Abstract
ATR is an essential protein that functions as a damage sensor and a proximal kinase in the DNA damage checkpoint response in mammalian cells. It is a member of the phosphoinositide 3-kinase-like kinase (PIKK) family, which includes ATM, ATR, and DNA-dependent protein kinase. Recently, it was found that ATM is an oligomeric protein that is converted to an active monomeric form by phosphorylation in trans upon DNA damage, and this raised the possibility that other members of the PIKK family may be regulated in a similar manner. Here we show that ATR is a monomeric protein associated with a smaller protein called ATRIP with moderate affinity. The ATR protein by itself or in the form of the ATR-ATRIP heterodimer binds to naked or replication protein A (RPA)-covered DNAs with comparable affinities. However, the phosphorylation of RPA by ATR is dependent on single-stranded DNA and is stimulated by ATRIP. These findings suggest that the regulation and mechanism of action of ATR are fundamentally different from those of the other PIKK proteins.
The DNA damage checkpoint response is the slowing down or arrest of cell cycle progression in response to DNA damage (1, 18, 29, 42). Conceptually, this stress response pathway encompasses three components, damage sensors, signal transducers, and effectors. At least two groups of proteins are thought to function as sensors in mammalian cells: ATM and ATR, which belong in the phosphoinositide 3-kinase-like kinase (PIKK) family, and the Rad17-RFC-9-1-1 complex, which belongs in the RFC/PCNA family of clamp loader-DNA clamp complexes (26).
The PIKK family encompasses three proteins, DNA-dependent protein kinase (DNA-PK), ATM, and ATR (1, 6, 30). These are large proteins with molecular masses in the range of 301 to 469 kDa. Although PIKKs exhibit sequence similarities in their carboxy-terminal domains to phosphoinositide 3-kinsases, they have no lipid kinase activities. Instead, all three PIKK members are Ser/Thr kinases with preference for amino acids within the SQ/TQ context (11). The prototype of the family, DNA-PK, is a heterotrimeric complex of a 469-kDa catalytic subunit, DNA-PKcs, and a dimer of Ku70 and Ku86 (8). Ku70-Ku86 is a rather stable dimer which binds to DNA ends and recruits DNA-PKcs to the sites of double-strand breaks. DNA-PK plays an essential role in double-strand break repair necessary for V(D)J recombination and in nonhomologous end joining of double-strand breaks induced by ionizing radiation and radiomimetic drugs (32). Whether DNA-PK signals double-strand breaks to the checkpoint machinery still remains controversial, but DNA-PK mutants do not have an overt DNA damage checkpoint defect (24). In contrast, ATM and ATR seem to function almost exclusively in the DNA damage checkpoint response, although it has been found that ATM does contribute to double-strand break repair during V(D)J recombination (22). Both ATM and ATR are DNA binding proteins. ATM binds preferentially to DNA termini, apparently in a monomeric form (33), whereas ATR binds to the DNA backbone with no preference for ends but with some preference for UV-induced (6-4) photoproducts (34). Consistent with these properties, ATM is the main kinase that initiates the immediate checkpoint response to ionizing radiation, and ATR is the predominant initiator of the checkpoint response to UV-induced DNA damage (1, 29, 30).
Because DNA-PKcs is recruited to DNA by the Ku complex, it has been suggested that ATM and ATR may be recruited to DNA in a similar manner, that is, through the intermediacy of smaller subunits (6). Indeed, in Schizosaccharomyces pombe spRad3 (ATR/ATM ortholog) forms a complex with spRad26 (7), and in Saccharomyces cerevisiae, scMec1 (ATR ortholog) forms a complex with scDdc2 (20, 25, 35); some evidence was obtained that these smaller proteins play a role in regulating the kinase and DNA binding activities, respectively, of the corresponding PIKK members (27, 37). Search for similar proteins in mammalian cells led to the discovery of an ATR-interacting protein, ATRIP (5). However, the role of ATRIP in regulating ATR, especially recruiting it to DNA, is uncertain because ATRIP-free ATR binds to DNA relatively efficiently (34). So far, no small subunit for ATM has been found, but it is thought that the ATM activity is regulated by the change in the oligomeric state of the protein (2).
Using immunoprecipitation assays coupled with site-specific mutagenesis, it was found that in undamaged human cells ATM was in the form of a homodimer or a higher-order oligomer that upon DNA damage became phosphorylated by an intermolecular reaction and dissociated to monomers, which appear to be the active form of the enzyme (2). In support of oligomerization as a general regulatory mechanism for PIKK family proteins, it was found that these proteins contain many HEAT repeats, which are known to be involved in protein-protein interactions (23). Because ATR, like ATM, catalyzes autophosphorylation, as we show below, we reasoned that ATR might similarly be an oligomeric protein whose activity is regulated by a change in its oligomerization state induced by DNA damage. Indeed, a preliminary study using gel filtration chromatography concluded that ATR was found predominantly in complexes with an average molecular mass of 1,000 kDa (40). We wished to expand upon this finding and to determine if the ATR activity may be modulated by a change in its quaternary structure, as has been found for ATM.
In this study we used hydrodynamic methods to determine the molecular weight of native ATR protein and the other members of the PIKK family. In agreement with earlier reports, we find that DNA-PKcs is a monomer (13). We also found that ATM is predominantly a dimer that is in equilibrium with monomeric and larger oligomeric forms. Importantly, in contrast to an earlier report (40), ATR behaves as a monomer, as analyzed both by gel filtration chromatography and by glycerol gradient sedimentation velocity centrifugation. Moreover, ATRIP, which has been found in ATR immunoprecipitates and is considered to be essential for the stability of ATR in vivo (5), readily dissociates from ATR during gel filtration or sedimentation under relatively mild conditions. Analyses of DNA binding and kinase activities of ATR and the ATR-ATRIP complex revealed that both forms bound to naked and replication protein A (RPA)-covered DNA with comparable affinities. The ATR-ATRIP complex also binds to RPA and phosphorylates the RPA32 subunit in a DNA-dependent manner. The binding to RPA is mediated by ATRIP, which also stimulates the kinase activity of ATR. Taken together, our data indicate that ATR has a distinct quaternary structure, unrelated to the other members of the PIKK family, and that it can directly bind to DNA without the intermediacy of another protein.
MATERIALS AND METHODS
Plasmids and antibodies.
Myc(3x)-tagged ATRIP cDNA (a kind gift from Stephen J. Elledge, Department of Biochemistry, Baylor College of Medicine) was used as a template for amplification of ATRIP by PCR. The PCR product was digested and ligated into pFastBacHTb to generate pFast-His6-Flag-ATRIP. The Flag-ATR construct containing the full-length cDNA clone of human ATR in the pcDNA3 (Invitrogen) expression vector used for the expression and purification of ATR from human embryonic kidney 293T (HEK293T) cells has been described (34).
Mouse monoclonal anti-Flag antibody was obtained from Sigma. Rabbit polyclonal ATRIP-N antibody was a gift from Stephen J. Elledge (5). Mouse monoclonal antibodies against RPA32 and RPA70 were obtained from Oncogene Research Products. Rabbit polyclonal antibody against ATR was obtained from Affinity Bioreagents. Mouse monoclonal antibody against DNA-PKcs was a kind gift from Dale A. Ramsden (Department of Biochemistry and Biophysics, University of North Carolina). Rabbit polyclonal antibody against ATM was a kind gift from Richard S. Paules (National Institute of Environmental Health Sciences).
