer of the shoot apical meristem, whereas KN1 protein is in the L1 and L2 layers of the meristem. However, KN1 is not found in differentiating leaf primordia or in meristem regions corresponding to incipient primordia. Smith et al. (1992) speculated that KN1 maintains the meristem in an undifferentiated state and that it is down-regulated on the flank of the meristem where the incipient leaf will form. In wild-type plants, RS1 mRNA is present in a ring around the young segment coincident with the position just below the incipient leaf but not in the differentiating leaf primordia. Thus, rs1 may be involved in determining the boundaries in the segment. The presence of LG3 mRNA in apical regions suggests that lg3 may also be involved in meristem function.
To determine more precisely the role of lg3 in maize development, we are characterizing three recessive lg3 alleles, each of which carries a Mu element in an exon. Preliminary work on these recessive mutations has not revealed an obvious effect on the phenotype, indicating that the phenotype of the recessive lg3 mutation may be subtle (R. Tyers, G.J. Muehlbauer, and M. Freeling, unpublished results). Alternatively, the maize genome is proposed to be an ancient allotetraploid (Gaut and Doebley, 1997), and it is very possible that an lg3 homolog exists and provides genetically redundant functions. The lg3 gene maps to the short arm of chromosome 3. On the short arm of chromosome 8, which is considered to be the duplicate region for chromosome 3, there are two other tightly linked knox genes, knox5 and knox11 (P. Bauer and M. Freeling, unpublished results; Kerstetter et al., 1994). Thus lg3, knox5, and knox11 may be genes that were duplicated during evolution. The dominant Lg4-O mutation, which also causes a blade-to-sheath transformation phenotype, maps to the same region as knox5 and knox11, suggesting that one of these genes is lg4 (Fowler and Freeling, 1996). It is possible that some developmental functions eliminated by recessive loss-of-function lg3 alleles are covered by the action of knox5 and/or knox11. This possibility is being investigated by identifying recessive mutations in the knox5 and knox11 genes and making double- and triple-mutant combinations with all three genes.
Characterization of other knox genes has led to the general conclusion that these genes are involved in meristem maintenance and in preventing differentiation. Recessive, null alleles of the kn1 gene cause a range of phenotypes, including fewer branches and spiklets on the tassel, ears that are often absent and when present are small with few spikelets, extra carpels in female florets, abnormally proliferating ovule tissue, and extra leaves in the axils of vegetative leaves. These phenotypes are in general agreement with the notion that the role of KN1 is to prevent premature differentiation (Kerstetter et al., 1997). Transgenic tobacco plants expressing kn1 create meristematic regions on the leaf blade (Sinha et al., 1993). Furthermore, the dominant Hooded mutation in barley causes floral meristems to develop on the awn (Müller et al., 1995). In addition, transgenic Arabidopsis overexpressing the Arabidopsis knox gene knat1 exhibits ectopic meristems on leaves (Chuck et al., 1996). Finally, loss-of function mutations in the Arabidopsis STM gene (also in the knox family) restrict shoot meristem function so that mutant plants make only cotyledons, indicating that STM is necessary to initiate or maintain the shoot meristem (Barton and Poethig, 1993; Clark et al., 1996; Endrizzi et al., 1996). Taken together, these studies suggest that the role of knox genes is to maintain meristem cell identities and prevent differentiation. The dominant Lg3 mutations appear to fit this pattern because ectopic expression leads to more basal phenotypes such as sheath tissue.
Ectopic Expression of lg3 Causes the Dominant Lg3 Mutant Phenotype
RNA expression of lg3 could not be detected by either northern or RT-PCR analysis in leaves of wild-type plants, indicating that lg3 is not expressed in any region of the leaf. However, in Lg3-O mutant leaves, LG3 message is present in the sheath, the ligular region, the transformed region of the blade, and, to a lesser extent, in untransformed blade (Fig. 7). In addition, expression of the LG3 message is detectable in the sheaths of plants carrying two other dominant Lg3 alleles, Lg3-Mlg and Lg3-347. These data strongly suggest that ectopic lg3 expression in the maize leaf causes the dominant Lg3 phenotype of blade-to-sheath transformation. It is somewhat surprising, given that Lg3 dominant mutations cause blade cells to adopt a sheath cell fate, that LG3 mRNA is not expressed in wild-type sheaths. However, our results indicate that the wild-type lg3 gene does not function to specify sheath identity, but when ectopically expressed in leaf development it specifies sheath-like identity in the leaf blade.
