Abstract
We report the expression of the barley (Hordeum vulgare L.) COR (cold-regulated) gene cor14b (formerly pt59) and the accumulation of its chloroplast-localized protein product. A polyclonal antibody raised against the cor14b-encoded protein detected two chloroplast COR proteins: COR14a and COR14b. N-terminal sequencing of COR14a and expression of cor14b in Arabidopsis plants showed that COR14a is not encoded by the cor14b sequence, but it shared homology with the wheat (Triticum aestivum L.) WCS19 COR protein. The expression of cor14b was strongly impaired in the barley albino mutant an, suggesting the involvement of a plastidial factor in the control of gene expression. Low-level accumulation of COR14b was induced by cold treatment in etiolated plants, although cor14b expression and protein accumulation were enhanced after a short light pulse. Light quality was a determining factor in regulating gene expression: red or blue but not far-red or green light pulses were able to promote COR14b accumulation in etiolated plants, suggesting that phytochrome and blue light photoreceptors may be involved in the control of cor14b gene expression. Maximum accumulation of COR14b was reached only when plants were grown and/or hardened under the standard photoperiod. The effect of light on the COR14b stability was demonstrated by using transgenic Arabidopsis. These plants constitutively expressed cor14b mRNAs regardless of temperature and light conditions; nevertheless, green plants accumulated about twice as much COR14b protein as etiolated plants.
Growth at low, nonfreezing temperature induces an adaptive response known as cold acclimation or hardening that improves the frost resistance of overwintering plants. In addition to resistance to freezing, cold-acclimated plants also show resistance to photoinhibition and an increased rate of photosynthesis (Oquist and Huner, 1993). Although the molecular basis of chloroplast cold acclimation remains unclear, these results suggest that the low-temperature-induced modifications in the photosynthetic apparatus are important in the cold-acclimation process (Gray et al., 1997).
The molecular dissection of cold hardening revealed a very complex situation, in which the coordinated and timely expression of a series of COR genes is associated with increased hardening. Analysis of the signal transduction pathways leading to the expression of COR genes has shown that in addition to cold, the accumulation of COR mRNAs can be induced or modified by ABA (Nordin et al., 1993), calcium influx (Monroy and Dhindsa, 1995), or drought stress (Yamaguchi-Shinozaki, 1994; Grossi et al., 1995). Also, light can enhance the expression of COR mRNAs coding for chloroplast-localized proteins (Chauvin et al., 1993; Crosatti et al., 1995). To date, three COR genes whose expression is linked to cold acclimation have been found to encode for proteins localized in the chloroplasts: cor15 in Arabidopsis (Lin and Thomashow, 1992), wcs19 in wheat (Chauvin et al., 1993; Gray et al., 1997), and cor14b (formerly pt59) in barley (Hordeum vulgare L.; Cattivelli and Bartels, 1990; Crosatti et al., 1995). The expression of the wcs19 and cor14b genes was found to be regulated by light.
The expression of other nuclear genes coding for chloroplast proteins has also been shown to be modified by light. The transcription of the genes coding for the small subunit of Rubisco and for the chlorophyll a/b-binding proteins is fully controlled by light, whereas the genes encoding chloroplast-localized Clp protease and RNA-binding protein are constitutively expressed but enhanced by light (Cheng et al., 1994; Ostersetzer and Adam, 1996). Similarly, the early light-inducible proteins, which normally accumulate during greening, are posttranscriptionally up-regulated in barley leaves during cold acclimation as a result of the photooxidative stress caused by light and cold (Montane et al., 1997). Finally, the 23-kD chloroplast heat-shock protein gene of Chenopodium rubrum is induced by heat stress and posttranslationally controlled by light (Debel et al., 1994). The expression of wcs19, a COR gene coding for chloroplast protein, is also stimulated by light through the reduction state of the plastoquinone pool, an indicator of PSII overexcitation (Gray et al., 1997).
In most cases the involvement of light in gene regulation requires the presence of specific photoreceptors. Plants contain three different photoreceptor systems: (a) the phytochromes, which primarily adsorb red and far-red light, (b) the blue light photoreceptor, and (c) the UV-B photoreceptor. The light signal transduction works as an integrated network in which light with synergetic or antagonistic effects interacts to control the expression of particular genes (Terzaghi and Cashmore, 1995). For example, blue light photoreceptors and phytochrome-mediated pathways control the expression of the nuclear genes encoding chloroplast glyceraldehyde-3-P dehydrogenase of Arabidopsis (Dewdney et al., 1993).
We have analyzed the regulation mechanisms that control the expression of the barley gene cor14b (formerly pt59; Cattivelli and Bartels, 1990) and the accumulation of the corresponding protein. This gene is specifically induced by low temperature (no ABA or drought induction; Grossi et al., 1992) and it encodes a polypeptide accumulated in the stroma fraction of the chloroplasts (Crosatti et al., 1995). In the present study we show that three different mechanisms affect the expression of cor14b and the accumulation of its protein: (a) the expression of the gene is strongly impaired in a barley albino mutant, suggesting the involvement of a plastidial factor in the control of gene expression; (b) a basal level of COR14b is present after cold treatment in etiolated plants, although protein accumulation can be enhanced by a short red or blue light pulse, suggesting that phytochrome and the blue light photoreceptors are also involved; and (c) by using transgenic Arabidopsis that constitutively expressed cor14b, we show that plant exposure to standard photoperiod increases the stability of COR14b.
MATERIALS AND METHODS
Plant Material, Growing Conditions, and Light Sources
The experiments were performed using two barley (Hordeum vulgare L.) genotypes, the winter cv Onice and the albino mutant an (accession no. 112 of the barley genetic stock of the Colorado State University, Fort Collins; Burnham et al., 1971) and either wild-type or transgenic Arabidopsis (ecotype Columbia), as described below.
All experiments were performed using plants grown in sterile conditions. Barley seeds were sterilized for 5 min in 70% ethanol and for 20 min in 4% sodium hypochlorite, then rinsed in sterile distilled water, and sown on 0.8% agar in tissue culture vessels. Arabidopsis seeds were sterilized for 20 min in 4% sodium hypochlorite, then rinsed in sterile distilled water, and germinated in Petri dishes on germination medium (Valvekens et al., 1988) with or without 50 μg mL−1 kanamycin for transgenic and wild-type plants, respectively.
