Abstract
In humans, inclusion or exclusion of the fibronectin EDA exon is mainly regulated by a polypurinic enhancer element (exonic splicing enhancer [ESE]) and a nearby silencer element (exonic splicing silencer [ESS]). While human and mouse ESEs behave identically, mutations introduced into the homologous mouse ESS sequence result either in no change in splicing efficiency or in complete exclusion of the exon. Here, we show that this apparently contradictory behavior cannot be simply accounted for by a localized sequence variation between the two species. Rather, the nucleotide differences as a whole determine several changes in the respective RNA secondary structures. By comparing how the two different structures respond to homologous deletions in their putative ESS sequences, we show that changes in splicing behavior can be accounted for by a differential ESE display in the two RNAs. This is confirmed by RNA-protein interaction analysis of levels of SR protein binding to each exon. The immunoprecipitation patterns show the presence of complex multi-SR protein-RNA interactions that are lost with secondary-structure variations after the introduction of ESE and ESS variations. Taken together, our results demonstrate that the sequence context, in addition to the primary sequence identity, can heavily contribute to the making of functional units capable of influencing pre-mRNA splicing.
The splicing process is a very flexible and critical step in gene expression. In fact, selected removal or inclusion of individual exons from nascent mRNA molecules allows a single gene to generate multiple proteins with different primary structures; in these cases, the process is known as alternative splicing (1, 38). Constitutive and alternative splicing processes are catalyzed by the spliceosome, a very complex RNA-protein aggregate that has been recently estimated to contain approximately 145 different proteins in addition to the five spliceosomal snRNAs (1, 38, 41, 68). These factors are responsible for accurate positioning of the spliceosome on the 5′ and 3′ splice sequences that define the exon. However, correct positioning of the spliceosome is a very complex process owing to the degeneracy of the splice site consensus sequences, the presence of cryptic splice sites in large introns, and the fact that most pre-mRNAs contain multiple introns (26). Therefore, the action of several different proteins is required to achieve accurate positioning of the spliceosome on the splice site. Not surprisingly, alterations in the splicing process have been increasingly reported as being involved in many genetic diseases (5, 8, 13, 23, 58). Among the well-known factors that may heavily influence the identification of intron-exon boundaries by the spliceosome are the exon length (3, 65), the presence of splicing enhancer and silencer elements (5, 38), the strength of splicing signals (26), and the promoter architecture (19, 33). In addition to these factors, it has been proposed that the natural tendency of RNAs to fold in highly stable secondary and tertiary structures can potentially influence splicing activity (2, 4, 17, 21, 43, 49, 50). Recently, changes in RNA structure have been suggested to play a role in pathogenetic processes involving the tau gene (27, 31, 66) and the dystrophin gene (48).
In the fibronectin (FN) gene, the influence of secondary structure has been suggested to play an important role in determining alternative splicing efficiency (52). This gene is a useful system for investigation of alternative splicing because it occurs in three developmentally regulated but noncoordinated sites (EDA and EDB exons and the IIICS region), giving rise to at least 20 different polypeptides in humans (24, 37, 47). Furthermore, the functional relevance of these alternative splicing events has been recently shown in a genetically engineered mouse model devoid of regulated EDA splicing (53). In particular, inclusion of the EDA exon is totally dependent on the presence of an 81-nucleotide (nt) sequence located within its central region (42) that was subsequently found to contain two cis-acting elements, a purine-rich human exonic splicing enhancer (hESE [5′ GAAGAAGA 3′]) and a human exonic splicing silencer (hESS [5′ CAAGG 3′]), located 13 nt downstream of the ESE (10). Aside from their enhancing or silencing function, these two elements are also very different in context dependence. In fact, while the function of the hESE element can be exported to at least some different exonic contexts, the hESS function was observed to strictly depend on the complete EDA exonic context (52). Even more strikingly, we have reported that in a hybrid exon containing both EDA and EDB exon sequences, the hESS element acquires the properties of an enhancer sequence (52). Our results thus raised the possibility that the function of the hESS element in the EDA context might be to maintain the correct display of the ESE sequence rather than to bind specific splicing regulatory proteins (52). Indeed, the importance of RNA secondary structure in the regulation of FN alternative splicing has also been proposed for another region of the EDA exon (64) and a report investigating the splicing of the EDB exon has indicated the presence of an exonic splicing enhancer element(s) that may be context dependent (39) in addition to the well-characterized intronic enhancer element (29).
Comparative study of functional sequences through evolution has been a very useful tool with which to understand mechanisms and structures that otherwise elude interpretation (25, 36). Recently, this approach has yielded important information regarding the evolution of the FGF-R2 gene through alternative splicing of the mutually exclusive IIIb and IIIc units (49). Hence, our interest in analyzing the EDA ESE function in a highly homologous system, such as the mouse. As expected, the mouse and human exons have a very high level of nucleotide sequence similarity and the difference between the two sequences consists of only eight nucleotide substitutions. This similarity is also reflected at the functional level, as the hESE and mouse ESE (mESE) sequences have already been reported to be interchangeable (54).
In the present study, we have characterized the effects of introducing similar mutations into the mouse and hESS homologous regions. Our results show that these mutations introduced into the mouse and human sequences do not have the same effect on the regulation of alternative splicing, with the putative mouse ESS (mESS) sequence behaving as a splicing enhancer. This unexpected finding can be accounted for by the existence of significant differences in the two RNA secondary structures that, following mutation, can differentially affect ESE display. These results were confirmed by immunoprecipitation (IP) analysis, which showed a correlation between SR protein occupancy of the ESE sequences and RNA secondary-structure predictions.
MATERIALS AND METHODS
Plasmid construction.
Constructs hTot, hΔ2e, hΔ4, mTot, and mΔA have been previously described (10, 54). The mΔB5, mΔB6, AhΔB5, AhΔB5AhTh, AhΔB53h, AmΔB53h, and m/hΔ4Cm mutations were introduced by PCR-directed mutagenesis starting from the mouse EDA minigene as previously described (54). Plasmid ESEx2 was obtained by cloning into the PstI site of pBluescript II SK+ the sequence 5′ CTGATGGTGAAGAAGACACTGCACCTGATGGTGAAGAAGACACTGCAG 3′ in such a way as to allow multiple insertions. Sequence analysis confirmed the presence of two inserts in the correct orientation. Plasmid NF-1 IVS37 was obtained by amplification of a 170-nt-long intronic region from nt 131 to nt 305 of IVS37 in the NF-1 gene by using sense and antisense primers 5′ CCTCTTAAAGTTTAGTTGTT 3′ and 5′ ACAGTACTTGGCAATAGCAGATAA 3′.
Cell culture, transfection, and RT-PCR analysis.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 50 mg of gentamicin per ml, and 4 mM l-glutamine. Approximately 1.6 × 106 cells were transfected with 5 μg of specific plasmid purified by CsCl gradient centrifugation and 3 μg of T antigen expression plasmid (pb5′sVBglII) by means of a modified calcium phosphate precipitation method (32) or DOTAP reagent (Boehringer Mannheim) as suggested by the manufacturer. Cells were harvested 36 h after transfection. Total RNA was then prepared from the transfected cells by a single-step extraction method with RNAzol TMB (TEL-TEST Inc.). A standard reverse transcription (RT) protocol with Moloney murine leukemia virus from Gibco/BRL was used. To perform the RT-PCR assay for analysis of the alternatively spliced products, the cDNA was synthesized with an oligo(dT) primer and then amplified by PCR with the primers previously described for the human and mouse minigenes (10, 54). Four independent transfections were carried out for each construct.
