Abstract
The chloroplast gene psbD encodes D2, a chlorophyll-binding protein located in the photosystem II reaction center. Transcription of psbD in higher plants involves at least three promoters, one of which is regulated by blue light. The psbD blue-light-regulated promoter (BLRP) consists of a −10 promoter element and an activating complex, AGF, that binds immediately upstream of −35. A second sequence-specific DNA-binding complex, PGTF, binds upstream of AGF between −71 and −100 in the barley (Hordeum vulgare) psbD BLRP. In this study we report that ADP-dependent phosphorylation selectively inhibits the binding of PGTF to the barley psbD BLRP. ATP at high concentrations (1–5 mm) inhibits PGTF binding, but in the presence of phosphocreatine and phosphocreatine kinase, this capacity is lost, presumably due to scavenging of ADP. ADP inhibits PGTF binding at relatively low concentrations (0.1 mm), whereas other nucleotides are unable to mediate this response. ADP-mediated inhibition of PGTF binding is reduced in the presence of the protein kinase inhibitor K252a. This and other results suggest that ADP-dependent phosphorylation of PGTF (or some associated protein) inhibits binding of PGTF to the psbD BLRP and reduces transcription. ADP-dependent phosphorylation is expected to increase in darkness in parallel with the rise in ADP levels in chloroplasts. ADP-dependent phosphorylation in chloroplasts may, therefore, in coordination, inactivate enzymes involved in carbon assimilation, protein synthesis, and transcription during diurnal light/dark cycles.
The chloroplast genes psbA and psbD encode D1 and D2, chlorophyll-binding proteins that comprise the reaction center of PSII. Expression of these and other plastid and nuclear genes involved in photosynthesis increases coordinately with leaf and chloroplast development (for review, see Mullet, 1988, 1993). Light regulates leaf and chloroplast development, together with overall chloroplast gene expression, through the action of red-light photoreceptors such as phytochrome (for review, see Pratt et al., 1997; Quail, 1997), and blue-light photoreceptors (cryptochromes) (Cashmore, 1997). Hence, mutation of genes in the photoreceptor signal transduction pathways, such as det1 (Chory and Peto, 1990), modifies chloroplast gene expression, including transcription of psbD (Christopher and Hoffer, 1998). In addition, light regulates the initial accumulation of D1, P700, and CP43 during development in higher plants through the light-dependent accumulation of chlorophyll, which is needed to stabilize chlorophyll-binding proteins (Mullet et al., 1990; Kim et al., 1994).
Once leaf and chloroplast biogenesis is complete, expression of most plastid genes decreases to levels needed for maintenance of the photosynthetic apparatus (Gamble et al., 1988). However, synthesis of D1 and D2 is maintained at relatively high levels in mature chloroplasts of developed leaves (Gamble et al., 1988; Christopher and Mullet, 1994). Maintenance of high rates of synthesis of these proteins is needed to replace D1 and D2 subunits that are damaged and turned over in illuminated plants (Mattoo et al., 1984, 1989; Ohad et al., 1985; Schuster et al., 1988; Christopher and Mullet, 1994). Elevated D1 synthesis in mature chloroplasts is paralleled by high levels of psbA RNA (Mullet and Klein, 1987; Gamble et al., 1988; Baumgartner et al., 1993). The relatively high abundance of psbA mRNA is due primarily to the unusual stability of these transcripts (Kim et al., 1993) and to a smaller extent to light-induced transcription (Klein and Mullet, 1990). Even though psbA mRNA levels are relatively constant in dark and light in mature chloroplasts, D1 synthesis is light dependent and regulated at the level of translation (Fromm et al., 1985; Malnoë et al., 1988).
Recent studies suggest that light regulates D1 synthesis by ADP-dependent phosphorylation (Danon and Mayfield, 1994a) and redox-regulated association of an RNA-binding protein complex with the psbA RNA 5′-untranslated region (Danon and Mayfield, 1994b). It is interesting that ADP-dependent phosphorylation was found much earlier to modulate the activity of enzymes involved in carbon assimilation in chloroplasts (Ashton and Hatch, 1983). For example, ADP-dependent phosphorylation of PPDK inactivates this enzyme when plants are transferred to darkness (Budde et al., 1986; Smith et al., 1994). Similarly, redox state is well known to regulate the organization of the light-harvesting chlorophyll proteins (for review, see Bennett, 1991; Allen and Nilsson, 1997; Gal et al., 1997). In this case, a redox-responsive protein kinase associated with the chloroplast thylakoid membrane phosphorylates PSII proteins to modify light harvesting and energy transfer among the photosystems.
The capacity for high rates of D2 synthesis is retained in mature chloroplasts primarily due to a psbD BLRP (Gamble and Mullet, 1989; Sexton et al., 1990). Transcription from this promoter requires a prokaryotic −10 promoter element but not a −35 element (Kim et al., 1999). The function of the −35 element is replaced by an activating complex, AGF, which binds to an AAG-box motif immediately upstream of −35 (Kim and Mullet, 1995). An additional DNA region located between −71 and −100 has also been shown to contribute to promoter activity in vivo (Allison and Maliga, 1995). Sequences in this region resemble the GT-motif found in light-regulated nuclear genes and the region was therefore named the PGT-box (Kim and Mullet, 1995). In this study we further investigate a protein complex that binds to the PGT-box and report that binding is inhibited by ADP-dependent phosphorylation. These data suggest that ADP-dependent phosphorylation in higher-plant chloroplasts modulates transcription in addition to translation and enzyme activity.