It is noteworthy that in mutant leaves LG3 mRNA is expressed not only in transformed regions but also in the sheath, which does not display an obvious mutant phenotype. In addition to the transformation phenotype, plants displaying a severe Lg3 phenotype have shortened internodes and smaller leaves in the upper nodes (Fowler and Freeling, 1996). These phenotypes were initially hypothesized to be secondary effects of the leaf-transformation phenotype, because mutant leaves in the lower portion of the plant fail to unroll properly during plant growth and physically restrain the growth of upper leaves. Alternatively, these phenotypes could be due to LG3 mRNA expression in the sheath, resulting in a reduction in sheath length. The presence of LG3 mRNA in the sheath could also result in subtle and as-yet-undetected phenotypes.
Of potential interest for understanding the mechanism that causes the dominant Lg3 phenotype is the Lg3-Or671 allele. This allele contains a Mu element insertion in an exon encoding the carboxy terminus of the homeobox region, but it still exhibits a mild dominant phenotype. Our RT-PCR data indicate that the Lg3-Or671 allele causes a very low level of ectopic LG3 message expression (Fig. 8). Therefore, the Mu insertion in the homeobox region disrupts the mRNA levels conditioned by the Lg3-O allele. Several possibilities exist for obtaining a dominant phenotype from this allele. One possibility is that ectopic expression of the Mu-truncated LG3 protein could be sufficient to confer the dominant Lg3 phenotype. Another possibility is that the Mu element could be spliced out of the LG3 message at a low frequency, producing enough full-length message (and therefore full-length protein) to confer the dominant phenotype. Alternatively, somatic excision of the Mu element from the Lg3 gene could also produce a full-length message and the dominant phenotype. The appearance of an RT-PCR product (which spans the Mu insertion site) of the correct size in the leaves of Lg3-Or671 plants supports either of the two latter alternatives.
Transformation Phenotypes Caused by Dominant knox-Like Genes Can Be Explained by the Maturation Schedule Hypothesis
Expression patterns of kn1 and rs1 are similar to those of lg3. In wild-type plants, Kn1, Rs1, and Lg3 mRNA expression is confined to the apical regions (Smith et al., 1992; Jackson et al., 1994; Schneeberger et al., 1995). However, in Kn1-O and Rs1-O mutant plants, ectopic expression of Kn1 and Rs1 mRNA is found in leaves, where the mutant phenotype is manifest (Smith et al., 1992; Schneeberger et al., 1995). The ectopic Lg3 expression pattern is consistent with these other KNOX mRNAs, suggesting that the Kn1-O, Rs1-O, and Lg3-O dominant mutant phenotypes are caused by a similar mechanism.
Several models to explain the blade-to-sheath transformation phenotype of the dominant Kn1-like mutations can be envisioned (Freeling, 1992). Our current working model, referred to as the maturation schedule hypothesis (Muehlbauer et al., 1997), is that ectopic expression of the kn1-like genes in the leaf interferes with the time-dependent process by which regions adopt their specific identity, an identity that will later manifest itself in specific differentiated cell types. This model is supported by the observation that Lg3 mutant activity (i.e. ectopic expression of LG3 mRNA) can induce several distinct transformation phenotypes in the blade (e.g. not solely transformation to sheath), and that the distinct phenotypic classes correlate with the timing of ectopic Lg3 expression in the leaf (Muehlbauer et al., 1997). The maturation schedule hypothesis states that groups of cells in the early leaf primordium possess the competency to express the sheath cell fate. Over time, distal cells that will become ligule, auricle, and blade progress to the ligule/auricle competency stage, whereas more proximal cells remain in the sheath competency stage. Next, the most distal cells progress to the blade competency stage, whereas cells that will be ligule or auricle remain in the ligule/auricle competency stage. Cell fates are determined by the competency stage each region of the leaf is in when signals to differentiate are received. We hypothesize that the ectopic expression of the kn1-like homeobox genes in the leaf blade retards the correct progression through this schedule, restricting cells in the blade region to an earlier competency stage (e.g. sheath), resulting in the observed transformation phenotypes. This model also helps to explain the apparent lack of phenotype in Lg3 sheath tissue, despite the presence of ectopic LG3 mRNA. Sheath cells never progress past the earliest (sheath) competency stage in wild-type leaves and thus cannot be affected by ectopic expression of an activity that interferes with progression through the schedule.