Expression and purification of recombinant proteins.
HEK293T cells were grown in Dulbecco's modified Eagle medium (GIBCO/BRL) supplemented with 10% fetal bovine serum and 100 U of penicillin and streptomycin/ml. Cells (8 × 106/225-cm2 flask) were transfected either with 25 μg of pcDNA3-Flag-ATR plasmid alone or cotransfected with 15 μg of Flag-ATR and 10 μg of Myc(3x)-tagged ATRIP plasmids by using a calcium phosphate transfection method (10). Forty-eight hours after transfection cells were washed with phosphate-buffered saline and lysed in 5 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM β-glycerophosphate, 10% glycerol, 1% Tween 20, 0.1% NP-40, 1 mM Na3VO4, 1 mM NaF, and protease inhibitors [Roche Molecular Biochemicals]) for 30 min on ice. After centrifugation at 32,000 × g for 30 min, the supernatants were incubated with 40 μl of anti-Flag M2 affinity gel (Sigma) at 4°C overnight. For Flag-ATR-transfected cells, the Flag-ATR-bound beads were washed three times with 1.5 ml of washing buffer that contained 50 mM Tris-HCl (pH 7.5) and either 150 mM or 1 M NaCl and the proteins were eluted in 200 μl of elution buffer that contained 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol [DTT], and 200 μg of Flag-peptide/ml. For Flag-ATR- and Myc(3x)-ATRIP-cotransfected cells, washing was performed only under low salt conditions (150 mM NaCl).
Baculovirus for expression of His6-Flag-ATRIP was generated with the BAC-TO-BAC baculovirus expression system (GIBCO/BRL) and protocols suggested by the manufacturer. The optimal multiplicity of infection for recombinant virus was empirically determined. A monolayer of Sf21 insect cells (Invitrogen) grown in Grace's insect medium (GIBCO/BRL) supplemented with 10% fetal bovine serum and 100 U of penicillin and streptomycin/ml was infected with virus and harvested 48 h later. The cells were lysed and centrifuged as described for HEK293T cells, and protein-bound Flag-beads were washed three times in a buffer containing 50 mM Tris-HCl (pH 7.5) and 300 mM NaCl, and the proteins were eluted in elution buffer with 200 μg of Flag-peptide/ml. The 9-1-1 complex containing Flag-Hus1 was purified from High Five insect cells as previously described (15). The recombinant RPA was expressed and purified from BL21(DE3) cells as described previously (9).
Gel filtration analysis.
HEK293T whole-cell extract was prepared in hypotonic buffer (20 mM HEPES [pH 7.9], 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT) as described previously (14). A Superdex-200 PC 3.2/30 (2.4-ml) gel filtration column was equilibrated at 4°C in a buffer with 20 mM HEPES (pH 7.9), 250 mM NaCl, 1.5 mM MgCl2, and 2 mM DTT. Forty microliters of whole-cell extracts (25 mg/ml) which contain approximately 1.5 to 2 pmol of ATR and 0.35 to 0.5 pmol of ATRIP (as determined by Western blotting using recombinant proteins as standards) was loaded onto the column, and the column was developed with the same buffer. After determining the excluded volume (Vo = 0.87 ml) and included volume (Vi = 1.708) of the column, the elution volume (Ve) was measured for standard proteins with a known Stokes radius. For each protein, Kav was calculated from the equation that follows, and a linear relationship was obtained by plotting (−logKav)1/2 against the Stokes radius: Kav = (Ve − Vo)/(Vi − Vo). The standard proteins used were bovine serum albumin (67 kDa, 1.46 ml), rabbit muscle aldalose (158 kDa, 1.33 ml), bovine liver catalase (232 kDa, 1.28 ml), horse spleen ferritin (440 kDa, 1.12 ml), and bovine thyroid thyroglobulin (669 kDa, 0.98 ml). Elution profiles of the proteins of interest were determined by immunoblot analysis.
Glycerol gradient centrifugation.
Whole-cell extracts (500 μg) obtained from nonirradiated or UV-irradiated (50 J/m2, extract made 1 h after irradiation) HEK293T cells in a total volume of 20 μl (25 mg/ml) were layered on top of 2.4-ml 15 to 35% glycerol gradients in buffer containing 20 mM HEPES (pH 7.9), 50 mM KCl, 7 mM MgCl2, and 0.5 mM DTT. The gradients were spun for 6 h at 55,000 rpm in an SW60 rotor at 4°C. Fractions of 200 μl were collected from the bottom of the tube, and 75-μl aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To determine the sedimentation coefficient of ATRIP in the absence of ATR, 4 μg of His6-Flag-tagged ATRIP (purified from insect cells) in a total volume of 40 μl was loaded onto the glycerol gradient and processed as for the whole-cell extract. The elution positions of the proteins of interest were determined by Western blot analysis. Reference proteins with known sedimentation coefficients were used to calibrate the gradients: chicken ovalbumin (3.55 S), rabbit muscle aldalose (7.3 S), bovine liver catalase (11.3 S), bovine thyroid thyroglobulin (19 S).
Biotinylated DNA binding assay.
Streptavidin-conjugated Dynabeads M280 (Dynal) were washed three times with Tris-EDTA (TE) buffer plus 1 M NaCl. A 5′-biotinylated 80-nucleotide (nt) oligomer was added to the beads in the same buffer and incubated at room temperature for 1 h. The sequence of the 5′-biotinylated oligonucleotide used in this study is 5′TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCTCCCTTCTTCTCCTCCCTCTCCCTTCCCTTTTTTTTTTTTTTTTTTTTT 3′.
For each reaction, 7 pmol of DNA and 5 μl of Dynabeads were used. The beads and DNA mixture were washed three times with TE buffer plus 1 M NaCl followed by two washes with TE buffer. The DNA-bound beads were incubated with purified proteins at 30°C for 30 min in a reaction volume of 400 μl with a binding buffer (10 mM MgCl2, 2 mM MnCl2, 10 mM Tris-Cl [pH 7.5], 50 mM NaCl, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 10 μg of bovine serum albumin/ml). For each binding reaction, 7 pmol of RPA, 4 pmol of ATRIP, and 4 pmol of ATR were used. These amounts of DNA and protein were chosen for the binding assays so as to have concentrations of reactants comparable to those used in a recent study (43). After the beads were washed three times with washing buffer (binding buffer with 100 mM NaCl and 0.01% NP-40), the bound proteins were analyzed by Western blotting. In reaction mixtures containing RPA-covered DNA, the 7 pmol of 5′-biotinylated 80-nt DNA was first incubated with RPA at 30°C for 30 min and then either ATR, ATRIP, or ATR-ATRIP complex reconstituted by preincubating the purified ATR and ATRIP on ice for 30 min was added to RPA-coated DNA and further incubated for 30 min at 30°C. After the beads were washed to remove the unbound proteins, the bound proteins were eluted by boiling the beads in SDS buffer and analyzed by Western blotting.