MATERIALS AND METHODS
Plant Growth and Plastid Isolation
Barley (Hordeum vulgare L. var Morex) seedlings were grown in controlled environmental chambers at 23°C, as described by Kim et al. (1993). Seedlings were germinated and grown in complete darkness. After 4.5 d the dark-grown seedlings were harvested under a dim-green safelight. Plastids were isolated from the top 4 cm of the primary leaves of barley seedlings by Percoll gradient (35%–75%) centrifugation (Klein and Mullet, 1986). The concentration of plastids was quantitated (plastids per microliter) by phase-contrast microscopy using a hemacytometer.
Preparation of Radiolabeled DNA Probes, Competitor Fragments, and Plastid Extracts
The plastid extracts and DNA fragments used as either radiolabeled probes or competitor fragments were prepared as described by Kim and Mullet (1995).
Gel-Retardation Assay
Experiments were done as described by Kim and Mullet (1995). Some of the binding reactions were performed after preincubation of plastid extracts with various chemicals or enzymes described in the figure legends (Ap5A was obtained from Boehringer Mannheim, K252a was from Calbiochem, and all of the other chemicals were from Sigma) for 10 min at room temperature. However, preincubation of plastid extracts with K252a was done in complete darkness due to the instability of K252a in the light. The concentration of the chemicals used is specified in the figure legends. Phosphocreatine kinase (type III) was diluted with DEAE buffer supplemented with 10 mm MgCl2 and 0.1% albumin, according to the supplier's instructions.
RESULTS
Analysis of psbD PGT-Box Binding Proteins
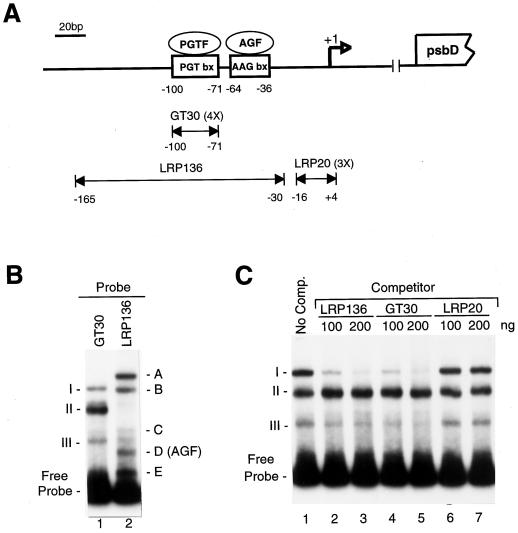
A diagram of the psbD BLRP appears in Figure 1A. We have previously shown that protein complexes bind specifically to the AAG-box and PGT-box sequences present in the psbD BRLP (Kim and Mullet, 1995). The protein complex that bound to the AAG-box, labeled AGF, was required for in vitro transcription from the psbD BLRP (Kim and Mullet, 1995). This was one of five protein complexes formed when LRP136, which contained the −30 to −165 region of the psbD BLRP, was radiolabeled and used in gel-shift assays (Fig. 1B, lane 2, [AGF corresponds to complex D]). All of the gel-shift bands were removed by protease treatment, indicating that they were formed by protein-DNA interactions (data not shown).
Figure 1.
Schematic representation of the barley chloroplast psbD BLRP. A, The top portion of the figure shows the location of the AAG-box and PGT-box present upstream of the psbD transcription initiation site (marked by an open arrow and designated +1). The psbD open reading frame is shown at the far right. The double-headed arrows represent DNA fragments from the psbD light-responsive promoter used for gel-retardation and competition-binding experiments. GT30 and LRP20 represent four (4X) and three (3X) tandem copies of each region indicated in the figure. B, Gel-shift assays using a GT30 tetramer (lane 1) or the LRP136 DNA fragment (lane 2). All gel-shift assays contained 1 μg of poly (dI–dC)·(dI–dC). Gel-shift complexes A to E and I to III are noted. Band D was previously identified as an AGF, which binds specifically to the AAG-box shown in A. C, Gel-shift assays using the radiolabeled tetramer of GT30 and various amounts (100–200 ng) of unlabeled LRP136, GT30, or LRP20 DNA. The migration of complexes I, II, and III and the free probe is indicated. No Comp., No competitor.
In a previous study we noted that the formation of complex A with LRP136 could be prevented by the addition of a tetramer of the sequence from −71 to −100 (Kim and Mullet, 1995). This 30-bp DNA sequence corresponds to the PGT-box and is referred to as GT30. Results in Figure 1B show that three protein complexes were formed when a tetramer of GT30 was added to plastid extracts from 4.5-d-old, dark-grown barley plants (Fig. 1B, lane 1). The addition of unlabeled LRP136 or GT30 before the gel-shift assays effectively competed for the binding of complex I and the GT30 tetramer (Fig. 1C, lanes 2–5). A nonspecific competitor DNA, LRP20, did not reduce the abundance of complex I (Fig. 1C, lanes 6 and 7). When a similar experiment was carried out using radiolabeled LRP136, GT30 and LRP136 were able to compete for binding to complex A (Kim and Mullet, 1995). Based on these results, we concluded that complex A formed with LRP136 and complex I formed with GT30 contained similar DNA-binding proteins that allowed specific binding to the PGT-box. The protein-binding complex that interacted with the PGT-box is labeled as PGTF in Figure 1A. Figure 1C also indicates that unlabeled LRP136 and GT30 did not significantly reduce the abundance of complex II or III, suggesting that binding in these complexes was nonspecific.