Barley plants were grown for 7, 8, 10, or 12 d at 12-h light/12-h dark (20°C/15°C) and hardened for 7, 10, or 12 d at 8-h light/16-h dark (3°C/1.5°C). With respect to light conditions, plants grown under standard photoperiods were subjected to 160 μmol m−2 s−1, whereas other samples were grown completely in the dark (12-h light/12-h dark [20°C/15°C]). Fully etiolated plants were hardened in the dark (8-h light/16-h dark [3°C/1.5°C]) with or without being subjected to a single short period of light (5 min or less). To ensure that dark-grown plants were never exposed to light, the in vitro culture vessels were wrapped with aluminum foil and closed in black boxes. All manipulations of dark-grown plants were performed in complete darkness. Short light exposures were applied to each sample by using a lamp (model LS2, Hansatech, King's Lynn, UK) attenuated with neutral density filters with a white light (about 200 μmol m−2 s−1). Blue, green, red, and far-red lights of equal intensity (about 10 μmol m−2 s−1) were obtained by using additional optical interference filters with peak transmissions of 400-, 450-, 500-, 660-, and 730-nm wavelengths, respectively. Arabidopsis plants were grown at standard photoperiods of 16-h light/8-h dark (both at 22°C) and hardened for 7 d at 8-h light/16-h dark (3°C/1.5°C). Plants were harvested in a dark room and frozen in liquid nitrogen.
Transformation
The cor14b sequence was cleaved using the RsaI and RsaI restriction sites from the pUC9 vector and inserted into the SmaI restriction site of a pUC19-derived vector between the CaMV 35S promoter and the nopaline synthase (NOS) terminator. The EcoRI-EcoRI fragment containing the chimeric construct 35SCaMV-cor14b-NOS was subcloned into the EcoRI site of pBIN19 (Hoekema et al., 1983). The resulting binary vector was sequenced to verify the correct insertion and introduced into Agrobacterium tumefaciens strain LBA4404. Transformants 35SCaMV-cor14b-NOS A. tumefaciens were selected on yeast extract beef medium containing 100 μg mL−1 streptomycin, 100 μg mL−1 rifampicin, and 25 μg mL−1 kanamycin. The chimeric gene was introduced into plants of Arabidopsis (ecotype Columbia) by A. tumefaciens-mediated transformation of root explants. The procedure used was as described previously (Valvekens et al., 1988).
Chloroplast Preparation
For chloroplast preparation barley and Arabidopsis plants were grown in pots at 12-h light/12-h dark (20°C/15°C) in 50% sand/50% soil. Seven days of cold hardening treatment was given to barley plants only. Leaves were homogenized in 400 mm sorbitol, 0.1 m Tricine-HCl, pH 7.8, 0.5% (w/v) skim milk, and 1 mm PMSF, according to the method of Bassi et al. (1985). The homogenate was filtered through two layers of Miracloth (Calbiochem) and centrifuged at 1,400g for 10 min. Intact chloroplasts were gently resuspended in 25 mm Hepes-KOH, pH 7.5, 10 mm EDTA with a brush. Thylakoids and soluble proteins were separated by centrifugation at 10,000g for 15 min. All steps were carried out at 4°C.
Purification and N-Terminal Sequencing of COR14
COR14 proteins for N-terminal sequencing were isolated from about 200 g of cold-acclimated barley leaves. The soluble protein fraction from the chloroplasts was obtained as described above and subjected to further centrifugation at 40,000g for 30 min (4°C). The supernatant (100 mL) was dialyzed overnight against 1% Gly and the protease inhibitors PMSF (1 mm), aminocaproic acid (3 mm), and benzamidine (1 mm), and was then loaded onto a free-flow recycling IEF apparatus upon addition of 1% ampholytes (pH 3.0–10.0). Focusing was performed for 3 h at 25 W in constant power conditions. Thirty fractions were eluted with IEF and aliquots were analyzed by SDS-PAGE and immunoblotting. Eight fractions in the pH range between 3.0 and 5.0, containing COR14a and COR14b, were pooled, diluted to 100 mL, and subjected to a further preparative IEF in the 3.0 to 6.0 pH range; fractions were analyzed as above. COR14a and COR14b proteins focused at approximately pH 4.5, and the corresponding fractions were pooled and subjected to preparative electroendosmotic electrophoresis (Curioni et al., 1988) using the buffer system of Schagger and von Jagow (1987). Fractions containing COR14a and COR14b, as detected by immunoblotting, were pooled, concentrated, loaded onto a SDS-Tricine gel, and blotted onto a PVDF membrane. N-terminal sequencing of intact proteins was according to the method of Edman (1950) using a sequencer (model 477A, Perkin Elmer), equipped with an on-line phenylthiohydantoin-amino acid analyzer (model 120A, Perkin Elmer) according to the protocol of the manufacturer.
Protein Extraction, Electrophoresis, and Immunoblot Analysis
Barley and Arabidopsis leaves were ground to a fine powder in liquid nitrogen. The powder was suspended in acetone containing 10% (w/v) TCA and 0.07% β-mercaptoethanol and stored at −20°C for 1 h to allow protein precipitation. After the sample was centrifuged (20 min, 3000g, 4°C), the pellet was suspended in acetone containing 0.07% β-mercaptoethanol, stored at −20°C for 1 h, and centrifuged as before. This step was repeated twice, and then the pellet was dried completely under a vacuum. Five milligrams of dried powder was solubilized using 280 μL of loading buffer (4% SDS, 12% glycerol, 50 mm Tris, pH 6.8, 2% β-mercaptoethanol, and 0.01% bromphenol blue). The samples were boiled for 2 min, centrifuged for 5 min at 10,000g, and 30 μL of supernatant was loaded onto a 10% Tricine-SDS-PAGE gel overlaid with a 4% stacking gel, according to the method of Schagger and von Jagow (1987). Proteins were electroblotted onto a nitrocellulose membrane (BA83, Schleicher & Schuell) according to the method of Szewczyk and Kozloff (1985) and probed with the COR14 polyclonal antibody. The preparation of the antibody was previously described (Crosatti et al., 1995). The AluI-AluI 458-bp DNA fragment isolated from the cDNA clone cor14b was subcloned into the SmaI site of the pGEX-3X expression vector. The fusion protein encoded by the chimeric gene glutathione S-transferase-cor14b was expressed in Escherichia coli, purified by affinity chromatography, and used to raise the corresponding antibody (Crosatti et al., 1995).
Western blotting was performed with an enhanced chemiluminescence kit (ECL, Amersham) and the working dilution of the antibody was 1:3500. The following modifications were introduced to the ECL manufacturer's protocol: 20 mm Tris, 137 mm NaCl (TBS) buffer, pH 9.6, and 5% skim milk (w/v) were added to the TBS buffer during the blocking step and also during incubation with primary and secondary antibodies. Tween 20 (0.3%, v/v) was added to TBS buffer during incubation with antibodies. The membranes were treated with 50 mm Tris-HCl, pH 7.5, before chemoluminescent detection. In addition to the COR14 proteins, the COR14 polyclonal antibody cross-reacted with an additional polypeptide of about 29 kD, the expression of which was not affected by either cold or chloroplast development. This anonymous protein was used as a loading control for all western experiments (an example is shown in Fig. 3). In all experiments filters were exposed to Kodak X-Omat film for about 60 s; the western blots shown in Figure 6 were exposed for an additional 10 and 30 s to record only the strongest signals. Densitometric scanning of films after antibody exposure was performed with Molecular Analyst software (version 1.5, Bio-Rad).