Enzymatic analysis of RNA secondary structure.
Single-strand-specific (S1 nuclease and T1 RNase) and double-strand-specific (V1 RNase) enzymes were used to analyze the secondary structure of the in vitro-transcribed RNAs. Single-stranded regions were identified by cleavage with RNase T1 (which cleaves in correspondence to guanosine residues present in single-strand regions) and nuclease S1 (which cleaves single-stranded nucleic acids without base specificity). Double-stranded or stacked regions were also identified by cleavage with RNase V1 (which does not possess any base specificity). A sequencing reaction performed with the same primer used in the RT analysis was used to fit all of the observed cleavage sites in the primary nucleotide sequence. The complete mouse sequences originally inserted into the pSVED-A vectors were cloned into the SmaI site of the pBluescript II SK+ plasmid and transcribed with T7 RNA polymerase (Pharmacia Biotech). Enzymatic digestion was performed essentially as previously described (52). Briefly, reaction mixtures contained 1 μg of RNA and 0.02 U of RNase V1 (Pharmacia Biotech), 0.5 U of RNase T1 (Sigma), or 20 U of S1 nuclease (Pharmacia Biotech) and were incubated at 30°C for 15 min. A control aliquot of RNA without the addition of RNases was processed simultaneously with the digested samples. The RNase cleavage sites were identified by primer extension with a 32P-end-labeled oligonucleotide primer (5′ GGGTGGTGGGTGCGTTAA 3′), and the RT reaction products were loaded onto a 6% polyacrylamide sequencing gel and exposed to Kodak X-Omat AR film for 12 to 24 h. The results were then used to optimize the RNA secondary-structure predictions performed by the mFold program (available at The European mFold Server [http://bibiserv.techfak.uni-bielefeld.de/mfold/]) (70).
IP of SR proteins following UV cross-linking.
The RNA probes used in this study (hTot, hΔ2e, hΔ4, mTot, mΔA, mΔB5, mΔB6, and NF-1 IVS37) were obtained by cloning into pBluescript KS+ the human and mouse sequences both wild type and carrying the different mutations. In order to avoid possible interference with other factors binding to the first half of the mouse and human EDA exon, only the sequences from nt 107 to nt 270 of the human exon and from nt 103 to nt 270 of the mouse exon were cloned. Each plasmid was then linearized with the appropriate restriction enzyme and transcribed with T7 RNA polymerase in accordance with standard procedures. Plasmid ESEx2 was transcribed with T3 RNA polymerase and T7 RNA polymerase (to obtain a control RNA). UV cross-linking of [α-32P]UTP-labeled RNAs with commercial HeLa nuclear extract (C4; Biotech) was performed as described before (7). To each sample we added 150 μl of IP buffer (20 mM Tris [pH 8.0], 300 mM NaCl, 1 mM EDTA, 0.25% NP-40) together with 1 μg of monoclonal antibodies (MAbs) and incubated the mixture for 2 h at 4°C on a rotator wheel. Anti-SF2/ASF (MAb 96) and anti-SR phosphorylated domain (MAb 1H4) MAbs were purchased from Zymed Laboratories Inc., while anti-SC35 MAb was purchased from Sigma. After the 2-h incubation, we added to each sample 30 μl of Protein A/G-Plus Agarose (Santa Cruz Biotechnologies) and incubated the mixture at 4°C overnight. On the following morning, the beads were subjected to four washing cycles with 1.5 ml of IP buffer and then loaded onto a sodium dodecyl sulfate (SDS)-11% polyacrylamide gel electrophoresis (PAGE) gel. Gels were run at a constant 30 mA for approximately 3.5 h, dried, and then exposed for 4 to 6 days with a BioMax Screen (Kodak). Three independent IP assays were carried out for each antibody. Competition experiments were performed by adding unlabeled RNA in a 5 M excess to the reaction mixture before addition of the labeled hTot RNA. IPs were then performed as already described.
RESULTS
Comparison of alternative splicing regulation in the mouse and human EDA sequences following homologous deletions.
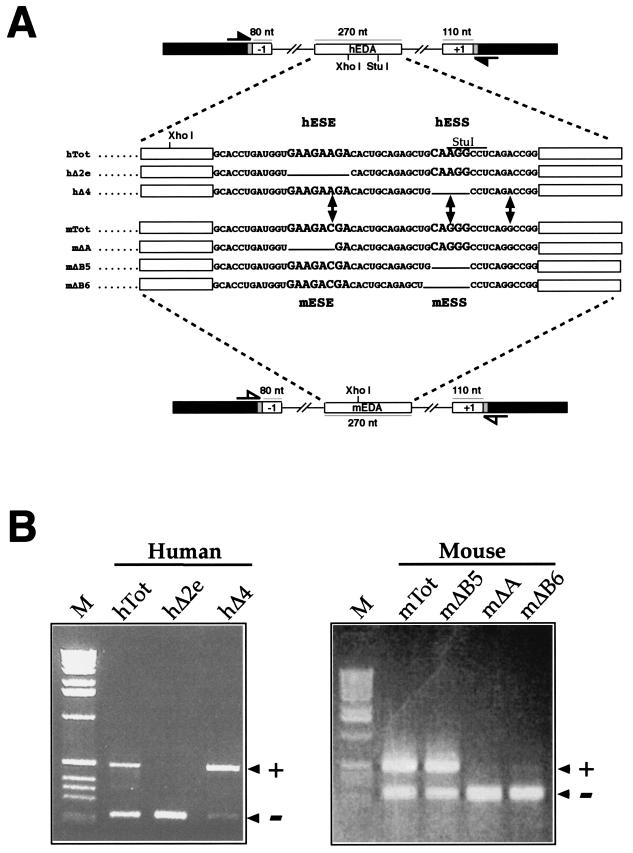
To compare the alternative splicing regulation of the human and mouse FN EDA exons, we used an FN-α-globin hybrid minigene, named pSVαGlob (10), into which we inserted the mouse and human EDA exons (mTot and hTot, respectively) together with flanking introns and exons (Fig. 1A). This is a well-proven system for measurement of the levels of in vivo alternative splicing of the FN EDA exon in different cellular systems and has been used extensively in previous studies of both human and mouse EDA exons (9, 10, 12, 52, 54). Figure 1B shows the effect on the alternative splicing of the EDA exon of analogous deletions in the region surrounding the mESE, mESS, hESE, and hESS elements. It can be seen that deletion of both hESE and mESE polypurinic sequences (hΔ2e and mΔA mutants) causes complete exon skipping from the resulting mRNA. Thus, both hESE and mESE act as exonic splicing enhancers, a result that has been previously described by our laboratory (10, 54). However, when we deleted the putative mouse silencer element (identified solely on the basis of homology with the human sequence), we did not observe the same effects. In fact, while a deletion in the hESS element (hΔ4 mutant) causes almost total exon inclusion, the identical mutation in mESS (mΔB5 mutant) does not appear to have any effect on the levels of exon inclusion (Fig. 1B). Even more strikingly, deletion of a single additional nucleotide in the mESS element (mΔB6) causes total exon exclusion. This result was identical to that observed when the mESE element had been deleted (mΔA mutant), thus identifying the putative mESS sequence as possessing enhancer characteristics.