Kinase assay.
Kinase assays were performed in 50-μl reaction mixtures by using 50 ng of ATR, 200 ng of RPA, 200 ng of ATRIP, and 25 μg of single-stranded DNA (ssDNA) in a kinase buffer containing 25 mM HEPES (pH 7.9), 50 mM KCl, 10 mM MgCl2, 2 mM MnCl2, 20% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM Na3VO4, and 1 mM NaF. The reaction mixtures were incubated on ice for 5 min before ATP addition. ATP was added to a final concentration of 50 μM along with 5 μCi of [γ-32P]ATP. Reaction mixtures were incubated at 30°C for the indicated times. Phosphorylated proteins were separated on SDS-10% polyacrylamide gels, visualized by silver staining, and exposed to a Molecular Dynamics PhosphorImager screen for quantitation by IMAGEQUANT software.
Immunoprecipitation assays.
HEK293T cells were either singly transfected or cotransfected with the indicated plasmids by a calcium phosphate method. After 16 h of incubation at 37°C, the cells were washed twice in serum-free Dulbecco's modified Eagle medium, fresh medium was added, and the cells were incubated for another 48 h. The cells were lysed in ice-cold lysis buffer for 30 min. The cell lysates were centrifuged for 30 min at 30,000 × g, and the supernatants were incubated with anti-Flag M2 affinity gel (Sigma) at 4°C for 4 h. The beads were then washed three times with TBS buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl), and bound proteins were eluted with TBS buffer containing 200 μg of Flag-peptide (Sigma)/ml.
RESULTS
Determination of native molecular weight of ATR by hydrodynamic methods.
We determined the molecular weight of ATR in whole-cell extract by the method of Siegel and Monty (31). In this method the Stokes radius is obtained by gel filtration chromatography and the sedimentation coefficient is determined by sedimentation velocity in a density gradient. The molecular weight (Mr) is then calculated from the formula Mr = 6πηoNaS/(1 − υρ), where S is the sedimentation coefficient and a is the Stokes radius of the protein (see Table 1, footnote a, for other definitions).
TABLE 1.
Hydrodynamic properties of PIKK family membersa
| Protein | S value | Stokes radius (Å) | Mr
|
|
|---|---|---|---|---|
| Hydrodynamic | Sequence | |||
| ATR | 12.61 | 72.8 | 385,500 | 301,491 |
| 361,000 | ||||
| ATRIP | 6.40 | 105,000 | 89,891 | |
| DNA-PKcs | 15.72 | 503,000 | 469,146 | |
| ATM | 21.93 | 830,000 | 350,730 | |
Sequence Mr values were calculated from the polypeptide sequences. Hydrodynamic Mr values were calculated from S values and Stokes radii obtained from the equation (31) Mr = 6πη0NaS/(1 − υρ), where N is Avogadro's number (6.02 × 1023), η0 is the viscosity coefficient (1.00 × 10−2 g·s−1·cm−1), ρ is the solution density (1 g/cm3), and υ is the partial specific volume (0.72 cm3/g). Mr values from sedimentation coefficients were obtained from the equation (16) Mr1 = Mr2(S1/S2)3/2.
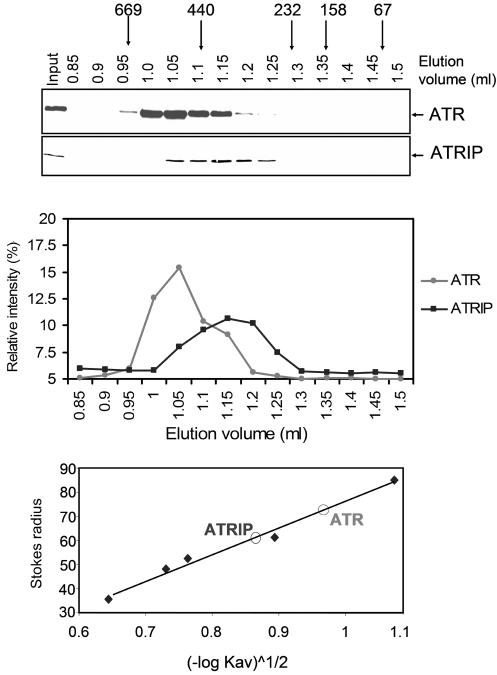
Figure 1 shows the elution profiles of ATR and ATRIP when the whole-cell extract is applied to a gel filtration column. Two points of interest emerge from these profiles. First, the elution profiles of the two proteins do not coincide, indicating that ATR and ATRIP are not in a tight complex. Second, the Stokes radius of ATR is estimated to be 72.8 Å, which is only slightly larger than that of a spherical protein with an Mr of 301,491 (the theoretical molecular weight of ATR). Similarly, the Stokes radius of ATRIP is calculated to be 60 Å, which is considerably larger than that of a spherical protein with an Mr of 89,891 (the theoretical molecular weight of ATRIP). This is due to ATR-ATRIP interaction, as will be shown by sedimentation analysis.
FIG. 1.
Analysis of ATR and ATRIP by gel filtration chromatography. (Top) HEK293T whole-cell extract was loaded onto a Superdex-200 PC 3.2/30 (2.4-ml) column, and fractions were analyzed by Western blotting with anti-ATR and anti-ATRIP antibodies. The column was calibrated with bovine serum albumin (67 kDa, 35.5 Å), rabbit muscle aldalose (158 kDa, 48.1 Å), bovine liver catalase (232 kDa, 52.2 Å), horse spleen ferritin (440 kDa, 61 Å), and bovine thyroid thyroglobulin (669 kDa, 85 Å). The input lane contains 1/10 of the load. (Middle) Quantitative analysis of data shown in the top panel. (Bottom) Determination of the Stokes radii of ATR and ATRIP (indicated as open circles) from the gel filtration data by plotting (−logKav)1/2 against the Stokes radius.
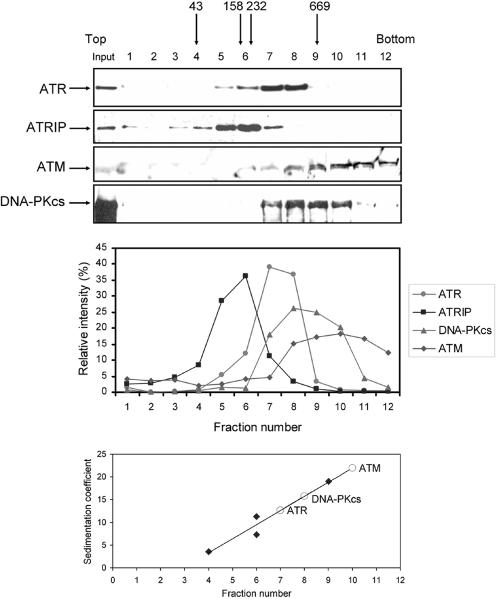
To obtain the sedimentation coefficient of ATR, we subjected a HEK293T whole-cell extract to sedimentation through a 15 to 35% glycerol gradient. The gradient was calibrated with a set of standard proteins of known S values. However, because most of these proteins have masses less than that of ATR, we also analyzed the migration of ATM and DNA-PKcs in the same gradient so that the native molecular weights of the three PIKK member enzymes could be directly compared to one another. Assuming spherical shapes and monomeric forms for all three enzymes, it would be expected that the DNA-PKcs monomer (Mr = 469,146) would sediment faster than ATM (Mr = 350,730), which would sediment faster than ATR (Mr = 301,491). Figure 2 shows that ATM sediments over a wide range but mostly at a position with an S value of 21.9, which is consistent with that of an ATM dimer. DNA-PKcs, which is known to be a monomer, has an S value of 15.7, in reasonable agreement with the theoretical value of a monomeric protein of that size. ATR sediments with an S value of 12.6, from which an Mr of 361,000 is calculated. This is larger than the monomer of ATR (Mr = 301,491) and somewhat smaller than that of the ATR-ATRIP heterodimer (301,491 + 89,891 = 391,382), suggesting that the association constant of ATR-ATRIP is relatively low, resulting in partial dissociation of the two subunits during the centrifugation. This is evident in Fig. 2, in which the peaks of the sedimentation profiles of ATR and ATRIP are clearly separated. When the Mr of ATR is calculated from the Siegel-Monty equation (31), which incorporates both the sedimentation coefficient and the Stokes radius, we obtain an Mr of 385, 500 for ATR (Table 1), which is reasonably close to the theoretical molecular weight of the ATR-ATRIP heterodimer.