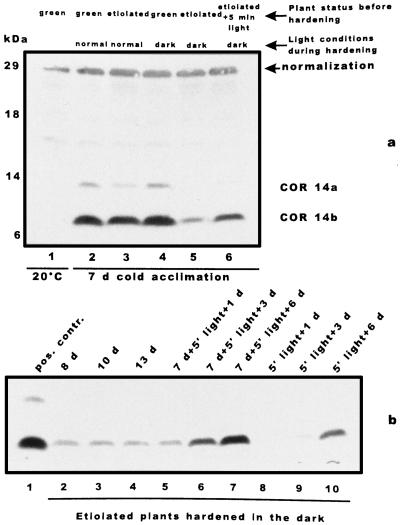
Figure 3.
Light-dependent accumulation of COR14. a, Barley plants (cv Onice) were grown and hardened for 7 d under different light conditions. Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody; the 29-kD anonymous protein band used for western blot normalization is also indicated. Lane 1, Green plants grown at 20°C; lane 2, green plants grown and hardened under the standard photoperiod; lane 3, etiolated plants hardened under the standard photoperiod; lane 4, green plants hardened in the dark; lane 5, etiolated plants hardened in the dark; and lane 6, etiolated plants exposed for 5 min to light (200 μmol m−2 s−1) and then hardened in the dark. b, Etiolated barley plants (cv Onice) were hardened in the dark for 13 d and compared with plants grown and hardened in the same conditions, except for a pulse of white light (5 min at 200 μmol m−2 s−1) after 7 d of hardening or before hardening. Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. Lane 1, Green plants hardened under the standard photoperiod; lane 2, etiolated plants hardened in the dark for 8 d; lane 3, etiolated plants hardened in the dark for 10 d; lane 4, etiolated plants hardened in the dark for 13 d; lane 5, etiolated plants hardened in the dark for 7 d, exposed to light, and further hardened in the dark for 1 d; lane 6, etiolated plants hardened in the dark for 7 d, exposed to light, and further hardened in the dark for 3 d; lane 7, etiolated plants hardened in the dark for 7 d, exposed to light, and further hardened in the dark for 6 d; lane 8, etiolated plants exposed to light before 1 d of hardening in the dark; lane 9, etiolated plants exposed to light before 3 d of hardening in the dark; and lane 10, etiolated plants exposed to light before 6 d of hardening in the dark.
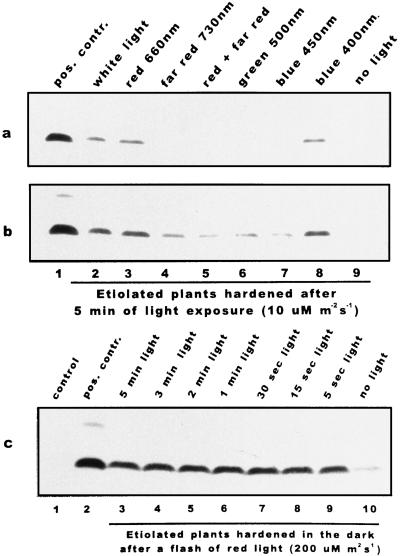
Figure 6.
Red (660 nm) and blue (400 nm) light were the most effective in stimulating COR14b accumulation. Etiolated barley plants (cv Onice) were exposed to 5 min of different light spectra (about 10 μmol m−2 s−1) and then hardened for 7 d in the dark. a, Result obtained after about 10 s of autoradiography exposure so that only the strongest signals were recorded. b, A 30-s exposure of the same western blot is presented to allow the detection of a basal level of COR14b induced by far-red, green, and 450 nm of light, whereas the effect of the cold signal alone was even lower (see also c, no light sample). Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. Lane 1, Green plants hardened under the standard photoperiod; lane 2, etiolated plants hardened in the dark after 5 min of white light; lane 3, etiolated plants hardened in the dark after 5 min of red (660 nm) light; lane 4, etiolated plants hardened in the dark after 5 min of far-red (730 nm) light; lane 5, etiolated plants hardened in the dark after 5 min of red (660 nm) plus 5 min of far red (730 nm) light; lane 6, etiolated plants hardened in the dark after 5 min of green (500 nm) light; lane 7, etiolated plants hardened in the dark after 5 min of blue (450 nm) light; lane 8, etiolated plants hardened in the dark after 5 min of blue (400 nm) light; and lane 9, etiolated plants hardened in the dark. c, Etiolated barley plants (cv Onice) were exposed to a short period of red light (200 μmol m−2 s−1) and then hardened for 7 d in the dark. Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. Lane 1, Green plants grown at 20°C; lane 2, green plants hardened under the standard photoperiod; lane 3, etiolated plants hardened in the dark after 5 min of light; lane 4, etiolated plants hardened in the dark after 3 min of light; lane 5, etiolated plants hardened in the dark after 2 min of light; lane 6, etiolated plants hardened in the dark after 1 min of light; lane 7, etiolated plants hardened in the dark after 30 s of light; lane 8, etiolated plants hardened in the dark after 15 s of light; lane 9, etiolated plants hardened in the dark after 5 s of light; and lane 10, etiolated plants hardened in the dark.
RNA Extraction and Northern Analysis
Frozen leaves were ground in liquid nitrogen and suspended in 0.05 m Tris, 0.01 m EDTA, 0.1 m NaCl, and 2% (w/v) SDS. After three phenol-chloroform (1:1, v/v) extractions the poly(A+) RNAs were isolated by chromatography on oligo(dT)-cellulose (Boehringer Mannheim) according to published methods (Sambrook et al., 1989). Equal amounts (0.8 μg) of poly(A+) RNAs for each sample were separated on an agarose formaldehyde gel and transferred to a positively charged nylon filter (Hybond N+, Amersham). Radioactive probes were obtained by oligo-labeling of cDNA clones, and the hybridization was performed at 68°C in 6× SSC, 2× Denhardt's solution (Sambrook et al., 1989), 0.1% SDS, and 100 g mL−1 of denatured herring-sperm DNA. Filters were then washed at 68°C with 1× SSC, 0.1% SDS (3 × 20 min). For each northern experiment a single filter was produced and subsequently hybridized with all of the probes required. DNA probes were removed from filters by washing them in 0.5% (w/v) SDS at 100°C. To control the integrity and the amount of poly(A+) RNA loaded in each lane of the northern blot of Figure 4, the filter was hybridized with α-32P-labeled 20-mer oligo(dT) (Capel et al., 1997). The filter presented in Figure 7 was hybridized with paf93, a COR gene whose expression is not affected by light (Grossi et al., 1998), and which therefore represents the internal control of the experiment.