FIG.1.
Changes in mouse and human FN EDA alternative splicing caused by the introduction of deletions into the ESE and ESS regulatory regions. (A) Schematic representation of the human and mouse minigenes. The FN exons and introns are labeled by white boxes and lines, linker sequences are shadowed, and α-globin exon 3 is indicated by black boxes. The locations of the primers used in the RT-PCR assay are shown: closed arrows indicate human-specific primers, while open arrows indicate mouse-specific primers. The sequence of the exonic region involved in EDA splicing regulation is reported for the human wild-type minigene (hTot) and mutants carrying a deletion of the ESE (hΔ2e), ESS (hΔ4) or for the mouse wild-type minigene (mTot) and mutants carrying a deletion of the mESE (mΔA) or mESS (mΔB5 and mΔB6). The respective ESE (hESE and mESE) and ESS (hESS and mESS) sequences are in bigger characters. (B) RT-PCR analysis of total RNA from cells expressing each of the indicated constructs in the NIH 3T3 cell line. PCR products either containing (+) or lacking (−) the FN EDA exon in the messenger transcribed from the transfected minigene are indicated. M, molecular weight markers (1 kb; BRL).
Effects of stepwise changes in the mΔB5 sequences toward their hΔ4 homologue.
The observed changes in splicing behavior may be caused by the effects of one (or more) nucleotide substitutions on putative SR binding sites. In fact, in many experimental systems, single nucleotide substitutions have been known to affect ESE elements (11, 13, 14, 44). This may be particularly relevant for the EDA exon, as proteins belonging to the SR protein family have a well-known ability to alter EDA inclusion in the mature mRNA (19, 40, 42). The use of SR scoring matrices to identify potential SR binding sites has been very useful in functionally characterizing several disease-causing mutations in the BRCA1 and SMN1 genes, as recently reviewed (13). Therefore, we decided to apply the available scoring matrices for SF2/ASF, SC35, SRp40, and SRp55 to the human and mouse sequences (scoring matrices are available at http://exon.cshl.org/ESE/ESEmatrix.html) (15). The results showed that the 8-nt differences between the human and mouse EDA exon sequences may have resulted in changes in either the positioning or scoring result of several putative SR protein binding sites (data not shown).
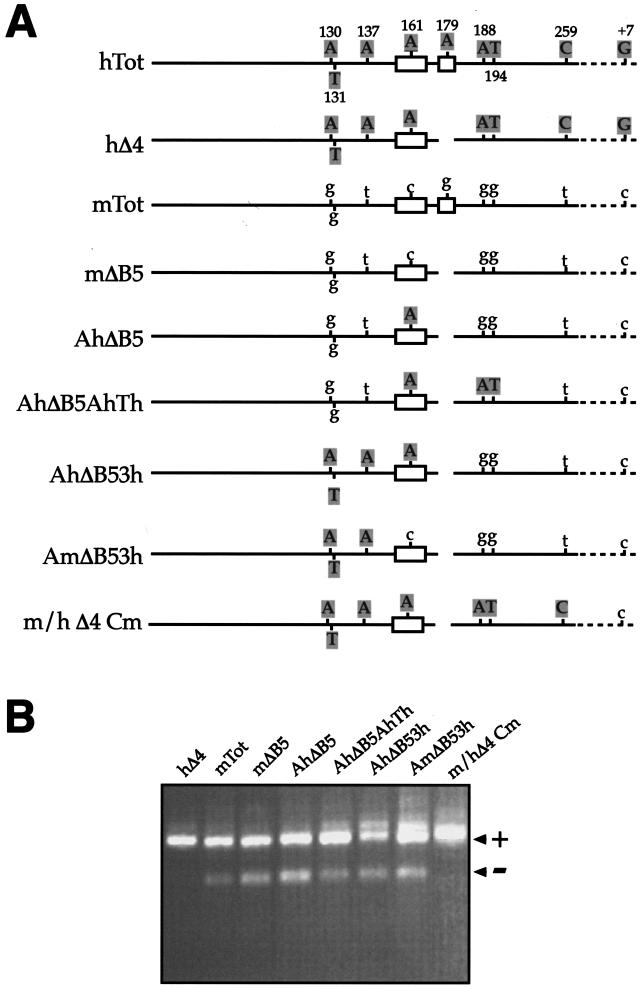
In order to evaluate experimentally the importance of these potential binding site changes, we decided to progressively “humanize” the exon sequence of mΔB5 toward the hΔ4 sequence. The rationale for this mutagenesis approach was that if any of these exonic single-nucleotide substitutions was responsible for the observed changes in splicing behavior (through either creation or deletion of SR protein binding sites), then we would expect some mutants to display the hΔ4 splicing pattern (Fig. 1B). Figure 2A shows a schematic diagram of the mutants engineered to test this hypothesis (AhΔB5, AhΔB5AhTh, AhΔB53h, and AmΔB53h). These mutants were transfected in NIH 3T3 cells, and by RT-PCR, we analyzed the effect on EDA exon inclusion. Figure 2B shows that none of the mutants acquires the hΔ4 splicing pattern (all included) and that the nucleotide substitutions, at most, cause small shifts in the EDA ± ratio. Only a complete change of all of the exonic nucleotides to the human sequence restores the hΔ4 pattern, as shown for the m/hΔ4Cm mutant. It should be noted that in this construct the human G-to-C substitution at position +7 of the 5′ splice site is kept unchanged, also ruling out any potential effect of differing 5′ splice site relative strengths in the generation of the hΔ4 splicing pattern. It should also be noted that this mutational analysis also rules out the possible influence of additional SR proteins (for which no matrices are currently available) or the presence of more elusive regulatory sequences such as the composite enhancer and silencer elements recently identified in the cystic fibrosis transmembrane regulator (57, 59). It is thus clear that the reason why mESS and hESS behave differently must lie elsewhere.
FIG.2.
Effects on the splicing process of gradually humanizing the mΔB5 mouse sequence to the hΔ4 sequence. (A) Schematic diagram of each mutant analyzed with the minigene system in NIH 3T3 cells. Open boxes represent the ESE and ESS sequences, respectively. Uppercase letters represent human nucleotides, while lowercase letters denote mouse nucleotides. On the human sequence, nucleotides are numbered. Straight lines represent exonic sequences, while broken lines represent intronic sequences. For clarity, the diagrams have not been drawn to scale. (B) RT-PCR analysis of total RNA from cells expressing each of the indicated constructs in the NIH 3T3 cell line. PCR products either containing (+) or lacking (−) the FN EDA exon in the messenger transcribed from the transfected minigene are indicated.
Determining the RNA secondary structure of the mouse EDA exon.