FIG. 2.
Analysis of PIKK family kinases by glycerol gradient sedimentation. Whole-cell extract from HEK293T cells was sedimented through a 15 to 35% (wt/wt) glycerol gradient. (Top) Fractions were collected and analyzed by Western blotting with ATR, ATRIP, ATM, and DNA-PKcs antibodies. The input lane contains 1/10 of the material loaded onto the gradient. Gradient calibration was carried out with hen ovalbumin (43 kDa, 3.55 S), rabbit muscle aldalose (158 kDa, 7.3 S), bovine liver catalase (232 kDa, 11.3 S), and bovine thyroid thyroglobulin (669 kDa, 19 S). Arrows indicate the positions of the peak fractions of these reference proteins. (Middle) Quantitative analysis of data from the top panel. (Bottom) Estimation of the sedimentation coefficients of the PIKK proteins (indicated as open circles) determined from those of standard proteins run in a parallel gradient.
In conclusion, hydrodynamic analysis reveals that ATR is mostly in the form of an ATR-ATRIP heterodimer and, in contrast to ATM, does not form a homodimer and higher-order oligomers, and thus its activity must be regulated by a different mechanism.
Effect of UV irradiation on ATR.
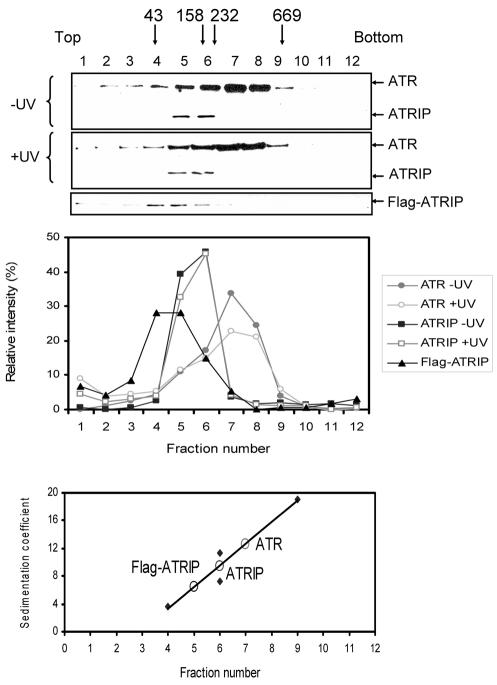
It was shown that ionizing radiation drastically alters ATM quaternary structure by causing transphosphorylation followed by dissociation to the monomeric form (2). We wished to find out if DNA damage affected ATR by changing its interactions with other cellular proteins, in particular with ATRIP. Since ATR is the principal sensor in the UV-induced checkpoint response, we used UV irradiation as the genotoxicant and compared the behavior of the protein on a glycerol gradient before and after UV irradiation. In particular, we were interested in finding out if there might be a change in ATR-ATRIP interaction following DNA-damage. Figure 3 shows that the sedimentation of ATR and ATRIP in the glycerol gradient before and after UV irradiation is essentially the same. Importantly, there does not appear to be any change in the strength of the ATR-ATRIP interaction. ATRIP sediments faster than would be expected from its native molecular weight under both conditions, indicative of ATR-ATRIP ⇋ ATR + ATRIP equilibrium that is not affected by DNA damage. It should be noted that the faster sedimentation of ATRIP is not caused by an unusual shape, as purified ATRIP, free of ATR, has an S value consistent with its monomeric molecular weight (Fig. 3 and Table 1). It must be also noted that under the irradiation conditions we used for Fig. 3 there is an ATR-mediated checkpoint activation, as evidenced by Chk1-S345 phosphorylation (data not shown). Taken together, these data lead us to conclude that after UV damage there is no long-lasting measurable change in the interaction of ATR with its smaller partner, ATRIP, or any other cellular protein.
FIG. 3.
Lack of effect of UV damage on hydrodynamic properties of ATR. (Top) HEK293T whole-cell extracts from unirradiated or UV-irradiated (50 J/m2) cells were separated on 15 to 35% (wt/wt) glycerol gradient gel, and the fractions were probed with anti-ATR and anti-ATRIP antibodies by Western blotting. Positions of marker proteins are indicated by arrows: hen ovalbumin (43 kDa), 3.55 S; rabbit muscle aldalose (158 kDa), 7.3 S; bovine liver catalase (232 kDa), 11.3 S; bovine thyroid thyroglobulin (669 kDa), 19 S. We also included the sedimentation profile of His6-Flag-ATRIP purified from insect cells and centrifuged in an identical density gradient (Flag-ATRIP immunoblot) to obtain the sedimentation coefficient of this subunit in isolation. (Middle) Quantitative analysis of data from the top panel. (Bottom) Estimation of the sedimentation coefficients of ATR and ATRIP.
Binding of ATR, ATRIP and ATR-ATRIP to DNA.