Figure 4.
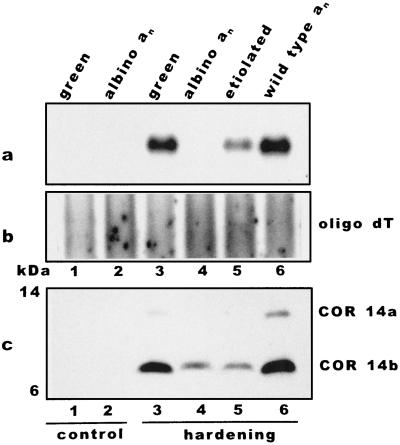
Albino plants show a minimal cor14b expression. Barley plants carrying the homozygote albino mutation an, the corresponding heterozygotes, and plants of the barley cv Onice were hardened for 7 d under different light conditions. A Northern blot made with poly(A+) RNAs was probed with cor14b (a) and normalized with [32P]dATP-labeled oligo(dT) (b). Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody (c). Lane 1, Green plants grown at 20°C; lane 2, albino mutant an grown at 20°C; lane 3, green plants (cv Onice) hardened under the standard photoperiod; lane 4, albino mutant an hardened under the standard photoperiod; lane 5, etiolated plants hardened in the dark; and lane 6, green heterozygote an plants hardened under the standard photoperiod.
Figure 7.
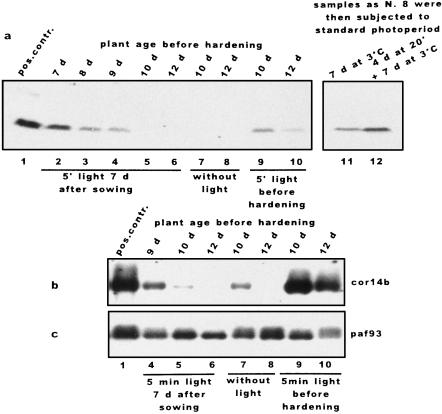
Etiolated plants were exposed to white light for 5 min and kept in the dark at 20°C for a few days before 7 d of hardening in the dark. Total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody (a). A northern blot made with poly(A+) RNAs was probed with the COR genes cor14b (b) and paf93 (c). Lane 1, Green plants hardened under the standard photoperiod; lane 2, 7-d-old etiolated plants exposed to light before hardening in the dark; lane 3, 7-d-old etiolated plants exposed to light, kept in the dark for 1 d, and hardened in the dark; lane 4, 7-d-old etiolated plants exposed to light, kept in the dark for 2 d, and hardened in the dark; lane 5, 7-d-old etiolated plants exposed to light, kept in the dark for 3 d, and hardened in the dark; lane 6, 7-d-old etiolated plants exposed to light, kept in the dark for 5 d, and hardened in the dark; lane 7, same as lane 5 without light treatment; lane 8, same as lane 6 without light treatment; lane 9, 10-d-old etiolated plants exposed to light and then hardened in the dark; lane 10, 12-d-old etiolated plants exposed to light and then hardened in the dark; lane 11, same as lane 8 plus 7 d of hardening under the standard photoperiod; lane 12, same as lane 8 plus 4 d at 20°C and 7 d of hardening under the standard photoperiod.
Total RNA was isolated from 100 mg of Arabidopsis leaves by using the Trizol reagent (GIBCO-BRL). Samples were homogenized in 1 mL of reagent and incubated for 10 min at room temperature before the addition of 0.2 mL of chloroform. The tubes were vortexed and centrifuged at 12,000g for 10 min. The aqueous phase was transferred to a fresh tube from which the RNA was precipitated with 0.5 mL of isopropyl alcohol. Equal amounts of total RNAs for each sample (15 μg) were separated in a 1% agarose/formaldehyde gel, visualized with ethidium bromide to test for equal loading and to assess RNA integrity (Bradford and Chandler, 1992), and transferred to a nylon filter (Hybond N+, Amersham).
RESULTS
Identification of the cor14b-Encoded Chloroplast Protein
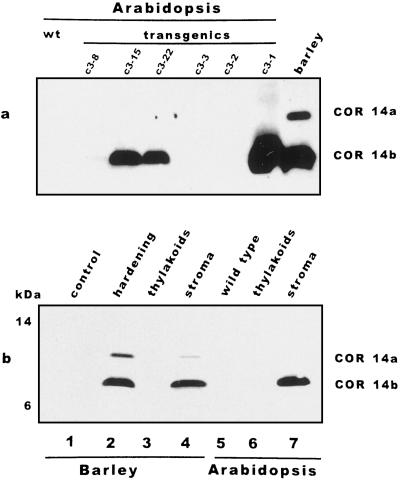
The cDNA clone cor14b (formerly pt59) was previously isolated from leaves of barley upon exposure to low temperature (Cattivelli and Bartels, 1990). Immunoblotting with a polyclonal antibody raised against a fusion protein expressed in vitro using the cor14b sequence led to detection of two SDS-PAGE bands with slightly different electrophoretic mobilities. These were named COR14a and COR14b, respectively, according to their high (a) and low (b) relative molecular masses (Crosatti et al., 1995, 1996). The detection of multiple bands with an antibody raised against a single gene product may be due to either primary sequence homology, leading to epitope sharing, or to maturation of a single precursor protein at multiple sites (Johansson and Forsman, 1992). To verify the relationship between the two SDS-PAGE bands with the cor14b gene, the separation between COR14a and COR14b was first improved by screening several SDS-PAGE methods, leading to optimal separation with the Tris-Tricine buffer system (Schagger and von Jagow, 1987). Two different experiments were then performed. With the first approach, transgenic Arabidopsis plants were obtained by transformation with the chimeric construct 35SCaMV-cor14b, thus leading to the constitutive expression of cor14b. Six independent transgenic Arabidopsis lines were obtained and analyzed for integration of the chimeric gene (Southern analysis) and tested for their ability to accumulate COR14b (Fig. 1a). Plants C3–15 and C3–22 were selfed and the T3 generation was used for the experiments. Analysis of chloroplast extracts and subchloroplast stromal and thylakoidal fractions from transgenic Arabidopsis showed that a single SDS-PAGE immunoreactive band was detected by the anti-COR14b antibody having a mobility corresponding to COR14b. Moreover, the transgene product was correctly targeted to the chloroplast stroma compartment, as in the cold-treated barley (Fig. 1b).