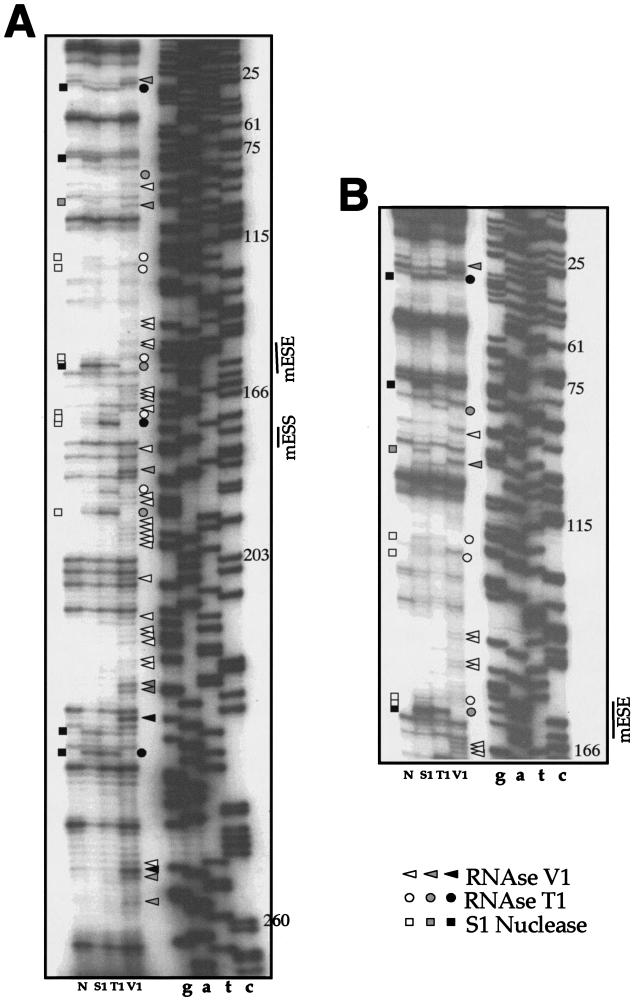
Previous studies performed in our laboratory had already indicated that the function of the hESS element in the human EDA exon is totally dependent on its original human context (52). Thus, the potential existence of structural differences between the human and mouse sequences might well result in different behaviors of the two RNAs when they are subjected to similar deletions. To further analyze this issue, it was first necessary to determine the wild-type mouse secondary structure. To experimentally determine which of the mouse EDA secondary structures predicted by computer analysis with the mFold program (69-71) was closer to reality, we performed an RNase mapping analysis of the mouse EDA exon (Fig. 3A and B). Single-stranded regions were identified by cleavage with RNase T1 and nuclease S1, while double-stranded or stacked regions were identified by cleavage with RNase V1. The results (Fig. 4B) were then compared with our previously published secondary structure of the human EDA exon (52) (Fig. 4A).
FIG.3.
Enzymatic determination of the RNA secondary structure of the mouse EDA exon. In vitro-transcribed mTot RNA was enzymatically digested with S1 nuclease and RNases T1 and V1 and reverse transcribed with an antisense primer. The RT products were separated on a sequencing polyacrylamide gel. A sequencing reaction (numbered according to the exon length) performed with the same primer was run in parallel with the cleavages in order to precisely determine the cleavage sites. In order to cover the whole exon length, short (A) and long (B) runs were performed. Squares, circles, and triangles indicate S1 nuclease and RNase T1 and V1 cleavage sites, respectively. Black, shaded, and white symbols indicate high, medium, and low cleavage intensities, respectively. No enzyme was added to the reaction mixture in lane N. The mESS and mESE positions are marked on the right of each sequencing reaction.
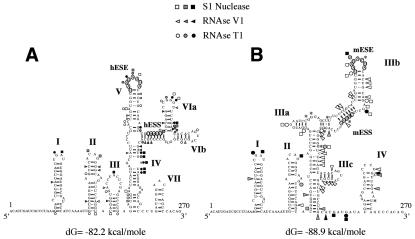
FIG. 4.
Comparison of the RNA secondary structure of the human (A) and mouse (B) wild-type EDA sequences. The two structures were optimized by computer-assisted RNA modeling, and the respective ESE and ESS elements are circled to facilitate their localization. Squares, circles, and triangles indicate S1 nuclease and RNase T1 and V1 cleavage sites, respectively. Black, shaded, and white symbols indicate high, medium, and low cleavage intensities, respectively. The asterisks in the mouse secondary-structure model mark the nucleotide differences between the mouse and human nucleotide sequences.
Figure 4A and B show that the cleavages in the first half of the exon are very well conserved, a result that is in keeping with the absence of nucleotide differences in the first half of the mouse and human EDA exons. This is also consistent with a previous phylogenetic analysis that indicated that during evolution, the first two stem-loop domains (I and II) are absolutely conserved between different species, including mice and humans (64). On the other hand, the eight nucleotide substitutions between the human and mouse sequences in the second half of the exon result in distinct changes in the digestion pattern, indicating that structural changes may be present. In fact, the secondary-structure predictions for this region indicated several changes, such as the loss of single-stranded cuts in the human CCGGGU loop (VIa) with respect to the homologous CCGGGG* sequence that is predicted to form part of the main stem of mouse domain III (as indicated by the V1 cleavages on the two C's). Overall, what is observed is a change concerning the general disposition of the stem-loop elements in the mouse and human sequences. However, it is nonetheless important to note that the mESE sequence is localized in an apical stem-loop position while the putative mESS sequence is also part of a double-stranded region (thus mimicking the hESE-hESS sequence disposition).
Analysis of the changes in the mouse and human RNA secondary structures following homologous deletions.
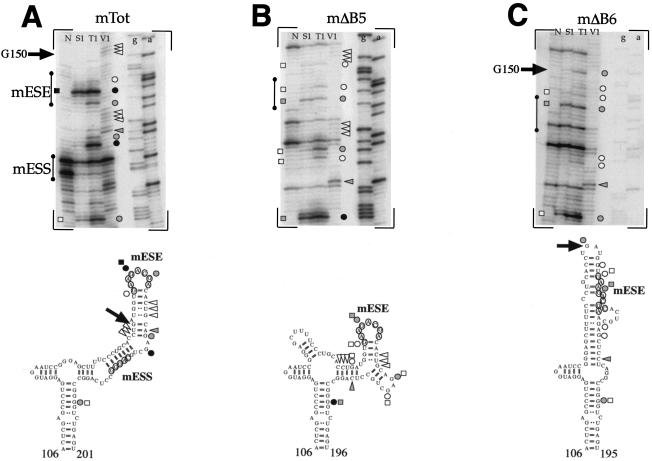
It was then of interest to analyze the changes in secondary structure following the introduction of the mΔB5 and mΔB6 mutations (in particular regarding mESE display). Figure 5B and C show the secondary-structure analysis of the mΔB5 and mΔB6 mutants compared to the mTot sequence (Fig. 5A). This analysis shows that the mESE element in the mΔB5 RNA remains in a stem-loop configuration, as demonstrated by the presence of single-strand-specific cleavages in its location flanked by double-stranded cleavage sites (Fig. 5B, top). In keeping with this observation, the secondary-structure predictions indicate a local change in the secondary structure which nonetheless maintains the mESE region in a stem-loop configuration (Fig. 5B, bottom). On the other hand, deletion of the mΔB6 region results in more extensive changes in the restriction cleavage sites with respect to the mTot structural analysis (Fig. 5C, top). In particular, it is worth noting the appearance of a strong T1 cleavage site in correspondence to guanosine G150 (indicated by an arrow) which is totally absent in the mTot RNA, where this G150 is predicted to be part of a stem region and is part of a region cut by V1 RNase (Fig. 5A, top). This is consistent with the secondary-structure predictions performed for the mΔB6 mutant, which place the G150 nucleotide in a loop region at the apex of a new structure while, as previously said, in the mTot RNA this nucleotide was predicted to be part of a stem region. In this structure, an important feature is represented by the loss of a stem-loop configuration for the mESE region and its partial inclusion in a stem configuration (Fig. 5C, bottom), a situation very different from the mTot and mΔB5 results.