ADP Inhibits PGTF Binding to the PGT-Box
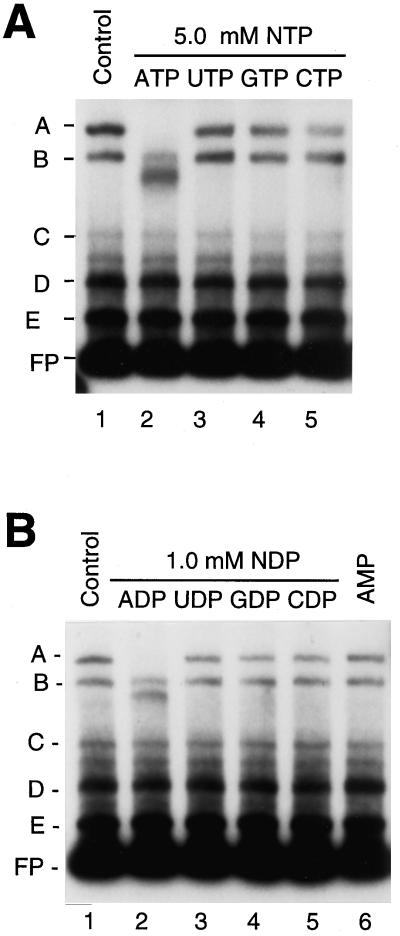
We tested the modulation of protein association with the psbD BLRP by protein phosphorylation or nucleotide binding by the addition of ATP and other ribonucleotides to plastid extracts before the gel-shift assays. We incubated plastid high-salt extracts obtained from 4.5-d-old, dark-grown barley with 5.0 mm ATP, UTP, GTP, or CTP for 10 min at room temperature, and then carried out gel-retardation assays using LRP136 as the DNA probe (Fig. 2A). Preincubation of plastid extracts with 5.0 mm ATP reduced the abundance of complex A (PGTF) and led to the appearance of a new complex that migrated just below complex B (Fig. 2A, lane 2). Incubation with UTP, GTP, and CTP had little effect on the abundance of complex A (Fig. 2A, lanes 3–5). Preincubation of the protein extracts with 1.0 mm ADP also caused a loss of complex A (Fig. 2B, lane 2). Other ribonucleoside diphosphates and AMP had little effect on the abundance of complex A (Fig. 2B, lanes 3–6).
Figure 2.
Gel-retardation experiments with plastid extracts preincubated with various nucleotides. Experiments in A and B were done with radiolabeled LRP136 DNA fragments and plastid extracts that were preincubated with either 5.0 mm ATP, UTP, GTP, or CTP (A, lanes 2–5, respectively) or 1.0 mm ADP, UDP, GDP, CDP, or AMP (B, lanes 2–6, respectively). Plastid extracts were obtained from 4.5-d-old, dark-grown barley plants. The major complexes observed with the preincubation of plastid extracts without any exogenous nucleotides (control reaction, lanes 1) are labeled A, B, C, D, and E. Migration of the free probe (FP) is indicated.
The nature of the new complex migrating just below complex B, which forms with LRP136 when PGTF (complex A) binding is reduced, is presently unknown (Fig. 2, lane 2). Kim and Mullet (1995) observed this complex previously, when the abundance of complex A was reduced by the addition of DNA fragments (GT30 or LRP136) that competed for PGTF binding. Previous studies also showed that this complex could not compete with unlabeled LRP136 or GT30, at least with normal levels of DNA used in competition assays. This suggests that the new complex was formed by either an abundant specific-binding complex (i.e. RNA polymerase) or nonspecific-binding proteins. Similar complexes were not formed when ATP, ADP, or competitor DNAs induced a similar reduction in the abundance of complex I formed with GT30 (Figs. 1C and 3B; Kim and Mullet [1995]). This indicates that the complex binding to LRP136 when PGTF binding was reduced must have interacted with a region of LRP136 outside of the GT30 domain.
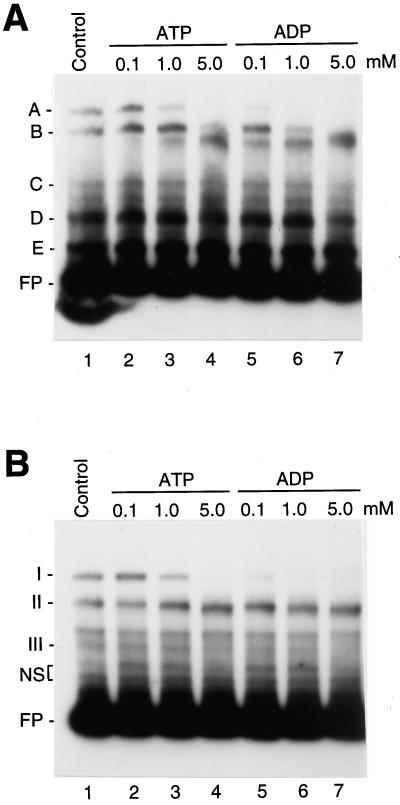
Figure 3.