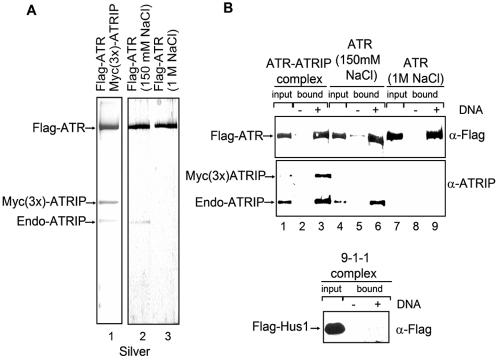
Flag-ATR purified under stringent conditions, in particular by Flag-peptide elution following extensive washing of the protein bound to the affinity resin with 1 M NaCl, contains trace amounts or no detectable ATRIP (34, 43). We wished to compare the DNA binding properties of ATR and the ATR-ATRIP complex to find out if ATRIP might play a role similar to that of scDdc2 (which reportedly recruits scMec1) in recruiting ATR to DNA. We prepared ATR and ATR-ATRIP by purifying the tagged protein under high-salt and low-salt wash conditions, respectively. Figure 4A shows the results for the ATR preparations obtained from cells transfected with vectors expressing either ATR alone or ATR and ATRIP and purified by high- and low-salt washes. Purification from cells cotransfected with ATR- and ATRIP-expressing vectors yielded a preparation containing both endogenous and Myc(3x)-tagged ATRIP (Fig. 4A, lane 1). As expected, ATR purified under low stringency contains ATRIP (Fig. 4A, lane 2), whereas ATR purified with high stringency is free of ATRIP (Fig. 4A, lane 3). We investigated the DNA binding properties of these proteins by using an 80-nt long 5′-biotinylated ssDNA attached to streptavidin-coated beads and identified the DNA-bound proteins by Western blotting. Figure 4B shows that, within the resolution of our assay, ATR binds with the same affinity to ssDNA whether it contains both endogenous and overexpressed ATRIP (Fig. 4A, lane 1, and Fig. 4B, lane 3), just the endogenous ATRIP (Fig. 4A, lane 2, and Fig. 4B, lane 6), or no ATRIP at all (Fig. 4A, lane 3, and Fig. 4B, lane 9). Thus, it appears that the function of ATRIP is not analogous to those of Ku and scDdc2, which recruit DNA-PKcs and scMec1, respectively, to DNA.
FIG. 4.
Binding of ATR and the ATR-ATRIP complex to DNA. (A) Purification of the ATR and ATR-ATRIP complex. Shown are silver-stained SDS-polyacrylamide gels of purified proteins. HEK293T cells were cotransfected with Flag-ATR- and Myc(3x)-ATRIP-expressing plasmids, and the complex was immunoaffinity purified with anti-Flag (α-Flag)-agarose under low-salt (150 mM NaCl) conditions (lane 1). The positions of ATR as well as the ectopically expressed and endogenous (Endo) ATRIP are indicated. Flag-tagged ATR purified from transiently transfected HEK293T cells with anti-Flag-agarose by a low-salt (150 mM NaCl) wash contains endogenous ATRIP (lane 2). Flag-ATR prepared by high-salt (1 M NaCl) wash has no detectable ATRIP (lane 3). (B) The proteins purified in panel A were used in a DNA binding assay with streptavidin-biotin-immobilized DNA. For each binding reaction, 7 pmol of 5′-biotinylated 80-nt oligomer was attached to 5 μl of streptavidin-coated beads and the ssDNA-coated beads were incubated with 5 pmol of the indicated protein samples at 30°C for 30 min. The protein samples from panel A in lanes 1, 2, and 3 were used for lanes 1 to 3, 4 to 6, and 7 to 9, respectively. After the unbound proteins were washed away from the beads, ATR and ATRIP associated with ssDNA were separated by SDS-PAGE and the proteins were detected by immunoblotting with corresponding antibodies. Input represents one-fourth of the total protein used in the experiment. At the bottom, the 9-1-1 complex was applied to the DNA-biotin-streptavidin-beads as a negative control for nonspecific adsorption to the beads. The Hus-1 subunit of the complex was Flag tagged and was used to locate the complex. Input is the total protein used in the experiment.
Binding of ATR-ATRIP to RPA.
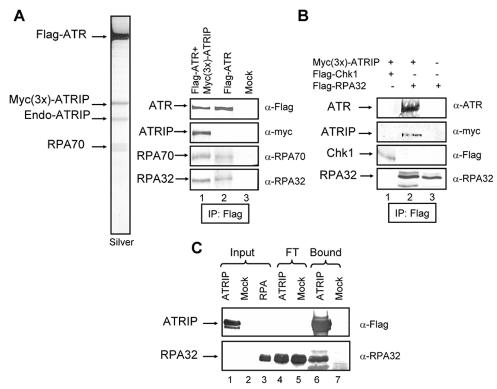
In our attempts to purify the ATR-ATRIP complex by ATR immunoprecipitation following published procedures (5), we noticed that many ATR preparations purified in this manner, in addition to ATRIP, contained another prominent band on SDS-polyacrylamide gels with an Mr of 70,000. Mass spectrometric analysis revealed this protein to be RPA70. Significantly, stringent washes of ATR immunoprecipitates that removed ATRIP also removed RPA, raising the possibility that ATRIP may mediate the ATR-RPA interaction. Initially, RPA was detected in immunoprecipitates of cells cotransfected with both ATR- and ATRIP-expressing plasmids. However, by immunoblotting we could demonstrate that both RPA70 and RPA32 were immunoprecipitated from extracts of cells transfected with a Flag-ATR-expressing plasmid only (Fig. 5A, right panel). Since Flag-ATR immunoprecipitates washed with low salt contain endogenous ATRIP, the data in Fig. 5A does not address the question of whether ATRIP is required for the ATR-RPA interaction. To address this issue, we transfected cells with either Flag-tagged RPA32 or with Flag-RPA32 plus Myc(3x)-ATRIP, immunoprecipitated RPA32, and immunoblotted the precipitate with antibodies to ATR and ATRIP. As shown in Fig. 5B, endogenous ATR coimmunoprecipitates with RPA32 only when ATRIP is overexpressed by transfection, suggesting that ATRIP may mediate the ATR-RPA interaction by binding to both proteins. The binding of ATR and ATRIP to one another is well-documented (5). To further test for direct ATRIP-RPA interaction, we expressed ATRIP in Sf21 insect cells, isolated it by immunoprecipitation, and then incubated it with recombinant RPA purified from Escherichia coli. As seen in Fig. 5C, RPA specifically binds ATRIP, leading us to conclude that ATRIP mediates the interaction of ATR with RPA. We wished to determine the functional significance of this interaction by performing DNA binding and kinase assays with various combinations of the three proteins as detailed below.
FIG. 5.
Interaction of RPA with the ATR-ATRIP complex. (A) Interaction among endogenous proteins. (Left) HEK293T cells were cotransfected with Flag-ATR and Myc(3x)-ATRIP expression vectors, and the ATR-ATRIP complex was purified on a Flag monoclonal antibody resin. The ATR-ATRIP complex separated by SDS-PAGE and visualized by silver staining contained, in addition to ATR and endogenous (Endo) and tagged ATRIP, a fourth band of an apparent molecular size of 70 kDa, which was identified as RPA70 by mass spectrometry. (Right) Coimmunoprecipitation of RPA with ATR and the ATR-ATRIP complex. Immunoprecipitations (IP) were carried out with anti-Flag (α-Flag)-agarose from transfected HEK293T cells and analyzed with the indicated antibodies. (B), ATR-RPA interaction is mediated by ATRIP. HEK293T cells were transfected with the indicated expression vectors, and the immunoprecipitates obtained with anti-Flag-agarose were analyzed by Western blotting with the indicated antibodies. Chk1 was used as a negative control for nonspecific binding to the resin. (C) Binding of ATRIP to purified RPA. Sf21 insect cells either mock treated or infected with His6-Flag-ATRIP-expressing baculovirus were lysed, and the lysates were mixed with Flag-agarose beads. Following extensive washing, 3 μg of RPA was added to the ATRIP-containing (∼ 4 μg) Flag-agarose beads. The protein mixture was incubated for 30 min on ice, and then the Flag-agarose beads were extensively washed three times. The bound and unbound proteins were identified by Western blotting with the indicated antibodies.