Figure 1.
cor14b encodes for COR14b. a, Total protein extracts from wild-type Arabidopsis (wt), transgenic Arabidopsis containing the chimeric construct 35SCaMV-cor14b-NOS, and cold-hardened (7 d at 3°C) barley (cv Onice) were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. b, Protein extracts isolated from different compartments of barley (cv Onice) and of the transgenic Arabidopsis line C3–15 were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. Lane 1, Total protein extract from barley grown at 20°C; lane 2, total protein extract from barley hardened 7 d at 3°C; lane 3, thylakoid protein extract from barley hardened 7 d at 3°C; lane 4, stroma protein extract from barley hardened 7 d at 3°C; lane 5, total protein extract from wild-type Arabidopsis; lane 6, thylakoid protein extract from transgenic Arabidopsis; and lane 7, stroma protein extract from transgenic Arabidopsis.
Although the above result suggests that the cor14b gene codes only for the low-molecular-mass COR14 (COR14b), no information was obtained concerning the origin of COR14a. Therefore, we proceeded to the alternative approach of isolating and characterizing the two immunoreactive bands isolated directly from cold-treated barley leaves. To this end, intact chloroplasts were isolated from cold-treated barley leaves. After osmotic shock and centrifugation, the stroma-soluble fraction was obtained and proteins therein were fractionated by preparative free-flow IEF, thus obtaining 30 fractions in the 3.0 to 12.0 pH range. Aliquots from IEF fraction were assayed by SDS-PAGE and immunoblotting. The pooled positive fractions were subjected to preparative electrophoresis (electroendosmotic electrophoresis), thus obtaining pure fractions of COR14a and COR14b. Aliquots were blotted onto a PVDF membrane and subjected to automated N-terminal amino acid-microsequence analysis. An 11-residue sequence was obtained from COR14a, allowing us to compare it with the amino acid sequence deduced from cor14b and from other COR genes.
Using the IEF step during COR14b purification, we determined the protein pI (4.5), which was very different from the value of 11 calculated from the cor14b-deduced sequence, suggesting a re-evaluation of the DNA sequence data. Sequencing of the cor14b cDNA clone indeed showed that thymine in position 313 was absent, leading to a frame shift from amino acid 77. The mistake has been corrected in GenBank (accession no. M60732). The amended sequence for COR14b is shown in Figure 2 as aligned with WCS19 (Chauvin et al., 1993) and the stretch obtained from microsequencing of COR14a. However, higher homology was obtained with the protein sequence deduced from WCS19 of wheat than from cor14b (Fig. 2), thus suggesting that COR14a could be the product of a WCS19 homologous barley gene rather than of cor14b. Since WCS19 is a chloroplast-localized protein (Chauvin et al., 1993), the alignment between the N-terminal sequence of the mature COR14a and the amino acid sequence deduced from WCS19 cDNA suggests a possible maturation site for WCS19 located at position 93. Identity was obtained over short antigenic sequences between COR14b and WCS19 (between residues 158 and 168 in the WCS19 sequence), leading to the conclusion that cross-reactivity was due to epitope sharing.
Figure 2.
Amino acid sequence alignment among WCS19 from wheat, COR14b, as deduced from the cDNA clones, and the N-terminal sequence of COR14a. “I” and “x” indicate a perfect match and homologous substitution, respectively, between WCS19 and COR14b. “#” and “*” indicate a perfect match and homologous substitution, respectively, between WCS19 and COR14a.
Effect of Light and Cold on the Expression of cor14b-mRNA and on the Accumulation of the Corresponding Protein
Since the expression of cor14b was shown to be light dependent (Crosatti et al., 1995), the following experiment was performed to further characterize the role of light. Barley plants grown for 7 d under day/night conditions (standard photoperiod) or in the dark (etiolated) were subjected to cold hardening (7 d) at a normal photoperiod or in the dark. Plants grown and hardened in the dark accumulated a reduced amount of COR14b (about 30%, as determined on the basis of 10 independent experiments), as compared with plants grown and hardened under the standard photoperiod (Fig. 3a, lane 5 versus lane 2). COR14a could not be detected in plants never exposed to light even with longer exposure. A short light exposure of etiolated plants before cold treatment (200 μmol m−2 s−1 for 5 min) enabled the plants to enhance COR14b accumulation during the following cold treatment to 60% of the level of fully greened leaves, whereas only traces of COR14a were detected (Fig. 3a, lane 6).
An additional experiment was designed to verify the relationship between the effects of light and cold on COR14b accumulation. Seven-day-old etiolated plants were cold acclimated for 8 to 13 d in the dark. Immunoblotting showed that longer hardening periods did not produce any further COR14b accumulation compared with plants that were hardened for 8 d only (Fig. 3b, lanes 2–4). When the cold plus dark period was interrupted after 7 d by a pulse of white light (200 μmol m−2 s−1 for 5 min), COR14b accumulated during the next 6 d to two-thirds of the control level (Fig. 3b, lanes 5–7). When the same light plus cold treatment was applied without previous hardening, COR14b was detected after 6 d at about two-thirds of the level (Fig. 3b, lanes 8–10). Therefore, the accumulation of COR14b is controlled by the cumulative effects of cold and light: The cold signal alone promotes COR14b accumulation to about one-third of the maximum level, whereas the cold signal and a light pulse allow COR14b accumulation to about two-thirds of the maximum amount.
cor14b Expression in the Albino Mutant
To assess the role of the chloroplast development on cor14b expression and protein accumulation, cold-hardening experiments were performed on albino plants carrying the mutation an, which blocks chloroplast development in the early stages (Fig. 4). Albino mutant plants were cold acclimated for 7 d under a standard photoperiod, and the cor14b mRNA level was compared with that found in etiolated plants hardened in the dark and with green plants hardened in the light. Northern blots showed that mRNA levels were about 5% and 30%, respectively, in albino and etiolated plants with respect to those detected in fully greened plants (Fig. 4a, lanes 4 and 5 versus lane 3).
When the plants described above were assayed for accumulation of COR14b, the amount of protein accumulated in the two samples was similar (Fig. 4c, lane 4 and 5) despite the lower cor14b mRNA levels in albino plants with respect to etiolated plants. Since two different cor14b mRNA levels support the same amount of protein accumulation, it could be suggested that posttranscriptional mechanism(s) are involved in the accumulation of COR14b. Western analysis made with protein extracted from either albino or etiolated plants did not allow the detection of COR14a even after longer autoradiography exposure. Normal COR14a accumulation was, however, detected in plants carrying the mutation an as a heterozygote (Fig. 4c, lane 6).