Gel-retardation experiments with plastid extracts preincubated with various concentrations of either ATP or ADP. Experiments were done with either radiolabeled LRP136 (A) or GT30 (B) DNA fragments and plastid extracts that were preincubated with ATP (A, B, lanes 2–4) or ADP (A, B, lanes 5–7). The concentration of ATP and ADP used is indicated above the lanes. Plastid extracts were obtained from 4.5-d-old, dark-grown barley seedlings. The major complexes observed in plastid extracts without exogenous nucleotides (control reaction, lanes 1) are labeled A, B, C, D, and E (A) and I, II, and III (B). Migration of the free probe (FP) is indicated and the binding activity of nonspecific-binding proteins to the GT30 probe is designated NS.
The concentration dependence of ATP and ADP-induced loss of complex A (probe LRP136) and complex I (probe GT30) appears in Figure 3. We preincubated the plastid extracts obtained from 4.5-d-old, dark-grown barley with concentrations of either ATP or ADP (0.1–5.0 mm), and then carried out gel-retardation assays. The abundance of complex A and complex I decreased when extracts were incubated with 0.1 mm ADP; however, 1.0 mm ATP was required to cause a similar decrease in abundance of either complex A or complex I (Fig. 3). The ATP or ADP-induced reduction of complex I abundance (GT30) resulted in an increase in complex II (Fig. 3B). In addition, at 5 mm ADP the abundance of complex B also decreased (Fig. 3A, lane 7). The abundance of the other protein complexes formed with LRP136 and GT30 was not altered by ATP or ADP, showing that the response to nucleotides was selective.
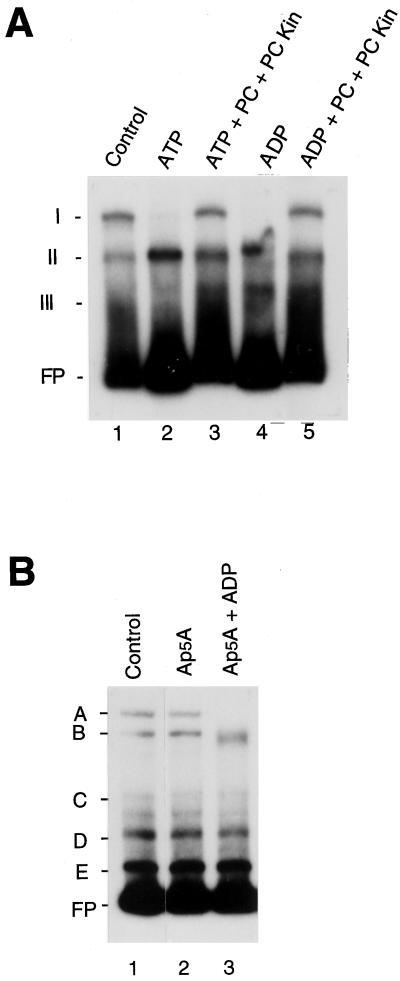
Plastid extracts probably have endogenous activity that converts nucleoside triphosphates to nucleoside diphosphates. Therefore, the ATP-induced loss of complex A binding could be due to ADP formed from ATP. We tested this possibility by adding phosphocreatine and phosphocreatine kinase to extracts to convert ADP into ATP. As shown in Figure 4, the addition of ATP to extracts in the presence of phosphocreatine and phosphocreatine kinase greatly reduced the ability of ATP to inhibit formation of complex I (Fig. 4, lane 2 versus lane 3). Preincubation of extracts with ADP in the presence of phosphocreatine and phosphocreatine kinase also reduced the ability of ADP to inhibit formation of complex I, presumably because ADP was converted into ATP. In addition, ADP-induced inhibition of complex A formation did not change when plastid extracts were preincubated with ADP in the presence of the adenylate kinase inhibitor, Ap5A (Fig. 4B). This inhibitor would have slowed the conversion of ADP to ATP by the endogenous adenylate kinase in plastid extracts. These results suggest that ADP rather than ATP was required to inhibit formation of complex A and complex I.
Figure 4.
Gel-retardation experiments showing the effects of the preincubation of plastid extracts with either ATP, ADP, phosphocreatine and phosphocreatine kinase, and/or an inhibitor of adenylate kinase. A, Gel-shift assays were done using radiolabeled GT30 DNA fragments and plastid extracts, which were preincubated with either 5.0 mm ATP or 1.0 mm ADP in the presence of 5.0 mm phosphocreatine (PC) and 2 units of phosphocreatine kinase (PC Kin) (lanes 3 and 5, respectively). Binding reactions using plastid extracts preincubated with 5.0 mm ATP or 1.0 mm ADP alone are shown in lanes 2 and 4, respectively. Plastid extracts were obtained from 4.5-d-old, dark-grown barley plants. The major complexes observed using the extracts without any additions (control reaction, lane 1) are labeled I, II, and III. Migration of the free probe (FP) is indicated. B, Gel-retardation experiments using radiolabeled LRP136 DNA probe testing the effect of preincubation of plastid extracts with ADP in the presence of the adenylate kinase inhibitor Ap5A. Plastid extracts were preincubated, before binding reactions, with 1.0 mm Ap5A in the presence (1.0 mm, lane 3) or absence (lane 2) of ADP. The major complexes observed in plastid extracts without additions are labeled A, B, C, D, and E (Control, lane 1). Migration of the free probe (FP) is indicated.