The Level of Ectopic Lg3 Expression Positively Correlates with the Extent of Blade Showing the Mutant Phenotype
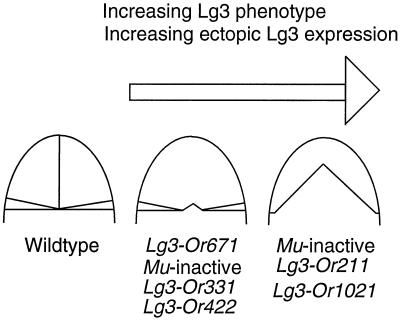
Our data show that the severity of the Lg3 phenotype is associated with the amount of LG3 message. Figure 9 shows a schematic diagram of the two classes of Lg3 partial revertant alleles. These alleles revealed two classes of blade-to-sheath transformations that showed levels of LG3 message that correlated well with the severity of the transformation phenotype. High LG3 message resulted in a severe Lg3 phenotype, whereas low LG3 message resulted in a mild Lg3 phenotype. In addition, Lg3-O homozygotes exhibit a more severe phenotype than Lg3-O heterozygotes, and the levels of LG3 message reflect the severity of the phenotype. Finally, heteroallelelic combinations of the partial revertant alleles show more extensive blade-to-sheath transformations in the heteroallelic combination than in plants carrying the single allele (G.J. Muehlbauer and M. Freeling, unpublished observations). Taken together, these data indicate that the amount of ectopic Lg3 expression controls the extent of the blade-to-sheath transformations in the Lg3 mutants.
Figure 9.
Lg3 leaf phenotypes and ectopic Lg3 expression are represented schematically. The severity connotated for each partially revertant Lg3 allele is shown along with a representation of the amount of ectopic Lg3 expression.
The maturation schedule hypothesis states that ectopic knox gene expression retards the acquisition of the leaf competency stages in a time-dependent manner. In a previous experiment in which we varied the timing of ectopic Lg3 expression, we found that early ectopic Lg3 gene expression results in a blade-to-sheath transformation, whereas later ectopic expression results in transformations to more distal phenotypes, such as a blade-to-auricle transformation (Muehlbauer et al., 1997). In the Lg3 alleles presented here, we were able to determine the effect of the amount of ectopic Lg3 expression. These Lg3 alleles show only blade-to-sheath transformations, even though the expression levels are quite different. Low and high ectopic Lg3 expression results in a blade-to-sheath transformation, indicating that the level of ectopic Lg3 expression alters only the extent of the phenotype, not the type of transformation. Based on data from varying the time of ectopic Lg3 expression (Muehlbauer et al., 1997), the maturation schedule model predicts that early ectopic expression will result in a blade-to-sheath transformation. Therefore, we propose that the timing of ectopic Lg3 expression in these Lg3 alleles is early in development, perhaps as early as the founder cells or young primordium. This early ectopic knox gene expression results in a blade-to-sheath transformation regardless of the level of message.
ACKNOWLEDGMENTS
The authors thank David A. Somers and Burle Gengenbach for critical reading of the manuscript.
Abbreviations:
- RFLP
restriction fragment-length polymorphism
- RT
reverse transcriptase
Footnotes
This work was supported by National Institutes of Health Postdoctoral grant no. 5 F32 GM-16619-02 to G.J.M., a Howard Hughes Medical Institute predoctoral fellowship to J.E.F., and a National Institutes of Health grant to M.F.