Posttranscriptional Control of COR14b Accumulation
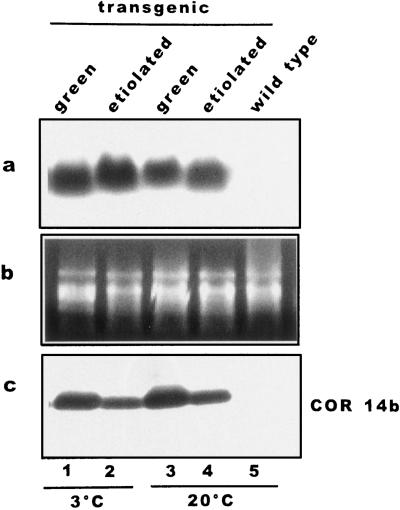
The role of fully developed chloroplasts on COR14b accumulation has been investigated in Arabidopsis plants transformed with the chimeric construct 35SCaMV-cor14b. These plants constitutively expressed cor14b mRNAs regardless of temperature and light conditions (Fig. 5a). Nevertheless, green plants accumulated about 2 times more COR14b than etiolated plants (Fig. 5c), confirming that the presence of light and/or fully developed chloroplasts stimulate further COR14b accumulation.
Figure 5.
Accumulation of COR14b in transgenic Arabidopsis plants. Green and etiolated Arabidopsis plants transformed with the chimeric construct 35SCaMV-cor14b were analyzed for the amount of cor14b mRNA (a) and of the corresponding COR14b protein (b). A northern blot made with total RNAs was stained with ethidium bromide (c) and probed with the cDNA clone cor14b; total protein extracts were separated by Tricine-SDS-PAGE and hybridized with COR14 antibody. Lane 1, Green transgenic plants hardened under the standard photoperiod; lane 2, etiolated transgenic plants hardened in the dark; lane 3, green transgenic plants grown at 22°C; lane 4, etiolated transgenic plants grown at 22°C; and lane 5, wild-type plants grown at 22°C.
Red (660 nm) and Blue (400 nm) Lights Are the Most Effective Wavelengths That Enhance COR14b Accumulation
Light can affect plant gene expression by either promoting photosynthesis or by activating specific photoreceptors, such as phytochrome or the blue-light receptor. To elucidate the pathways by which COR14b accumulation is affected, we applied a light pretreatment of different wavelengths, followed by standard cold hardening (7 d), and then verified protein accumulation by immunoblotting.
White light (10 μmol m−2 s−1 for 5 min) was enough to significantly enhance COR14b accumulation in etiolated plants, as shown in Figure 3, with respect to dark controls. Etiolated plants were exposed for 5 min to about 10 μmol m−2 s−1 of monochromatic light and then hardened for 1 week. Red light (660 nm) was as effective as white light but more effective than far-red light (730 nm). When far-red light followed red-light treatment, only traces of COR14b were detected, thus suggesting that phytochrome action was involved in COR14b accumulation. The possible involvement of a blue-light photoreceptor was investigated by treatment with two different blue-light sources of 400 and 450 nm, which are similarly absorbed by the photosynthetic apparatus, but the former is far more effective than the latter in eliciting a blue-light response. It was shown that 400 nm of light is as effective as 660 nm of red light in inducing COR14b accumulation, whereas 450 nm of light was the least effective. In Figure 6a, autoradiography following short exposure is shown so that only the strongest signals were recorded. A longer exposure (Fig. 6b) allowed detection of a basal level of COR14b induced by far-red, green, and 450 nm of light, whereas the effect of the cold signal alone was even lower (see also Fig. 6c, no light sample). Densitometric analysis revealed that the amount of COR14b obtained after white-, red 660-nm, and blue 400-nm light treatments was about twice that found after far-red 730-, blue 450-, and green 500-nm treatments.
An additional experiment was performed to determine the minimal duration of the light treatment effective in inducing COR14b accumulation: etiolated plants were treated with red light (660 nm) at 200 μmol m−2 s−1 for 5 s to 5 min, and then hardened in the dark for 1 week. The shortest treatment was already effective in inducing a significant accumulation of COR14b (Fig. 6c).
Lifetime of the Light Signal
The above results show that a short dim-light treatment is sufficient to induce COR14b accumulation through the action of phytochrome and/or blue-light photoreceptors, implying that one or more factors are released/activated that can influence cor14b gene transcription/translation. It is relevant, in this respect, to measure the lifetime of the light signal. To this aim, etiolated plants were grown and treated with 5 min of light as described above; however, a further dark period at room temperature for 1 to 4 d was applied before hardening. COR14b accumulation was detected after a 2- but not a 3-d delay in hardening, implying a 2- to 3-d lifetime for the light signal (Fig. 7a, lanes 2–6). This was due to decreased cor14b mRNA, as shown by northern analysis (Fig. 7b, lanes 4–6).
Effects of the Plant Age
Etiolated plants (10–12 d old) were almost unable (10 d old) or completely unable (12 d old) to accumulate cor14b mRNAs and the corresponding protein in the absence of light (Fig. 7, a and b, lanes 7 and 8). However, this effect was reversed by transferring plants to a standard photoperiod after which plants were able to undergo greening to normal conditions and restored their ability to accumulate COR14b when further acclimated in the presence of light (Fig. 7a, lanes 11 and 12). Moreover, this decline in mRNA induction was specific for cor14b, since 12-d-old, etiolated plants expressed other COR genes such as paf93 (Fig. 7c). The possibility of switching off the expression of cor14b without affecting the expression of paf93 shows that the expression of cor14b and paf93 is controlled by different signal transduction pathways.
DISCUSSION
The Signal Transduction Pathway Leading to COR14b Accumulation Is Independent from That Controlling the Expression of Other COR Genes
In fully etiolated leaves cold treatment promotes a limited expression of cor14b mRNA, which suggests a reduced level of COR14b. Notably, COR14b is most likely targeted to the proplastids, since the protein has the same electrophoretic mobility in both etiolated and green plants, implying that it was correctly processed. We were able to stop cor14b expression independently from the induction of other COR genes during cold treatment by using young, etiolated seedlings. The study of the water-stress response has demonstrated that several pathways, either ABA dependent or ABA independent, are involved in the control of drought-induced genes (Shinozaki and Yamaguchi-Shinozaki, 1997). It is well known that ABA can also control the expression of several COR genes (Hughes and Dunn, 1996) but not the expression of cor14b (Grossi et al., 1992). Nevertheless, in addition to the ABA-mediated cold induction, expression of COR genes can also be achieved through at least two ABA-independent pathways. The Arabidopsis gene rd29A was shown to have a cis-acting element responsible for its ABA-independent induction under cold and drought conditions (Yamaguchi-Shinozaki and Shinozaki, 1994), and the expression of the barley paf93 gene resembles very closely that of rd29A (Grossi et al., 1995). In this work we have identified a physiological condition in which the cold-induced expression of cor14b was blocked, whereas that of paf93 was not affected, demonstrating that the ABA-independent cold response can be separated into two pathways, one of which is effective in the control of gene coding for chloroplast-localized COR proteins.