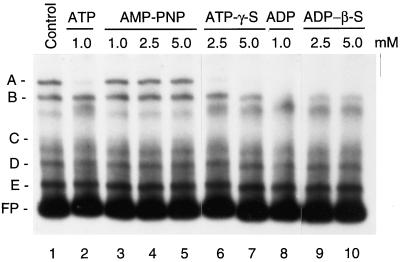
The nonhydrolyzable analog of ATP, AMP-PNP, did not inhibit formation of complex A (Fig. 5, lanes 3–5). In contrast, formation of complex A was reduced in the presence of ATP-γ-S and ADP-β-S (Fig. 5, lanes 6 and 7, and lanes 9 and 10, respectively). These results are consistent with a previous study that showed that these ATP/ADP analogs could be used as the substrates for ADP-dependent phosphorylation of the psbA RNA-binding complex (Danon and Mayfield, 1994a).
Figure 5.
Influence of ADP and ATP analogs on protein binding to the psbD BLRP. Plastid extracts were preincubated with ATP (lane 1), AMP-PNP (lanes 3–5), ATP-γ-S (lanes 6 and 7), ADP (lane 8), or ADP-β-S (lanes 9 and 10) prior to gel-shift assays using LRP136 as a probe. The concentration of the chemical used is indicated above the lanes. Binding experiments were done with plastid extracts obtained from 4.5-d-old, dark-grown barley plants. The major complexes observed without additions are labeled A, B, C, D, and E (control reaction, lane 1). Migration of the free probe (FP) is indicated.
An ADP-Dependent Protein Kinase Inhibits Formation of Complex A
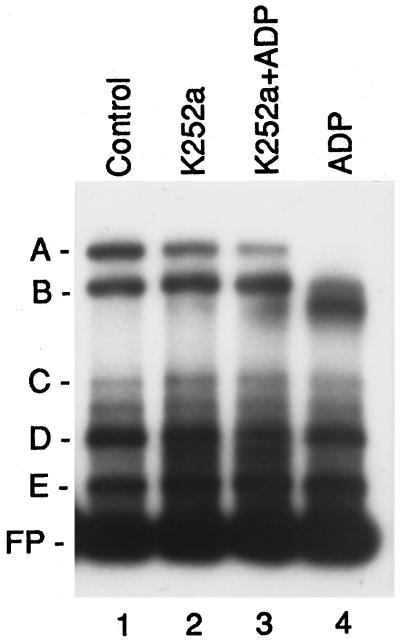
ADP could have inhibited complex A and complex I formation by binding or by the kinase-mediated phosphorylation of a protein that may have been associated with the complex. Dialysis of plastid extracts after incubation with ADP did not reverse the loss of complex A binding, suggesting that ADP was not acting by binding to complex A (data not shown). We tested the involvement of protein kinases by incubating plastid extracts with the general protein kinase inhibitor K252a. This inhibitor acts on various protein kinases, including Ser/Thr kinases, protein kinase C, cAMP- and cGMP-dependent protein kinases, and casein kinase II (Hashimoto, 1988). Treatment with inhibitor in the absence of ADP had no influence on the formation of complex A (Fig. 6, lane 2). However, K252a inhibited ADP-induced inhibition of complex A formation in plastid extracts (Fig. 6, lane 3 versus lane 4). This result indicates that an ADP-dependent protein kinase inhibited the formation of complex A.
Figure 6.
Gel-retardation experiments showing the effects of preincubation of plastid extracts with K252a. Gel-shift assays were carried out using radiolabeled LRP136 DNA fragment and plastid extracts obtained from 4.5-d-old, dark-grown barley plants. Before binding reactions, plastid extracts were preincubated either in the presence (lane 3) or absence (lane 4) of K252a (10 μm, final concentration), then further incubated with 1.0 mm ADP. The effect of the incubation of the plastid extracts with K252a alone was tested (lane 2). The major complexes observed with the preincubation of the extracts without additions are labeled A, B, C, D, and E (Control, lane 1). Migration of the free probe (FP) is indicated.
DISCUSSION
The results described here show that ADP-dependent phosphorylation inhibited the binding of PGTF to the chloroplast psbD BLRP in vitro. Inhibition of PGTF binding was caused by ADP rather than by ATP. This conclusion is based on several lines of evidence. First, ADP was able to reduce PGTF binding at a much lower concentration than ATP (threshold of 0.1 versus 1.0 mm). Second, the addition of phosphocreatine and phosphocreatine kinase, an ADP-scavenging system, to extracts in the presence of ATP inhibited the ability of ATP to modify PGTF binding. Third, ADP was able to inhibit PGTF binding in the presence of Ap5A, an inhibitor of adenylate kinase. Other ribonucleotides were unable to modify PGTF binding. Moreover, ADP only inhibited PGTF binding, suggesting that the modification was selective.