FIG. 5.
Secondary-structure analysis of the ESE region in the mTot (A), mΔB5 (B), and mΔB6 (C) constructs. The in vitro-transcribed RNAs were enzymatically digested with S1 nuclease and RNases T1 and V1 and reverse transcribed, and the RT products were separated on a sequencing polyacrylamide gel. No enzyme was added to the reaction mixture in lane N. The regions containing the ESE and ESS elements are shown by a vertical line. The upper part of each panel shows the enzymatic analysis of RNA templates of mTot, mΔB5, and mΔB6 constructs. The bottom part reports the cleavages on the optimized secondary-structure predictions. Squares, circles, and triangles indicate S1 nuclease and RNase T1 and V1 cleavage sites, respectively. Black, shaded, and white symbols indicate high, medium, and low cleavage intensities, respectively. The arrow indicates the position of G150, which is present in a stem position in the mTot RNA but shifts to a loop configuration in the mΔB6 mutant.
In conclusion, these findings correlate well with the transfection results (Fig. 1B), which have shown that the mΔB5 mutant displays a splicing pattern identical to that of mTot while the mΔB6 deletion results in exon skipping.
Recruitment of SR proteins on the human and mouse EDA exons following deletion of the hESE (hΔ2e) and mESE (mΔA) sequences.
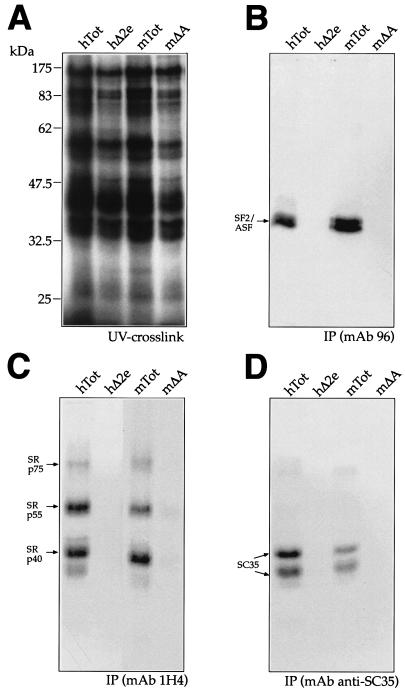
In order to functionally test the recruitment of the different SR proteins by the ESE elements of the mouse and human wild-type and mutant sequences, we performed UV cross-linking analysis, followed by IP with MAbs. Figure 6A shows the UV cross-linking profiles of hTot, hΔ2e, mTot, and mΔA with total nuclear extract from HeLa cells. The UV cross-linked samples were subsequently immunoprecipitated with MAbs against SF2/ASF (MAb 96) and the phosphorylated SR domain (MAb 1H4) that recognizes mainly Srp40, SRp55, and SRp75 and MAb anti-SC35 (Fig. 6B, C, and D, respectively). The results show that both the human and mouse wild-type RNAs (hTot and mTot) were able to immunoprecipitate SF2/ASF, SRp40, SRp55, and SRp75 at similar levels, with the only exception being represented by SC35, which shows lower affinity for the mouse sequence (Fig. 6D). Interestingly, consistent with these results, the SR scoring matrices predict the presence of a higher score in the human ESE (at position 161) as opposed to its homologous binding site in the mESE region (data not shown). On the other hand, neither the hΔ2e nor the mΔA RNA was able to immunoprecipitate any labeled SR protein, a result that is consistent with the identification of the ESE elements as the only target for SR protein recruitment to the EDA exon (42, 54).
FIG. 6.
UV cross-linking analysis of the wild-type human and mouse sequences (hTot and mTot) and ESE deletion-carrying mutants (hΔ2e and mΔA) with HeLa nuclear extract. Each RNA was labeled with [α-32P]UTP and then incubated with approximately 150 μg of HeLa nuclear extract before being subjected to UV cross-linking and digestion with RNase. Samples were then run on an SDS-11% PAGE gel and exposed to BioMax autoradiographic film. The electrophoretic mobility of prestained markers (Broad Range; New England Biolabs) is shown on the left (A). IP was then performed with equal amounts of each UV cross-linked sample shown in panel A with specific MAbs against different SR proteins: SF2/ASF (B, MAb 96), the phosphorylated RS domain (C, MAb 1H4), and SC35 (D, anti-SC35). The mobility of the SR proteins is indicated on the left. The SF2/ASF antibody immunoprecipitates more than one protein band owing to the presence of differently phosphorylated forms, as specified by the manufacturer.
Recruitment of SR proteins on the human and mouse EDA exons following introduction of the mΔB5 and mΔB6 mutations.
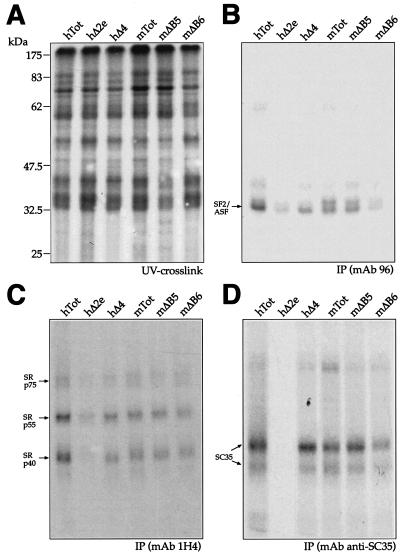
It was thus of interest to extend these analyses to the mΔB5 and mΔB6 mutations and determine whether they could affect SR recruitment by the mESE element. Figure 7 shows the UV cross-linking profiles of hTot, hΔ2e, hΔ4, mTot, mΔB5, and mΔB6 with total nuclear extract from HeLa cells before IP analysis (Fig. 7A) and after IP with MAb 96 (Fig. 7B), MAb 1H4 (Fig. 7C), and MAb anti-SC35 (Fig. 7D). The results show that the mΔB5 and hΔ4 mutants are very similar to the mTot IP pattern but the mΔB6 mutant could not immunoprecipitate SF2/ASF (Fig. 7B). This result is very similar to the observed effect of the hΔ2e and mΔA mutations (Fig. 6B and 7B), with the difference that the mΔB6 mutant contains the full mESE sequence, as opposed to hΔ2e, where the ESE sequence is deleted.
FIG. 7.
UV cross-linking analysis of human and mouse mutants with HeLa nuclear extract. Each RNA was labeled with [α-32P]UTP and then incubated with HeLa nuclear extract before being subjected to UV cross-linking and digestion with RNase. Samples were then run on an SDS-11% PAGE gel and exposed to BioMax autoradiographic film. The electrophoretic mobility of prestained molecular size markers (Broad Range; New England Biolabs) is shown on the left (A). IP was then performed with the same amount of each UV cross-linked sample shown in panel A with specific MAbs against different SR proteins: SF2/ASF (B, MAb 96), the phosphorylated RS domain (C, MAb 1H4), and SC35 (D, anti-SC35). The mobility of the SR proteins is indicated on the left. The SF2/ASF antibody immunoprecipitates more than one protein band owing to the presence of differently phosphorylated forms, as specified by the manufacturer.