Low temperature decreases photosynthetic electron transport rate, leading to an excess of light harvested by the antenna system and finally to an overreduction of the plastoquinone pool (Gray et al., 1997). In the short term, this condition leads to the reversible phosphorylation of the PSII subunit CP29 (Bergantino et al., 1995, 1998), causing an increase in the thermal dissipation of excess energy (Mauro et al., 1998) through a conformational change (Croce et al., 1996). The redox state of plastoquinone also affects the expression of nuclear genes coding for chloroplast proteins (Danon and Mayfield, 1995; Escoubas et al., 1995). In winter rye the expression of WCS19, a protein sharing homology with the N-terminal sequence of COR14a, has been correlated with the relative reduction state of PSII (Gray et al., 1997), demonstrating that the plastoquinone pool is a part of the signal transduction pathway leading to the expression of gene coding for chloroplast-localized COR proteins. By using the albino an mutant we showed that chloroplasts are required for the normal expression of the COR genes coding for chloroplast-localized proteins. The cor14b steady-state mRNA level was much lower in the mutant than in etiolated or green plants, although the former was grown under the standard photoperiod, a condition that generally promotes cor14b expression. This result proves that a signal deriving from proplastids or from chloroplasts is required for the normal expression of the gene, although our results indicate that the plastoquinone redox state is not the only factor regulating accumulation of COR plastid proteins. The cor14b gene can be expressed, although at lower level, when the photosynthetic apparatus is absent and the plastid development is blocked at very early stages. Furthermore, an enhancement of cor14b expression can be achieved through a photoreceptor-mediated signal.
Involvement of Photoreceptors
Although cold treatment alone induces low levels of COR14b accumulation, a short exposure of plants to light before cold treatment ensures higher cor14b expression and a corresponding increase in COR14b accumulation. The low intensity and the short duration of the light pulse needed for this effect suggest that a photoreceptor, rather than the photosynthetic electron chain, is involved. In fact, red (660 nm) and blue (400 nm) light were the most active light sources promoting COR14b accumulation, whereas the enhancing effect of red light can be reversed either by far-red treatment (Fig. 6a) or by keeping the plants at 20°C in the dark for a few days before cold acclimation (Fig. 7), suggesting phytochrome action. On the other hand, the contrasting effect of 400 versus 450 nm of light suggests that the blue-light receptor is also involved. Interactions between different wavelengths of light affecting the expression of particular genes are well known (Terzaghi and Cashmore, 1995). In etiolated Arabidopsis seedlings the steady-state mRNA levels for chloroplast glyceraldehyde-3-P dehydrogenase (GapA and GapB) and Lhc proteins increased after a 1-min exposure to white, red, or blue light (Dewdney et al., 1993), whereas red-light activation of the lhc gene (s) could be reversed by far-red light treatment (Karlin-Neuman et al., 1988; Dewdney et al., 1993). It is also known that the illumination of tomato seedlings with both blue and red light induces higher expression of nuclear-encoded thylakoid genes than does red light alone (Oelmuller et al., 1989), whereas the steady-state level of early light-induced protein transcripts in pea seedlings is regulated by a combined action of phytochrome and blue-light receptor systems (Adamska, 1995).
Regulation by Protein Stability
COR14b is more stable in albino leaves hardened in the presence of light than in etiolated leaves hardened in the dark. In fact, the amount of COR14b accumulated in the albino mutant was similar to that found in etiolated leaves, although the mutant expressed much less cor14b mRNA than etiolated plants. In transgenic Arabidopsis the same level of cor14b mRNA, under the control of 35SCaMV, supports different amounts of COR14b accumulation, depending on plant growth conditions (Fig. 5c). Similarly, fully greened plants hardened under the standard photoperiod accumulated a higher level of COR14b than etiolated plants exposed to short light pulses, despite the fact that in both conditions the plants showed the same cor14b mRNA steady-state level (Fig. 7b, lanes 1 and 9). These results are consistent with the hypothesis that degradation mechanisms active during growth in the dark reduce COR14b accumulation; on the contrary, growth under the standard photoperiod contributes to increasing the stability of COR14b. The degradation of chloroplast proteins can be caused by excess light (e.g. degradation of D1 protein), by targeting of the protein to a wrong compartment, or by the absence of cofactors or of other components interacting with the protein (Adam, 1996). Chlorophyll was found necessary for the stabilization of the CP43 and D1 proteins (Mullet et al., 1990), as well as Cu+ for the stabilization of plastocyanin (Merchant and Bogarad, 1986). The identical electrophoretic mobility of COR14b in green and etiolated leaves suggests that the protein is probably correctly targeted in etiolated tissues. Furthermore, the albino mutant grown under standard photoperiod showed an increased COR14b stability in comparison with etiolated plants. Therefore, degradation of COR14b in etiolated plants hardened in the dark may be due to the absence of a component that interacts with COR14b.
ACKNOWLEDGMENTS
The authors wish to thank Dr. F. Rizza for collaboration during the experiments with monochromatic light and Mrs. D. Pagani for skillful technical assistance.
Abbreviations:
- CaMV
cauliflower mosic virus
- COR
cold-regulated
Footnotes
This work was funded by Ministero per le Politiche Agricole, “Progetto Biotecnologie Vegetali” and by Consiglio Nazionale delle Richerche “Progetto Biotecnologie.”