Effect of ATRIP on DNA binding and kinase activities of ATR.
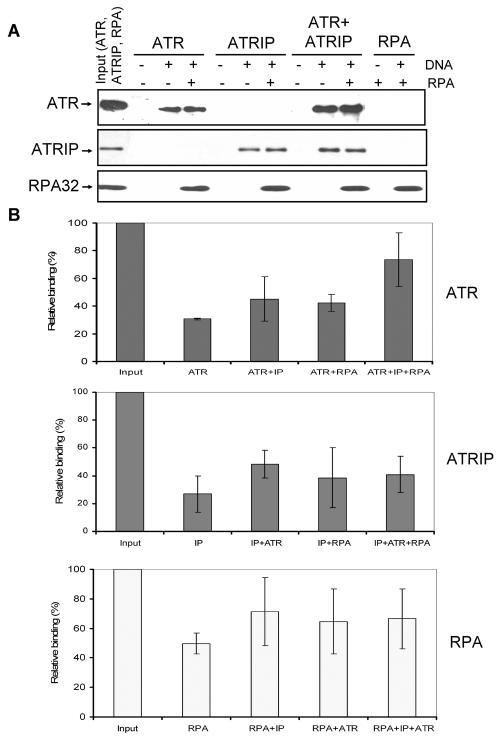
Recently, an in vivo study suggested that RPA played an important role in recruiting ATR to DNA (41) and an in vitro study found that ATRIP was essential for recruiting the ATR-ATRIP complex to RPA-covered DNA (43). Hence we tested the binding of ATR, ATRIP, and ATR-ATRIP to RPA-covered DNA in our system. We found that ATR and ATRIP bind to both naked and RPA-covered ssDNA with comparable affinities (Fig. 6A). The binding of ATR to RPA-covered DNA was enhanced by ATRIP by a small but statistically significant degree (Fig. 6B). However, clearly the ATR protein alone binds efficiently to RPA-covered DNA and the stimulation of its binding by ATRIP is modest.
FIG. 6.
Effects of RPA and ATRIP on binding of ATR to DNA. (A) An 80-nt 5′-biotinylated ssDNA (7 pmol) was immobilized on streptavidin beads, and where indicated RPA was added to the beads and incubated for 30 min at 30°C before the addition of other proteins as shown. For each binding reaction, 4 pmol of ATR, 4 pmol of ATRIP, and 7 pmol of RPA were used. Following addition of proteins to the beads and 30 min of incubation at 30°C, the beads were extensively washed and the bound proteins were identified by Western blotting. Input represents one-half of the total protein used in the experiment. The sources of proteins used in the binding assays were as follows. ATR is Flag-ATR purified from transfected HEK293T cells by a high-salt wash of the affinity resin and lacks detectable ATRIP by Western analysis; ATRIP was purified from baculovirus-infected Sf21 cells; the ATR + ATRIP complex was obtained by mixing and preincubating the two proteins prior to adding to the binding reaction mixture. Note that even though we used a mixture of ATR plus ATRIP to obtain the ATR-ATRIP complex for our binding assays as was done previously (43), we obtained similar properties with the ATR-ATRIP complex copurified from cells cotransfected with vectors expressing both proteins (data not shown). RPA was purified from an E. coli strain expressing all three subunits of the protein. (B) Quantitative analysis of the DNA binding data for ATR (top), ATRIP (middle), and RPA (bottom) from three experiments carried out under conditions described for panel A. The bound protein is expressed in terms of percentage of the input, and the bars indicate standard errors. There was statistically significant (P < 0.05) difference only between the binding of ATR in isolation and the binding of ATR in the ATR + ATRIP complex to RPA-covered DNA (top panel, last two columns).
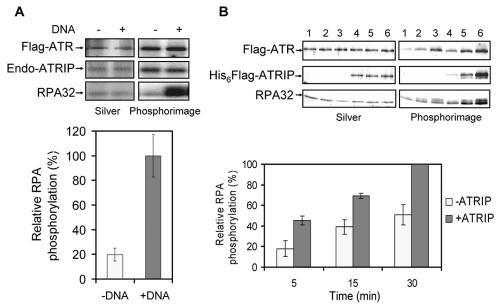
The kinase activity of ATR is essential for its checkpoint function (1). Many substrates are phosphorylated by ATR, including Chk1, Rad17, and RPA (3), and recent evidence demonstrated that the phosphorylation of Rad17 by ATR in vitro was dependent on both ssDNA and RPA (43) and that in vivo relocalization of RPA to ionizing-radiation-induced foci was dependent on the kinase activity of ATR (3). Therefore, we investigated the effects of ssDNA and ATRIP on the kinase activity of ATR, using RPA as a substrate. The RPA32 subunit of the trimeric RPA is phosphorylated by cdc2 and DNA-PK, and this phosphorylation modulates the RPA-DNA interactions (4, 19). Recently, it was shown that RPA32 is also phosphorylated by ATR (3). Hence, we investigated the effects of ssDNA and ATRIP on the phosphorylation of RPA32 by ATR. In agreement with a recent report (3) we find this phosphorylation is strongly stimulated by ssDNA (Fig. 7A). Similarly, we find that ATRIP affects the kinase activity of ATR on RPA32; however, the stimulation conferred by ATRIP is modest, in the range of a 1.5- to 2-fold increase, under our assay conditions (Fig. 7B). Thus, as for all other aspects of ATR activities tested in this study, it appears that there is no absolute requirement for ATRIP for ATR activity but ATRIP exerts a moderate modulatory effect on ATR functions.
FIG. 7.
Effects of DNA and ATRIP on phosphorylation of RPA32 by ATR. (A) Effect of DNA. Flag-tagged ATR was purified from HEK293T cells under low-salt conditions so that Flag-ATR contained endogenous (Endo) ATRIP; 0.2 μg of RPA was used as a substrate in the absence or presence of 25 μg of ssDNA/ml. (Top) Proteins in the reaction mixture were separated by SDS-PAGE and visualized by silver staining (left), and phosphorylated proteins were detected by a PhosphorImager screen (right). (Bottom) Quantitative analysis of RPA32 phosphorylation by ATR in the absence or presence of DNA. The mean values and standard errors were calculated from three independent experiments. The relative phosphorylation of RPA is normalized to the highest phosphorylation obtained, which was in the presence of DNA. (B) Effect of ATRIP. (Top) Flag-tagged ATR (50 ng) was purified from HEK293T cells under high-salt conditions and mixed with 0.2 μg of RPA and ssDNA (25 μg/ml) in either the absence (lanes 1 to 3) or presence (lanes 4 to 6) of ATRIP (0.2 μg) in a reaction volume of 50 μl. Kinase reactions were carried out at 30°C for either 5 min (lanes 1 and 4), 15 min (lanes 2 and 5), or 30 min (lanes 3 and 6). Proteins were separated by SDS-PAGE and visualized by silver staining (left), and phosphorylated proteins were detected by a PhosphorImager screen (right). (Bottom) Mean values and standard errors of RPA32 phosphorylation calculated from three independent experiments including the one for which results are shown in the top panel. The relative RPA32 phosphorylation is normalized against the highest phosphorylation obtained, which was at 30 min in the presence of ATRIP.