The ability of ADP to modify PGTF binding was inhibited by the protein kinase inhibitor K252a. This suggests that ADP-dependent protein phosphorylation modified PGTF binding either directly or indirectly. ADP-dependent phosphorylation of an active-site Thr was previously reported to have inactivated the stromal enzyme PPDK (Smith et al., 1994). In addition, ADP-dependent Thr phosphorylation inhibited the association of an RNA-binding complex with the psbA RNA 5′-untranslated region (Danon and Mayfield, 1994a). The psbA RNA-binding complex was modified by ADP-β-S and ATP-γ-S, but not by AMP-PNP (Danon and Mayfield, 1994a). Similarly, ADP-β-S and ATP-γ-S were both able to inhibit PGTF binding, whereas AMP-PNP was not (Fig. 5). It seems likely that in our plastid extracts ATP-γ-S is having its effect on PGTF binding after conversion to ADP. Danon and Mayfield (1994a) showed that a 60-kD protein in the psbA RNA-binding complex was phosphorylated on a Thr residue. Unfortunately, at present we do not have a method to isolate the PGTF and therefore have not been able to investigate the site and type of phosphorylation involved in the modulation of PGTF binding.
Diurnal regulation of PPDK activity was a consequence of ADP-dependent phosphorylation (inactivation) and phosphate-dependent dephosphorylation (activation) by a bifunctional kinase/phosphatase (Smith et al., 1994). Phosphorylation of PPDK increased in darkness in parallel with an increase in ADP concentration in chloroplasts, and dephosphorylation occurred upon plant illumination (Heber and Santarius, 1965; Hampp et al., 1982; Roeske and Chollet, 1989). Danon and Mayfield (1994a) also proposed that dark-induced, ADP-dependent phosphorylation of the psbA RNA-binding protein contributed to the inactivation of D1 translation in chloroplasts. In a similar way it seems likely that ADP-dependent phosphorylation of PGTF increased in darkness and caused a decrease in binding of this complex to the psbD BLRP. Deletion of the PGT-box in transgenic tobacco reduced the transcription from psbD BLRP 5-fold (Allison and Maliga, 1995). Therefore, it is reasonable to conclude that PGTF stimulated transcription in illuminated plants and ADP-dependent phosphorylation reduced binding and transcription from the psbD BLRP in darkness. The ability of K252a, an inhibitor of ADP-dependent phosphorylation, to stimulate transcription of psbD in dark-grown plants is consistent with this hypothesis (Christopher et al., 1997). Although this hypothesis is consistent with available information, we have not yet obtained consistent differences in the abundance of PGTF binding in extracts of dark-treated versus illuminated plants that confirm this idea. The action of kinases, phosphatases, and other factors during preparation of extracts or during binding assays may make this analysis subject to experimental variation. In the current study extracts from 4.5-d-old, dark-grown barley plants were used because they provided a relatively consistent amount of PGTF binding.
We expect that the ADP-dependent phosphorylation of PGTF increased in darkness, when levels of ADP were elevated (0.3–0.6 mm), relative to illuminated chloroplasts. The concentration of ADP in chloroplasts of dark-adapted plants (Heber and Santarius, 1965; Hampp et al., 1982) was sufficient to inhibit PGTF binding based on our in vitro analyses, which show that 0.1 mm ADP was required (Fig. 3). However, it is not clear that the 2-fold change in ADP concentration that normally occurs in chloroplasts during light/dark transitions is sufficient by itself to modulate PGTF binding in vivo. A similar question was raised concerning regulation of PPDK activity by ADP-dependent phosphorylation (Roeske and Chollet, 1989). Other factors such as metabolites, pH, redox potential, and other phosphorylation events may have modulated the susceptibility of PGTF to phosphorylation.
The psbD BLRP contains a −10 promoter element and an AAG-box sequence that binds the transcription activator, AGF (Kim and Mullet, 1995; Kim et al., 1999). This minimal 53-bp promoter domain was sufficient to drive transcription from the psbD BLRP in vitro. Sequences upstream of the AAG-box, including the PGT-box, had little influence on transcription from the psbD BLRP in vitro. The lack of influence of the PGT-box on transcription in vitro may have been due to the fact that binding of PGTF can be modulated by phosphorylation and other factors (i.e. phosphatases in the extracts) that are difficult to control under standard conditions used for in vitro transcription assays and during preparation of extracts. In addition, the influence of PGTF on transcription has only been assayed in extracts isolated from plants grown under a limited number of conditions.
In barley the PGT-box is located between −71 and −100 relative to the site of transcription initiation. The PGT-box sequence resembles regions (GT motifs) of several nuclear genes that are involved in light and circadian regulation (box II, box III, and I-box) (e.g. Green et al., 1988; Lam and Chua, 1990; Schindler and Cashmore, 1990; Borello et al., 1993; Kusnetsov et al., 1996; for review, see Gilmartin et al., 1990, 1991). However, nuclear GT motifs (i.e. box II; Green et al., 1988) and related sequences such as as-2, ga-1, and the CGF-binding elements (Gilmartin et al., 1991; Anderson et al., 1994) did not compete efficiently for PGTF binding to the PGT-box (data not shown).