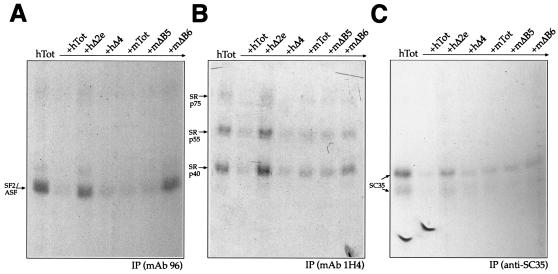
In order to complement and extend these observations, we also performed competition experiments with unlabeled RNAs to assess their ability to compete with the SR proteins for binding to the human RNA (hTot). The results, shown in Fig. 8, demonstrate that all of the RNAs behaved as expected from the direct IP analysis. In fact, both the hΔ2e and mΔB6 RNAs could not compete with labeled hTot RNA for SF2/ASF (Fig. 8A) while the mΔB5 and hΔ4 RNAs could compete for all of the SR proteins (Fig. 8A to C). Indeed, the mΔB6 mutant is the RNA that shows a general reduction in competition efficiency for all of the other SR proteins examined.
FIG. 8.
Competition and IP analysis of unlabeled mouse and human EDA sequences in the presence of labeled hTot RNA. Competition prior to UV cross-linking was performed by incubating labeled hTot RNA and HeLa nuclear extracts with equal amounts of unlabeled hTot, hΔ2e, hΔ4, mTot, mΔB5, and mΔB6 (in approximately fivefold molar excess) before IP analysis. IPs were performed with specific MAbs against SF2/ASF (A, MAb 96), the phosphorylated RS domain (B, MAb 1H4), and SC35 (C, anti-SC35). The mobility of the SR proteins is indicated on the left. Samples were run on an SDS-11% PAGE gel and exposed to BioMax autoradiographic film.
Recruitment of SR proteins by artificial and naturally occurring GAAGAAGA-containing sequences.
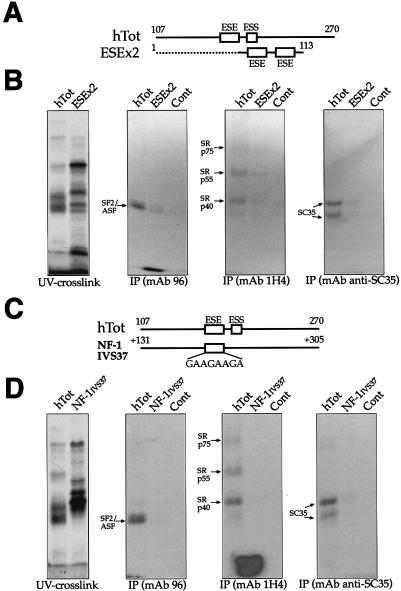
Finally, we wished to determine the contribution of the ESE sequence to exon SR protein recruitment with respect to the structural context. Therefore, we performed an IP analysis of two RNAs containing GAAGAAGA sequences in different contexts. First, we used a small artificial RNA containing two ESE sequence repeats (ESEX2) flanked by eight nucleotides of the EDA sequence on either side (Fig. 9A). As shown in Fig. 9B, this RNA was found to be a rather inefficient binder of SR proteins, especially considering the fact that it carried two tandem copies of the ESE sequence. This highlights the importance of the context in which the binding sequence is embedded.
FIG. 9.
IP analysis comparing the recruitment of SR proteins by human ESE sequences alone (GAAGAAGA) as opposed to the full EDA exon. (A) Comparison of the two RNAs used in this experiment: the hTot RNA contains the ESE within its original context (nt 107 to 270), while the ESEx2 RNA contains two copies of the ESE element (flanked only by 8 nt of the EDA sequence) at the 3′ end of pBluescript II SK+ sequences (dotted line). Numbering on the ESEx2 construct refers to the distance from the T3 RNA polymerase promoter on pBluescript II SK+. A control RNA is also included. IPs (B) were performed with equal amounts of the UV cross-linked samples and with the different specific MAbs against SR proteins: SF2/ASF (MAb 96), the phosphorylated RS domain (MAb 1H4), and SC35 (anti-SC35). The leftmost gel contains the total UV cross-linked proteins prior to IP. (C) Comparison of the hTot RNA with a naturally occurring, 170-nt-long intronic region containing a GAAGAAGA sequence from IVS37 of the NF-1 gene. The numbering on the IVS37 construct refers to intronic nucleotide positions starting from the 5′ splice site of NF-1 exon 37. (D) IP profile of these two sequences with the same antibodies used in panel B. As above, the leftmost gel contains the total UV cross-linked proteins prior to IP. In all of the gels, the mobility of the SR proteins is indicated on the left. Samples were run on an SDS-11% PAGE gel and exposed to BioMax autoradiographic film.
It was also of interest to check whether a naturally occurring GAAGAAGA sequence was capable of binding the same complement of SR proteins as the EDA exon. For this purpose, we chose an intronic region in IVS37 of the NF-1 gene that contains a GAAGAAGA sequence at position +167 and amplified the sequence surrounding it to mimic the hESE position in the human exon (Fig. 9C). It should be noted that, as expected, very little homology exists within the hTot and IVS37 sequences (data not shown). The IP analyses reported in Fig. 9D confirm that this naturally occurring GAAGAAGA sequence cannot bind any of the SR proteins detected with the hTot sequence. Taken together, these results provide an effective measurement of how important correct sequence display of the ESE in the EDA exon is for its recognition by SR protein factors.
DISCUSSION
In this study, we report that deletions in the putative mESS element localized in the FN EDA exon do not have the same effects on alternative splicing as deletions originally described for the hESS element (10). The sequences of the two exons are very similar, with only 8 nt substitutions out of a total 270 nt, and in this study we show that no single nucleotide (or subset) can account by itself for the observed differences in splicing behavior. However, when all specific nucleotides in the mouse EDA exon but not the 5′ splice site are replaced with their human counterparts, the mESS behaves the same way as the hESS. This raised the possibility that the nucleotide substitutions could be involved in some higher-order regulatory structure rather than in just the modification of the primary nucleotide sequence. In fact, the structural analysis performed in this study has shown that these eight substitutions induce distinct changes in the mouse and human RNA structures. However, before analyzing these changes in detail, it is worth discussing the similarities. First of all, our RNase digestion analysis shows a complete conservation of the first two stem-loops in the human and mouse exons. This is consistent with the fact that in the first part of the EDA exon the mouse and human sequences are identical and have the structure proposed by a previous phylogenetic study of this region (64). Most importantly, the detection of this expected similarity provides a good internal control for the structural differences that we observe in the 3′ half of the mouse and human exons. It is in this region, in fact, that the eight nucleotide differences between the human and mouse sequences are localized and may thus determine the changes in the overall stem-loop dispositions. The effect of these substitutions on RNA secondary structure is not an unexpected finding, as apparently benign single-nucleotide polymorphisms have already been reported to induce different structural folds in the pre mRNA structure both in vivo and in vitro (61). Despite these differences, an important similarity between the human and mouse structures is the presence of the ESE sequences within terminal stem-loop domains. The maintenance of this region in a single-stranded condition in both RNAs is consistent with the observation that single strandedness of RNA elements is a necessary requirement to obtain sequence-specific protein binding, such as in the case of the Drosophila doublesex splicing enhancer elements with the RNA-binding domain of Tra2 (28) or TDP-43 for UG repeated sequences (6).