DISCUSSION
ATM and ATR play key roles in the DNA damage checkpoint response (1, 29, 30) as damage sensors and as apical kinases that phosphorylate signal transducers and checkpoint effectors. How these proteins sense DNA damage and how they become activated are currently not well understood. Recent studies have raised some interesting possibilities for damage recognition. Evidence was obtained indicating that ATM is a dimer or a higher-order oligomer that becomes activated upon loss of chromatin compaction caused by double-strand breaks (2). In addition, it was found that all PIKK family members contained numerous HEAT motifs, which are known to mediate protein-protein interaction (23). These findings raised the possibility that ATR may also be an oligomer and that its activation may involve transition between various forms of oligomeric states. Indeed, it has been reported that, based on the results of gel filtration chromatography, ATR is in the form of a 1,000-kDa complex either as a homopolymer or a multiprotein complex (40). The present study was undertaken to address three interrelated questions: (i) the quaternary structure of ATR, (ii) the role of ATRIP in ATR function, and (iii) the effects of ATR and RPA on one another's activity. We recapitulate our main findings below.
Quaternary structure of ATR.
Hydrodynamic analysis of ATR in human whole-cell extract by gel filtration chromatography and glycerol gradient velocity sedimentation revealed that ATR behaved as a protein with a Stokes radius of 72.8 Å and a sedimentation coefficient of 12.6, from which an apparent molecular weight of 385,500 is calculated. This is in reasonable agreement with the theoretical molecular weight of 391,382 for the ATR-ATRIP complex. Thus, it appears that ATR is in the form of an ATR-ATRIP heterodimer in equilibrium with the individual components. Even though ATR is known to interact with many other proteins involved in the cell cycle checkpoint or DNA repair those interactions are probably transient and of not sufficient avidity for the proteins to be considered part of an ATR-containing multiprotein complex. This finding is in disagreement with an earlier report that concluded that ATR was predominantly in complexes with an average molecular mass of 1,000 kDa based on its elution profile in a gel filtration (40). The lysis procedure and the ionic strength of the gel filtration column used in the previous work were quite similar to ours, which excludes nonspecific aggregation as the cause of the abnormally high molecular mass found in the previous study. We have no satisfactory explanation for the discrepancy. However, as we obtain similar molecular weights both by gel filtration and sedimentation and by using the Siegel-Monty (31) formula, which is more rigorous than those obtained by either method alone, we consider ATR to be mostly in the form of an ATR-ATRIP heterodimer in vivo. We also find that ATM is in equilibrium among various oligomerization states, with the dimer form being the predominant species. This is the first hydrodynamic evidence for the recent report (2) that the native form of ATM in undamaged cells is a dimer or a higher-order oligomer.
It has been reported that DNA damage leads to transphosphorylation of ATM and its consequent conversion into a monomer, which is the active form that initiates the checkpoint cascade (2). We considered the possibility that ATR may undergo a similar change in its quaternary structure by a change in the interaction state of ATR and ATRIP. However, we did not detect any change in the ATR-ATRIP ⇋ ATR + ATRIP equilibrium following UV damage, which activates the DNA damage checkpoint pathway mainly through ATR. Even though our experimental design would have not detected a transient change in this equilibrium we believe that is unlikely and that ATR, in contrast to ATM, is not regulated by a change in quaternary structure caused by autophosphorylation. The different behavior of these two PIKK family members is not unexpected. Despite being members of the PIKK family and playing important roles in DNA damage checkpoint signaling, ATM and ATR do have some fundamental differences (1, 29, 30): ATM is most effectively activated by double-strand breaks, whereas ATR is the initiator of the checkpoint response for UV damage and replication stress. ATM is not essential for cell viability, but ATR is because of an apparently indispensable role during normal DNA replication. Finally, ATR has a smaller molecular weight partner called ATRIP, but ATM does not. Assuming that ATR is a monomeric protein bound to ATRIP only with moderate affinity, it might be asked why transfecting cells with the ATR-kinase-inactive mutant causes a dominant negative phenotype. The most common mechanism for the dominant negative phenotype is the formation of complexes between mutant and wild-type protomers of an oligomeric protein, as is the case for ATM (2). However, an equally plausible mechanism is the sequestration of either the upstream or the downstream proteins in a signal transduction pathway by the mutant protein. It is likely that kinase-inactive ATR exerts its dominant negative effect by both mechanisms. It may heterodimerize with ATRIP in competition with wild-type ATR and cause the degradation of the wild-type protein (5), and it may bind to downstream targets such as Claspin and Chk1 and sequester them from the wild-type ATR-initiated signaling pathway.
Finally, in comparing ATR with ATM some comment must be made on the role of autophosphorylation. DNA damage induces autophosphorylation of ATM (2). It has been shown that autophosphorylation in trans converts ATM to the active monomeric state. We find no change in hydrodynamic behavior of ATR upon DNA damage even though ATR is capable of autophosphorylation. Preliminary data with site-specific mutants of ATR suggest that elimination of major phosphorylation sites do not affect its known activities, including ATRIP binding, DNA binding, and kinase activity on other substrates (data not shown). Clearly, the significance of ATR autophosphorylation remains to be determined.
Role of ATRIP in ATR function.
Downregulation of ATRIP by siRNA in vivo causes a drastic decrease in ATR level, suggesting that ATRIP is necessary for the stability of ATR (5). This phenomenon is commonly observed with proteins that make very stable complexes such as the XPF and ERCC1 excision repair proteins (28, 39). In a previous study we found that ATRIP was readily separated from ATR by washing ATR immunoprecipitates with 1 M NaCl (34). Dissociating a heterodimer with such relatively high ionic strength was not unexpected. However, we were surprised to see that ATRIP partly separated from ATR under nearly physiological ionic strength in both a gel filtration column and during sedimentation in a glycerol gradient (Fig. 1 and 3). Lacking a concentration-dependent analysis of monomer-dimer equilibrium we are unable to calculate the equilibrium dissociation constant of the heterodimer. However, an approximate consideration of the available data would indicate that the equilibrium dissociation constant is in the micromolar range. These data strongly indicate that in vivo at any given time a significant fraction of ATR is not in complex with ATRIP. In this regard, the ATR-ATRIP interaction is more similar to DNA-PKcs-Ku interaction rather than those of other more stable heterodimers such as the XPF-ERCC1 repair/recombination endonuclease, which is quite stable in ionic strength up to 1 M and purifies through several columns as a complex (28, 39).