Deletion of the 30-bp sequence corresponding to the PGT-box in tobacco (labeled A-rich) decreased the output from the psbD BLRP promoter 5-fold in vivo in plants grown several days in cycling light conditions (16-h light:8-h dark) (Allison and Maliga, 1995). In contrast, the same deletion had only a small effect on white light (24 h) reactivation of psbD transcription in vivo in plants placed in darkness for 3 d. These results indicate that the PGT-box played an important role in regulating transcription from the psbD BLRP, but that the physiological role and optimal conditions to assay the influence of this element have not yet been established. It is possible that PGTF acts by blocking read-through transcription from the upstream psbD promoter that is active in dark-grown plants (Sexton et al., 1990). Similarly, PGTF may decrease transcription from trnS, which is located immediately upstream of the psbD BLRP, and read from the opposite DNA strand (Sexton et al., 1990). It is also possible that PGTF represents a more global regulator of chloroplast transcription that couples transcription of genes encoding proteins involved in photosynthesis in mature chloroplasts with the higher energy state present in illuminated plants. This activity may have complemented phosphorylation (inactivation) of sigma factors by an ATP-dependent kinase that occurred in dark-grown mustard seedlings (Tiller and Link, 1993). If PGTF is involved in global regulation, we would expect additional binding sites for PGTF in the chloroplast genome. This aspect of PGTF function is currently being analyzed through site-directed mutagenesis to better define the binding site and by searching other promoters for protein complexes that are modulated by ADP-dependent phosphorylation.
ACKNOWLEDGMENTS
The authors thank Ueli Klahre in Dr. Nam-Hai Chua's laboratory and Dr. Steve Kay for sending recombinant plasmids containing multimers of box II, as-2, ga-1, and CGF-binding elements.
Abbreviations:
- AMP-PNP
adenylyl-imidodiphosphate
- Ap5A
P1,P5-Di(adenosine-5′)pentaphosphate
- ATP-γ-S and ADP-β-S
γ and β forms of adenosine 5′-O-(3-thiotriphosphate), respectively
- BLRP
blue-light-regulated promoter
- PPDK
pyruvate orthophosphate dikinase
Footnotes
This research was supported by the National Institutes of Health (grant no. GM 37987 to J.E.M.) and by the Texas Agricultural Experiment Station.
LITERATURE CITED
- Allen JF, Nillson A. Redox signalling and the structural basis of regulation of photosynthesis by protein phosphorylation. Physiol Plant. 1997;100:863–868. [Google Scholar]
- Allison LA, Maliga P. Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 1995;14:3721–3730. doi: 10.1002/j.1460-2075.1995.tb00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA. Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 1994;6:457–470. doi: 10.1046/j.1365-313x.1994.6040457.x. [DOI] [PubMed] [Google Scholar]
- Hatch ARM, Ashton D. Regulation of C4 photosynthesis: regulation of pyruvate, Pi dikinase by ADP-dependent phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 1983;115:53–60. doi: 10.1016/0006-291x(83)90967-1. [DOI] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol. 1993;101:781–791. doi: 10.1104/pp.101.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. Protein phosphorylation in green plant chloroplasts. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:281–311. [Google Scholar]
- Borello U, Ceccarelli E, Giuliano G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different I box elements in plant light-regulated promoters. Plant J. 1993;4:611–619. doi: 10.1046/j.1365-313x.1993.04040611.x. [DOI] [PubMed] [Google Scholar]
- Budde RJA, Ernst SM, Chollet R. Substrate specificity and regulation of the maize (Zea mays) leaf ADP:protein phosphotransferase catalysing phosphorylation/inactivation of pyruvate, orthophosphate dikinase. Biochem J. 1986;236:579–584. doi: 10.1042/bj2360579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR. The cryptochrome family of photoreceptors. Plant Cell Environ. 1997;20:764–767. [Google Scholar]
- Chory J, Peto CA. Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA. 1990;87:8776–8780. doi: 10.1073/pnas.87.22.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Hoffer PH. DET1 represses a chloroplast blue light-responsive promoter in a developmental and tissue-specific manner in Arabidopsis thaliana. Plant J. 1998;14:1–11. doi: 10.1046/j.1365-313x.1998.00078.x. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. Separate photosensory pathways co-regulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Xinli L, Kim M, Mullet JE. Involvement of protein kinase and extraplastidic serine/threonine protein phosphatases in signaling pathways regulating plastid transcription and the psbD blue light-responsive promoter in barley. Plant Physiol. 1997;113:1273–1282. doi: 10.1104/pp.113.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J. 1994a;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994b;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Fromm H, Devic M, Fluhr R, Edelman M. Control of psbA gene expression: in mature Spirodelra chloroplasts light regulation of 32-kd protein synthesis is independent of transcript level. EMBO J. 1985;4:291–295. doi: 10.1002/j.1460-2075.1985.tb03628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Zer H, Ohad I. Redox-controlled thylakoid protein phosphorylation [news and views] Physiol Plant. 1997;100:869–885. [Google Scholar]
- Gamble PE, Mullet JE. Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 1989;8:2785–2794. doi: 10.1002/j.1460-2075.1989.tb08424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble PE, Sexton TB, Mullet JE. Light-dependent changes in psbD and psbC transcripts of barley chloroplasts: accumulation of two transcripts maintains psbD and psbC translation capability in mature chloroplasts. EMBO J. 1988;7:1289–1297. doi: 10.1002/j.1460-2075.1988.tb02943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Memelink J, Chua N-H (1991) Dissection of the light-responsive elements of pea RBCS3A. In B Thomas, CB Johnson, eds, Phytochrome Properties and Biological Action, Vol H-50. Springer-Verlag, Berlin, pp 141–155