At the same time, these structural differences also raise the possibility that deletion of the human and mESS sequences may not result in the same structural change in both RNAs (and thus in a different recruitment of splicing factors to the enhancers). In support of this hypothesis is the observation that RNA secondary structure can affect the recruitment of two nuclear factors belonging to the SR protein family of splicing regulators (B52 and SRp55) (55, 62). Moreover, even the binding of hnRNP A1 (a well-known splicing-antagonistic factor) has been recently observed to be affected by RNA secondary structure in the removal of the second intron in the human immunodeficiency virus type 1 rev and tat pre-mRNAs (20). Interestingly, it should be noted that this relationship goes both ways and proteins such as hnRNP A1 (56) and the polypyrimidine tract-binding protein (16) have been reported to bind on either side of exons and physically loop them out of the splicing process.
In this respect, it should be mentioned that overexpression of hnRNP A1 causes a slight but significant decrease in the inclusion of both mouse and human EDA exons (A. F. Muro, unpublished data). However, the comparison of the effects of hnRNP A1-mediated splicing inhibition on these exons has not revealed any significant difference between the two species. In addition, the degree of splicing inhibition detected in both systems is lower than the observed increase in EDA inclusion following similar levels of overexpression of SF2/ASF (19). Taken together, these data suggest that, in the EDA system, the inhibitory effect of hnRNP A1 may not be linked to a specific target sequence.
On a more general note, a question that also remains to be addressed is the presence of RNA secondary structure in vivo. There is still no definitive answer to this question. However, several recent lines of evidence indicate that RNA folding can occur in vivo (61) and that this process can influence biological processes. For example, binding of proteins to RNA trinucleotide repeats in vivo closely matches the in vitro results that predict these repeats to be folded in a characteristic hairpin shape (63). It could, of course, be argued that these structures are of a very localized and specific nature. However, statistical analysis of mRNA coding sequences has revealed that calculated mRNA folding is more stable than expected by chance, suggesting that codon bias may favor the existence of mRNA structures (60). Although these results have been challenged with a different set of statistical tools and genes (67), considerations analogous to those of Seffens and Digby (60) have been recently made concerning bacterial RNA (35). Thus, the picture that is beginning to emerge clearly favors the possibility that mRNA coding sequences are quite capable of harboring in vivo variable amounts of secondary structure that may affect biological processes.
The structural data reported in this study indicate that the mΔB6 deletion causes the mESE element to lose the original mTot RNA structure. It could then represent a situation analogous to that already reported to occur in a variety of 5′ and 3′ splice sites where RNA secondary structure prevents the U1 and U2 snRNP proteins from binding to the RNA (18, 22, 30, 45, 46, 51). In fact, inhibition of SR protein binding in the mΔB6 mutant has been confirmed by our IP analyses, which show that this mutant is unable to bind (or compete for binding of) the SF2/ASF protein and, with less efficiency, the other SR proteins. This binding profile correlates well with the splicing behavior of the mΔB6 mutant, which is identical to that of the hΔ2e and mΔA mutants. Interestingly, overexpression of this protein in a minigene assay has demonstrated that SF2/ASF possesses the greatest ability of any SR protein family member to stimulate EDA inclusion (19). This provides a likely explanation of why the maintenance of SC35, SRp40, SRp55, and SRp75 protein binding to mΔB6 cannot compensate for loss of SF2/ASF binding capacity. Analogously, the fact that the mΔB5 mutant possesses an SR protein IP profile identical to that of mTot is consistent with the observation that both RNAs are spliced to the same degree.
Nonetheless, it should be noted that the correlation between SR binding levels and splicing efficiency is not complete. It is clear that below an SR binding level threshold, functionality is lost, such as observed for the hΔ2e, mΔA, and mΔB6 mutants. However, in the case of the hΔ4 mutant (for which we observed full EDA inclusion), direct IP experiments detected a moderate reduction in SR binding levels, although the RNA preserves very efficient competitor activity (a factor distinguishing it from mΔB6 or hΔ2e RNA). It is thus clear that additional factors, other than the SR proteins, must contribute to the final outcome. One possibility is that the relative protein composition (“quality”) of the complex that binds the ESE might play a role besides the quantity of SR proteins bound to the RNA or, alternatively, that additional positive or negative splicing factors (34) besides those immunoprecipitated by our antibodies might play a role in hΔ4 splicing. Furthermore, other undetected structural changes near the 5′ and 3′ splice sites may have caused improvement of splicing even in the case of reduced SR binding efficiency.
In conclusion, our results show that changes in regions that do not directly participate in protein binding can affect ESE elements by inducing changes in mRNA conformation. The importance of the RNA secondary structure for recruitment of SR protein factors is also highlighted by the fact that a small RNA that contains two tandem copies of the human ESE sequence (GAAGAAGA) is considerably less efficient in binding the SR proteins detected by a single copy of the same sequence in its original context (the EDA exon). In this respect, an even more striking result is the inability of a naturally occurring intronic GAAGAAGA sequence to immunoprecipitate any of the SR proteins pulled down by the same human sequence in its original context. The results highlight the importance of the local context in determining a functional effect on mRNA processing. Taken together, our data demonstrate that the sequence context, in addition to primary sequence identity, can heavily contribute to the making of functional units capable of influencing pre-mRNA splicing.
Acknowledgments
This work was supported by grants from the Telethon Onlus Foundation (grant E1038 awarded to F.E.B.).
REFERENCES
- 1.Adams, M. D., D. Z. Rudner, and D. C. Rio. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8:331-339. [DOI] [PubMed] [Google Scholar]
- 2.Balvay, L., D. Libri, and M. Y. Fiszman. 1993. Pre-mRNA secondary structure and the regulation of splicing. Bioessays 15:165-169. [DOI] [PubMed] [Google Scholar]
- 3.Black, D. L. 1991. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5:389-402. [DOI] [PubMed] [Google Scholar]
- 4.Blanchette, M., and B. Chabot. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3:405-419. [PMC free article] [PubMed] [Google Scholar]
- 5.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 6.Buratti, E., and F. E. Baralle. 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276:36337-36343. [DOI] [PubMed] [Google Scholar]
- 7.Buratti, E., T. Dork, E. Zuccato, F. Pagani, M. Romano, and F. E. Baralle. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20:1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 9.Caputi, M., F. E. Baralle, and C. A. Melo. 1995. Analysis of the linkage between fibronectin alternative spliced sites during ageing in rat tissues. Biochim. Biophys. Acta 1263:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Caputi, M., G. Casari, S. Guenzi, R. Tagliabue, A. Sidoli, C. A. Melo, and F. E. Baralle. 1994. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 22:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi, M., R. J. Kendzior, Jr., and K. L. Beemon. 2002. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev. 16:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caputi, M., C. A. Melo, and F. E. Baralle. 1995. Regulation of fibronectin expression in rat regenerating liver. Nucleic Acids Res. 23:238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni, L., and A. R. Krainer. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30:377-384. [DOI] [PubMed] [Google Scholar]
- 15.Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang, and A. R. Krainer. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949-957. [DOI] [PubMed] [Google Scholar]
- 17.Clouet d'Orval, B., Y. d'Aubenton carafa, P. Sirand-Pugnet, M. Gallego, E. Brody, and J. Marie. 1991. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science 252:1823-1828. [DOI] [PubMed] [Google Scholar]
- 18.Coleman, T. P., and J. R. Roesser. 1998. RNA secondary structure: an important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry 37:15941-15950. [DOI] [PubMed] [Google Scholar]
- 19.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 20.Damgaard, C. K., T. O. Tange, and J. Kjems. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estes, P. A., N. E. Cooke, and S. A. Liebhaber. 1992. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 267:14902-14908. [PubMed] [Google Scholar]
- 23.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 24.Ffrench-Constant, C. 1995. Alternative splicing of fibronectin—many different proteins but few different functions. Exp. Cell Res. 221:261-271. [DOI] [PubMed] [Google Scholar]
- 25.Frazer, K. A., L. Elnitski, D. M. Church, I. Dubchak, and R. C. Hardison. 2003. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 13:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green, M. R. 1991. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell Biol. 7:559-599. [DOI] [PubMed] [Google Scholar]
- 27.Grover, A., H. Houlden, M. Baker, J. Adamson, J. Lewis, G. Prihar, S. Pickering-Brown, K. Duff, and M. Hutton. 1999. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274:15134-15143. [DOI] [PubMed] [Google Scholar]
- 28.Hertel, K. J., K. W. Lynch, E. C. Hsiao, E. H. Liu, and T. Maniatis. 1996. Structural and functional conservation of the Drosophila doublesex splicing enhancer repeat elements. RNA 2:969-981. [PMC free article] [PubMed] [Google Scholar]
- 29.Huh, G. S., and R. O. Hynes. 1994. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 8:1561-1574. [DOI] [PubMed] [Google Scholar]
- 30.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang, Z., J. Cote, J. M. Kwon, A. M. Goate, and J. Y. Wu. 2000. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20:4036-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan, M., A. Schallhorn, and F. M. Wurm. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadener, S., J. P. Fededa, M. Rosbash, and A. R. Kornblihtt. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. USA 99:8185-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34:460-463. [DOI] [PubMed] [Google Scholar]
- 35.Katz, L., and C. B. Burge. 2003. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 13:2042-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klaff, P., D. Riesner, and G. Steger. 1996. RNA structure and the regulation of gene expression. Plant Mol. Biol. 32:89-106. [DOI] [PubMed] [Google Scholar]
- 37.Kornblihtt, A. R., K. Umezawa, K. Vibe-Pedersen, and F. E. Baralle. 1985. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 4:1755-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krainer, A. R. 1997. Eukaryotic mRNA processing. Oxford University Press Inc., New York, N.Y.