LITERATURE CITED
- Adam Z. Protein stability and degradation in chloroplasts. Plant Mol Biol. 1996;32:773–783. doi: 10.1007/BF00020476. [DOI] [PubMed] [Google Scholar]
- Adamska I. Regulation of early light-inducible protein gene expression by blue and red light in etiolated seedlings involves nuclear and plastid factors. Plant Physiol. 1995;107:1167–1175. doi: 10.1104/pp.107.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R, Dal Belin Peruffo A, Barbato R, Ghisi R. Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheath and mesophyll cells in maize. Eur J Biochem. 1985;146:589–595. doi: 10.1111/j.1432-1033.1985.tb08692.x. [DOI] [PubMed] [Google Scholar]
- Bergantino E, Dainese P, Cerovic Z, Sechi S, Bassi R. A post-transcriptional modification of the photosystem II subunit CP29 protect maize from cold stress. J Biol Chem. 1995;265:8474–8481. doi: 10.1074/jbc.270.15.8474. [DOI] [PubMed] [Google Scholar]
- Bergantino E, Sandonà D, Cugini D, Bassi R. The photosystem II subunit CP29 can be phosphorylated in both C3 and C4 plants as suggested by sequence analysis. Plant Mol Biol. 1998;36:11–22. doi: 10.1023/a:1005904527408. [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Chandler PM. Expression of “dehydrin-like” proteins in embryos and seedlings of Zizania palustris and Oryza sativa during dehydration. Plant Physiol. 1992;99:488–494. doi: 10.1104/pp.99.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham CR, Eslick RF, Haus TE, Hockett EA, Jarvi AJ, McProud WL, Tsushiya T. Description of genetic stocks in the Barley Genetic Stock Center at Fort Collins, Colorado. Barley Genet News. 1971;1:103–193. [Google Scholar]
- Capel J, Jarillo JA, Salinas J, Martinez-Zapater JM. Two homologous low temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol. 1997;115:569–576. doi: 10.1104/pp.115.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattivelli L, Bartels D. Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol. 1990;93:1504–1510. doi: 10.1104/pp.93.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin LP, Houde M, Sarhan F. A leaf-specific gene stimulated by light during wheat acclimation to low temperature. Plant Mol Biol. 1993;23:255–265. doi: 10.1007/BF00029002. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Cline K, DeLisle AJ. An Arabidopsis chloroplast RNA-binding protein gene encodes multiple mRNAs with different 5′ ends. Plant Physiol. 1994;106:303–311. doi: 10.1104/pp.106.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce R, Breton J, Bassi R. Confomational changes induced by phosphorylation on the PSII subunit CP29. Biochemistry. 1996;35:11142–11148. doi: 10.1021/bi960652t. [DOI] [PubMed] [Google Scholar]
- Crosatti C, Nevo E, Stanca AM, Cattivelli L. Genetic analysis of the accumulation of COR14 proteins in wild (Hordeum spontaneum) and cultivated (Hordeum vulgare) barley. Theor Appl Genet. 1996;93:975–981. doi: 10.1007/BF00224101. [DOI] [PubMed] [Google Scholar]
- Crosatti C, Soncini C, Stanca AM, Cattivelli L. The accumulation of a cold-regulated chloroplastic protein is light-dependent. Planta. 1995;196:458–463. doi: 10.1007/BF00203644. [DOI] [PubMed] [Google Scholar]
- Curioni A, Dal Belin-Peruffo A, Nuti MP. Purification of cellulases from Streptomyces strain A20 by electroendoosmotic preparative electrophoresis. Electrophoresis. 1988;9:327–330. doi: 10.1002/elps.1150090708. [DOI] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light regulated translation of chloroplast messenger RNAs through redox potential. Science. 1995;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Debel K, Knack G, Kloppstech K. Accumulation of plastid HSP 23 of Chenopodium rubrum is controlled post-translationally by light. Plant J. 1994;6:79–85. [Google Scholar]
- Dewdney J, Conley TR, Shih MC, Goodman HM. Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol. 1993;103:1115–1121. doi: 10.1104/pp.103.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman P. A method for the determination of the amino acid sequence in peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GR, Chauvin LP, Sarhan F, Huner NPA. Cold acclimation and freezing tolerance. Plant Physiol. 1997;114:467–474. doi: 10.1104/pp.114.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi M, Cattivelli L, Terzi V, Stanca AM. Modification of gene expression induced by ABA, in relation to drought and cold stress in barley shoots. Plant Physiol Biochem. 1992;30:97–103. [Google Scholar]
- Grossi M, Giorni E, Stanca AM, Cattivelli L. Wild and cultivated barleys show differences in the expression pattern of a cold-regulated gene family under different light and temperature conditions. Plant Mol Biol. 1998;38:1061–1069. doi: 10.1023/a:1006079916917. [DOI] [PubMed] [Google Scholar]
- Grossi M, Gulli M, Stanca AM, Cattivelli L. Characterization of two barley genes that respond rapidly to dehydration stress. Plant Sci. 1995;105:71–80. [Google Scholar]
- Hoekema A, Hirsch P, Hooykaas P, Schilperoort R. A binary plant vector strategy based on segregation of vir and T region of the Agrobacterium tumefaciens Ti plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;47:291–305. [Google Scholar]
- Johansson IM, Forsman C. Processing of the chloroplast transit peptide of pea carbonic anhydrase in chloroplasts and in Escherichia coli. Indication of two cleavage sites. FEBS Lett. 1992;314:232–236. doi: 10.1016/0014-5793(92)81478-5. [DOI] [PubMed] [Google Scholar]
- Karlin-Neuman G, Sun L, Tobin EM. Expression of light harvesting chlorophyll a/b protein genes is phytochrome-regulated in etiolated Arabidopsis thaliana seedlings. Plant Physiol. 1988;88:1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Thomashow M. DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis COR15 and characterization of the COR15 polypeptide. Plant Physiol. 1992;99:519–525. doi: 10.1104/pp.99.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro S, Dainese P, Lannoye R, Bassi R. Cold-resistant and cold-sensitive maize lines differ in the phosphorylation of the photosystem II subunit, CP29. Plant Physiol. 1998;115:171–180. doi: 10.1104/pp.115.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogarad L. Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J Biol Chem. 1986;261:15850–15853. [PubMed] [Google Scholar]
- Montane MH, Dreyer S, Triantaphyllides C, Kloppstech K. Early light-inducible proteins during long-term acclimation of barley to photooxidative stress caused by light and cold: high level of accumulation by posttranscriptional regulation. Planta. 1997;202:293–302. [Google Scholar]
- Monroy AF, Dhindsa RS. Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25°C. Plant Cell. 1995;7:321–331. doi: 10.1105/tpc.7.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET. Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- Mullet JF, Klein PG, Klein RR. Chlorophyll regulates accumulation of the plastid encoded chlorophyll apoprotein CP43 and apoprotein D1 by increasing apoprotein stability. Proc Natl Acad Sci USA. 1990;87:4038–4042. doi: 10.1073/pnas.87.11.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmuller R, Kendrick RE, Briggs WR. Blue-light mediated accumulation of nuclear-encoded transcripts coding for proteins of the thylakoid membrane is absent in the phytochrome-deficient aurea mutant of tomato. Plant Mol Biol. 1989;13:223–232. doi: 10.1007/BF00016140. [DOI] [PubMed] [Google Scholar]
- Oquist G, Huner NPA. Cold-hardening-induced resistance to photoinhibition of photosynthesis in winter rye is dependent upon an increased capacity for photosynthesis. Planta. 1993;189:150–156. [Google Scholar]
- Ostersetzer O, Adam Z. Effects of light and temperature on expression of ClpC, the regulatory subunit of chloroplastic Clp protease, in pea seedlings. Plant Mol Biol. 1996;31:673–676. doi: 10.1007/BF00042238. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulphate-polyacrilamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk B, Kozloff LM. A method for the efficient blotting of strongly basic proteins from sodium dodecyl sulfate-polyacrylamide gels to nitrocellulose. Anal Biochem. 1985;150:403–407. doi: 10.1016/0003-2697(85)90528-7. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Valvekens D, van Montagu M, van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]