- Gilmartin PM, Sarokin L, Memelink J, Chua N-H. Molecular light switches for plant genes. Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Yong M-H, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua N-H. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988;7:4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp R, Goller M, Ziegler H. Adenylate levels, energy charge, and phosphorylation potential during dark-light and light-dark transition in chloroplasts, mitochondria, and cytosol of mesophyll protoplasts from Avena sativa L. Plant Physiol. 1982;69:448–455. doi: 10.1104/pp.69.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S. K252a, a potent protein kinase inhibitor, blocks nerve growth factor-induced neurite outgrowth and changes the phosphorylation of proteins in PC12h cells. J Cell Biol. 1988;107:1531–1539. doi: 10.1083/jcb.107.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber UW, Santarius KA. Compartmentation and reduction of pyridine nucleotides in relation to photosynthesis. Biochim Biophys Acta. 1965;109:390–408. doi: 10.1016/0926-6585(65)90166-4. [DOI] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol. 1993;22:447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- Kim J, Eichacker LA, Rudiger W, Mullet JE. Chlorophyll regulates accumulation of the plastid-encoded chlorophyll proteins P700 and D1 by increasing apoprotein stability. Plant Physiol. 1994;104:907–916. doi: 10.1104/pp.104.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Mullet JE. Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light-responsive psbD-psbC promoter. Plant Cell. 1995;7:1445–1457. doi: 10.1105/tpc.7.9.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Thum K, Morishige D, Mullet JM (1999) Detailed architecture of the barley chloroplast psbD-psbC blue light-responsive promoter. J Biol Chem (in press) [DOI] [PubMed]
- Klein RR, Mullet JE. Regulation of chloroplast-encoded chlorophyll-binding protein translation during higher plant chloroplast biogenesis. J Biol Chem. 1986;261:11138–11145. [PubMed] [Google Scholar]
- Klein RR, Mullet JE. Light-induced transcription of chloroplast genes: psbA transcription is differentially enhanced in illuminated barley. J Biol Chem. 1990;265:1895–1902. [PubMed] [Google Scholar]
- Kusnetsov V, Bolle C, Lübberstedt T, Sopory S, Hermann RG, Oelmüller R. Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol Gen Genet. 1996;252:631–639. doi: 10.1007/BF02173968. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua N-H. GT-1 binding site confers light responsive expression in transgenic tobacco. Science. 1990;248:471–474. doi: 10.1126/science.2330508. [DOI] [PubMed] [Google Scholar]
- Malnoë P, Mayfield SP, Rochaix J-D. Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J Cell Biol. 1988;106:609–616. doi: 10.1083/jcb.106.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M. Regulation of protein metabolism: coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc Natl Acad Sci USA. 1984;81:1380–1384. doi: 10.1073/pnas.81.5.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Marder JB, Edelman M. Dynamics of the photosystem II reaction center. Cell. 1989;56:241–246. doi: 10.1016/0092-8674(89)90897-0. [DOI] [PubMed] [Google Scholar]
- Mullet JE. Chloroplast development and gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. [Google Scholar]
- Mullet JE. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet JE, Klein PG, Klein RR. Chlorophyll regulates accumulation of the plastid-encoded chlorophyll apoproteins CP43 and D1 by increasing apoprotein stability. Proc Natl Acad Sci USA. 1990;87:4038–4042. doi: 10.1073/pnas.87.11.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet JE, Klein RR. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I, Kyle DJ, Hirschberg J. Light-dependent degrada-tion of the QB-protein in isolated pea thylakoids. EMBO J. 1985;4:1655–1659. doi: 10.1002/j.1460-2075.1985.tb03833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LH, Cordonnier-Pratt M-M, Kelmenson PM, Lazarova GI, Kubota T, Alba RM. The phytochrome gene family in tomato (Solanum lycopersicum L.) Plant Cell Environ. 1997;20:672–677. [Google Scholar]
- Quail PH. An emerging molecular map of the phytochromes. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- Roeske CA, Chollet R. Role of metabolites in the reversible light activation of pyruvate, orthophosphate dikinase in Zea mays mesophyll cells in vivo. Plant Physiol. 1989;90:330–337. doi: 10.1104/pp.90.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Cashmore AR. Photoregulated gene expression may involve ubiquitous DNA binding proteins. EMBO J. 1990;9:3415–3427. doi: 10.1002/j.1460-2075.1990.tb07549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G, Timberg R, Ohad I. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem. 1988;177:403–410. doi: 10.1111/j.1432-1033.1988.tb14389.x. [DOI] [PubMed] [Google Scholar]
- Sexton TB, Christopher DA, Mullet JE. Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 1990;9:4485–4494. doi: 10.1002/j.1460-2075.1990.tb07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Duff SMG, Chollet R. Partial purification and characterization of maize-leaf pyruvate, orthophosphate dikinase regulatory protein: a low-abundance, mesophyll-chloroplast stromal protein. Arch Biochem Biophys. 1994;308:200–206. doi: 10.1006/abbi.1994.1028. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapsis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]