- 39.Kuo, B. A., and P. A. Norton. 1999. Accurate selection of a 5′ splice site requires sequences within fibronectin alternative exon B. Nucleic Acids Res. 27:3945-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo, B. A., T. M. Uporova, H. Liang, V. D. Bennett, R. S. Tuan, and P. A. Norton. 2002. Alternative splicing during chondrogenesis: modulation of fibronectin exon EIIIA splicing by SR proteins. J. Cell. Biochem. 86:45-55. [DOI] [PubMed] [Google Scholar]
- 41.Lamond, A. I. 1993. The spliceosome. Bioessays 15:595-603. [DOI] [PubMed] [Google Scholar]
- 42.Lavigueur, A., H. La Branche, A. R. Kornblihtt, and B. Chabot. 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 7:2405-2417. [DOI] [PubMed] [Google Scholar]
- 43.Libri, D., A. Piseri, and M. Y. Fiszman. 1991. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science 252:1842-1845. [DOI] [PubMed] [Google Scholar]
- 44.Liu, H. X., L. Cartegni, M. Q. Zhang, and A. R. Krainer. 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 27:55-58. [DOI] [PubMed] [Google Scholar]
- 45.Liu, H. X., G. J. Goodall, R. Kole, and W. Filipowicz. 1995. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J. 14:377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loeb, D. D., A. A. Mack, and R. Tian. 2002. A secondary structure that contains the 5′ and 3′ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J. Virol. 76:10195-10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLeod, J. N., N. Burton-Wurster, D. N. Gu, and G. Lust. 1996. Fibronectin mRNA splice variant in articular cartilage lacks bases encoding the V, III-15, and I-10 protein segments. J. Biol. Chem. 271:18954-18960. [DOI] [PubMed] [Google Scholar]
- 48.Matsuo, M., H. Nishio, Y. Kitoh, U. Francke, and H. Nakamura. 1992. Partial deletion of a dystrophin gene leads to exon skipping and to loss of an intra-exon hairpin structure from the predicted mRNA precursor. Biochem. Biophys. Res. Commun. 182:495-500. [DOI] [PubMed] [Google Scholar]
- 49.Mistry, N., W. Harrington, E. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muh, S. J., R. H. Hovhannisyan, and R. P. Carstens. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277:50143-50154. [DOI] [PubMed] [Google Scholar]
- 51.Munroe, S. H. 1984. Secondary structure of splice sites in adenovirus mRNA precursors. Nucleic Acids Res. 12:8437-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 19:2657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muro, A. F., A. K. Chauhan, S. Gajovic, A. Iaconcig, F. Porro, G. Stanta, and F. E. Baralle. 2003. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J. Cell Biol. 162:149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muro, A. F., A. Iaconcig, and F. E. Baralle. 1998. Regulation of the fibronectin EDA exon alternative splicing. Cooperative role of the exonic enhancer element and the 5′ splicing site. FEBS Lett. 437:137-141. [DOI] [PubMed] [Google Scholar]
- 55.Nagel, R. J., A. M. Lancaster, and A. M. Zahler. 1998. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4:11-23. [PMC free article] [PubMed] [Google Scholar]
- 56.Nasim, F. U., S. Hutchison, M. Cordeau, and B. Chabot. 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8:1078-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pagani, F., E. Buratti, C. Stuani, and F. E. Baralle. 2003. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 278:26580-26588. [DOI] [PubMed] [Google Scholar]
- 58.Pagani, F., E. Buratti, C. Stuani, R. Bendix, T. Dork, and F. E. Baralle. 2002. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 30:426-429. [DOI] [PubMed] [Google Scholar]
- 59.Pagani, F., C. Stuani, M. Tzetis, E. Kanavakis, A. Efthymiadou, S. Doudounakis, T. Casals, and F. E. Baralle. 2003. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 12:1111-1120. [DOI] [PubMed] [Google Scholar]
- 60.Seffens, W., and D. Digby. 1999. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 27:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen, L. X., J. P. Basilion, and V. P. Stanton, Jr. 1999. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl. Acad. Sci. USA 96:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi, H., B. E. Hoffman, and J. T. Lis. 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 17:2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sobczak, K., M. De Mezer, G. Michlewski, J. Krol, and W. J. Krzyzosiak. 2003. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 31:5469-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staffa, A., N. H. Acheson, and A. Cochrane. 1997. Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J. Biol. Chem. 272:33394-33401. [DOI] [PubMed] [Google Scholar]
- 65.Stamm, S., J. Zhu, K. Nakai, P. Stoilov, O. Stoss, and M. Q. Zhang. 2000. An alternative-exon database and its statistical analysis. DNA Cell Biol. 19:739-756. [DOI] [PubMed] [Google Scholar]
- 66.Varani, L., M. Hasegawa, M. G. Spillantini, M. J. Smith, J. R. Murrell, B. Ghetti, A. Klug, M. Goedert, and G. Varani. 1999. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. USA 96:8229-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Workman, C., and A. Krogh. 1999. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 27:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 69.Zuker, M. 1989. Computer prediction of RNA structure. Methods Enzymol. 180:262-288. [DOI] [PubMed] [Google Scholar]
- 70.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In W. J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO ASI series. Kluwer Academic Publishers, Dordrecht, The Netherlands.