LITERATURE CITED
- Barton MK, Poethig S. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- Bauer P, Crespi MD, Szécsi J, Allison LA, Schultze M, Ratet P, Kondorosi E, Kondorosi A. Alfalfa Enod12 genes are differentially regulated during nodule development by Nod factors and Rhizobium invasion. Plant Physiol. 1994;105:585–592. doi: 10.1104/pp.105.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M. The liguleless1 gene acts tissue specifically in maize leaf development. Dev Biol. 1990;141:220–232. doi: 10.1016/0012-1606(90)90117-2. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Freeling M. Genetic analysis of Rough sheath1 developmental mutants of maize. Genetics. 1994;136:295–311. doi: 10.1093/genetics/136.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmann R, Werr W. Zmhox1a, the product of a novel maize homeobox gene, interacts with the Shrunken 26 bp feedback control element. EMBO J. 1992;11:3367–3374. doi: 10.1002/j.1460-2075.1992.tb05415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. Deficiency analysis. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 494–495. [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-J, Janssen B-J, Williams A, Sinha N. A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell. 1997;9:1289–1304. doi: 10.1105/tpc.9.8.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOTMERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Fowler J, Freeling M. Genetic characterization of the dominant liguleless mutations in maize. Dev Genet. 1996;18:198–222. doi: 10.1002/(SICI)1520-6408(1996)18:3<198::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fowler JE, Muehlbauer GJ, Freeling M. Mosaic analysis of the Liguleless3 mutant phenotype in maize by coordinate suppression of Mutator-induced alleles. Genetics. 1996;143:489–503. doi: 10.1093/genetics/143.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. A conceptual framework for maize leaf development. Dev Biol. 1992;153:44–58. doi: 10.1016/0012-1606(92)90090-4. [DOI] [PubMed] [Google Scholar]
- Freeling M, Hake S. Developmental genetics of mutants that specify knotted leaves in maize. Genetics. 1985;143:489–503. doi: 10.1093/genetics/111.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinat WC. The phytomer in relation to the floral homologies in the American Maydea. Bot Mus Leafl Harv Univ. 1959;19:1–32. [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Freeling M. Interactions of liguleless1 and liguleless2 function during ligule indiction in maize. Genetics. 1996;144:1871–1882. doi: 10.1093/genetics/144.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Scanlon M, Freeling M. Multiple-ligule (Mlg*-1) is probably an allele of lg3. Maize Genet Coop Newsl. 1995;69:22–23. [Google Scholar]
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell. 1994;6:1877–1887. doi: 10.1105/tpc.6.12.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfhage U, Trezzini GF, Meier I, Halbrock K, Somssich IE. Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell. 1994;6:695–708. doi: 10.1105/tpc.6.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Rasmussen I, Gausing K. DNA binding sites recognized in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare) FEBS Lett. 1997;408:25–29. doi: 10.1016/s0014-5793(97)00382-7. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi K, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D, Chomet P, Freeling M. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics. 1994;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Baron MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Ma HA, McMullen M, Finer JJ. Identification of a homeobox-containing gene with enhanced expression during soybean (Glycine max L.) somatic embryo development. Plant Mol Biol. 1994;24:465–473. doi: 10.1007/BF00024114. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Barkan A, Freeling M, Taylor WC. Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO J. 1989;8:1633–1639. doi: 10.1002/j.1460-2075.1989.tb03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell. 1993;5:1039–1048. doi: 10.1105/tpc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer GJ, Fowler JE, Freeling M. Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development. 1997;124:5097–5106. doi: 10.1242/dev.124.24.5097. [DOI] [PubMed] [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature. 1995;374:727–730. doi: 10.1038/374727a0. [DOI] [PubMed] [Google Scholar]
- Parnis A, Cohen O, Gutfinger T, Hareven D, Zamir D, Lifschitz E. The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell. 1997;9:2143–2158. doi: 10.1105/tpc.9.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeck P, Smeekens S. The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell. 1995;7:117–129. doi: 10.1105/tpc.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schichnes DE, Freeling M. Lax Midrib1-O, a systematic, heterochromic mutant of maize. Am J Bot. 1998;84:481–491. [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- Schneeberger RG, Becraft PW, Hake S, Freeling M. Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- Schena M, Davis RW. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Tamkun W, Hartzell GW., III The structure and function of the homeo domain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Martinez-Laborda A, Kim H-S, Zambryski PC. Localization of expression of KNAT3, a class 2 knotted1-like gene. Plant J. 1997;11:853–861. doi: 10.1046/j.1365-313x.1997.11040853.x. [DOI] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S. Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. A dominant mutation in the maize homeobox gene Knotted-1 causes its ectopic expression in leaf cell with altered fates. Development. 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M. Division and differentiation during normal and liguleless-1 maize leaf development. Development. 1990;110:9485–1000. doi: 10.1242/dev.110.3.985. [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Smith L, Freeling M. Acquisition of identity in the developing leaf. Annu Rev Cell Dev Biol. 1996;12:257–304. doi: 10.1146/annurev.cellbio.12.1.257. [DOI] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Phloem-specific expression of a plant homeobox gene during secondary phases of vascular development. Plant J. 1996;9:639–648. doi: 10.1046/j.1365-313x.1996.9050639.x. [DOI] [PubMed] [Google Scholar]
- Überlacker B, Klinge B, Werr W. Ectopic expression of the maize homeobox genes ZmHox1a and ZmHox1b causes pleiotropic alterations in the vegetative and floral development of transgenic tobacco. Plant Cell. 1996;8:349–362. doi: 10.1105/tpc.8.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]