The role of ATRIP in ATR functions is of particular interest because it was presumed that ATRIP was the functional analogue of Rad26 and Ddc2 in S. pombe and S. cerevisiae, respectively. However, there is some evidence that the roles of these Rad26-related proteins may differ among different organisms. It has been shown that spRad26 is required for spRad3 kinase activity in fission yeast (37), but scDdc2 is not required for scMec1 kinase activity in budding yeast (35) while ATRIP is required for stability of ATR in humans (5). Likewise, it was shown that scMec1 and scDdc2 colocalize to sites of DNA damage (12, 17) and that localization of scDdc2 in vivo requires scMec1 (17); however, fission yeast spRad26 seems to respond to DNA damage independently of spRad3 (38), as the localization pattern of spRad26 before and after DNA damage was unaffected by a null mutation in spRad3. Finally, it has been reported that although scDdc2 binds DNA independently of scMec1, both in vivo and in vitro binding of scMec1 to DNA requires scDdc2 (27). It was previously reported that, in contrast to scMec1, ATR was capable of binding to DNA without the aid of ATRIP (34). This finding has recently been confirmed and extended in a study that demonstrated that ATR free of ATRIP can bind to single- and double-stranded DNAs with comparable affinities (43). However, it was also reported that ATRIP was incapable of binding to ssDNA unless the DNA was covered with RPA and moreover that as a consequence of this property of ATRIP the ATR-ATRIP complex failed to bind ssDNA but bound to RPA-covered ssDNA (43). Therefore, it was suggested that binding of ATRIP to RPA-coated DNA enables the ATR-ATRIP complex to associate with RPA-covered ssDNA, which is a feature of a stalled replication fork (43). In the present study, we observed comparable bindings of ATR, ATRIP, and the ATR-ATRIP complex to both naked and RPA-covered DNA under our experimental conditions. However, we did observe a modest stimulation of ATR binding to RPA-covered DNA by ATRIP. Although there are some subtle differences between the reaction conditions used in the two studies, we believe our data strongly suggest that under physiological conditions ATRIP is not necessary for recruitment of ATR to a stalled replication fork. Considering our hydrodynamic analysis of ATR, which shows that a significant fraction of ATR in cells might be free of ATRIP, together with our DNA binding studies, we think it is possible that ATR and ATRIP in monomer forms may orchestrate different steps of the DNA damage checkpoint pathway. Clearly, more work is needed to deconvolute the ATRIP-dependent and independent functions of ATR.
Effect of ATR on RPA.
When ATR is immunopurified under relatively mild conditions such that ATRIP is retained, we routinely observe significant amounts of RPA in the immunoprecipitate as well. Therefore, we investigated the effects of ATR and RPA on one another's activity. First, we find that the RPA-ATR interaction is dependent on the presence of ATRIP and is most likely mediated by ATRIP. Second, ATR weakly phosphorylates RPA32 in the absence of DNA, but this is greatly stimulated by ssDNA, in agreement with a recent report (3). Finally, when the effect of ATRIP on RPA32 phosphorylation was tested it was found that ATR does phosphorylate RPA32 even in the absence of ATRIP but that ATRIP stimulated the kinase activity of ATR significantly both with regard to autophosphorylation of ATR and the phosphorylation of RPA32. The phosphorylation of RPA32 in a cell-cycle-dependent manner has been known for some time; however, its significance is unknown (36). Phosphorylation does modulate RPA-DNA interactions (4, 19). However, in vitro, phosphorylation of RPA32 does not affect the RPA activity in replication or in excision repair (21). The recent finding that the kinase activity of ATR is essential for irradiation-induced translocation of RPA to sites of DNA damage (3) seemed to be an important clue to the significance of RPA32 phosphorylation during S phase and DNA damage. Paradoxically, however, it was shown that when the phosphorylation sites of RPA32 were mutated, the nonphosphorylatable form of RPA32 did not affect the DNA damage checkpoint response (3). Thus, this issue remains unresolved. Nevertheless, the interactions of among ATR, ATRIP, RPA, and DNA that have been uncovered in this and other recent studies are likely to contribute to a better understanding of the damage-sensing step of the DNA damage checkpoint response.
Acknowledgments
This work was supported by NIH grant GM32833.
We thank Stephen J. Elledge (Baylor College of Medicine) and David Cortez (Vanderbilt University) for the ATRIP plasmid and ATRIP antibodies, Mark C. Hall (University of North Carolina) for mass spectroscopy, Dale A. Ramsden (University of North Carolina) for help with gel filtration chromatography, and Jerard Hurwitz (Sloan-Kettering Institute) and Laura Lindsey-Boltz (University of North Carolina) for critical comments on the manuscript.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 3.Barr, S. M., C. G. Leung, E. E. Chang, and K. A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047-1051. [DOI] [PubMed] [Google Scholar]
- 4.Binz, S. K., Y. Lao, D. F. Lowry, and M. S. Wold. 2003. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions: evidence for an intersubunuit interactions. J. Biol. Chem. 278:35584-35591. [DOI] [PubMed] [Google Scholar]
- 5.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 6.Durocher, D., and S. P. Jackson. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13:225-231. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb, T. M., and S. P. Jackson. 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72:131-142. [DOI] [PubMed] [Google Scholar]
- 9.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 10.Jordan, M., A. Schallhorn, and F. M. Wurm. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538-37543. [DOI] [PubMed] [Google Scholar]
- 12.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 13.Leuther, K. K., O. Hammarsten, R. D. Kornberg, and G. Chu. 1999. Structure of DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J. 18:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, J. J., and T. J. Kelly. 1985. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol. Cell. Biol. 5:1238-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindsey-Boltz, L. A., V. P. Bermudez, J. Hurwitz, and A. Sancar. 2001. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc. Natl. Acad. Sci. USA 98:11236-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, R. G., and B. N. Ames. 1961. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J. Biol. Chem. 236:1372-1379. [PubMed] [Google Scholar]
- 17.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 19.Oakley, G. G., S. M. Patrick, J. Yao, M. P. Carty, J. J. Turchi, and K. Dixon. 2003. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 42:3255-3264. [DOI] [PubMed] [Google Scholar]
- 20.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 21.Pan, Z. Q., C. H. Park, A. A. Amin, J. Hurwitz, and A. Sancar. 1995. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. Proc. Natl. Acad. Sci. USA 92:4636-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins, E. J., A. Nair, D. O. Cowley, T. Van Dyke, Y. Chang, and D. A. Ramsden. 2002. Sensing of intermediates in V(D)J. recombination by ATM. Genes Dev. 16:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry, J., and N. Kleckner. 2003. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112:151-155. [DOI] [PubMed] [Google Scholar]
- 24.Rathmell, W. K., W. K. Kaufmann, J. C. Hurt, L. L. Byrd, and G. Chu. 1997. DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer Res. 57:68-74. [PubMed] [Google Scholar]
- 25.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 27.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 28.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 29.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem., in press. [DOI] [PubMed]
- 30.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 31.Siegel, L. M., and K. J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346-362. [DOI] [PubMed] [Google Scholar]
- 32.Smith, G. C., and S. P. Jackson. 1999. The DNA-dependent protein kinase. Genes Dev. 13:916-934. [DOI] [PubMed] [Google Scholar]
- 33.Smith, G. C., R. B. Cary, N. D. Lakin, B. C. Hann, S. H. Teo, D. J. Chen, and S. P. Jackson. 1999. Purification and DNA binding properties of the ataxia-telangiectasia gene product ATM. Proc. Natl. Acad. Sci. USA 96:11134-11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unsal-Kacmaz, K., A. M. Makhov, J. D. Griffith, and A. Sancar. 2002. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. USA 99:6673-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 37.Wolkow, T. D., and T. Enoch. 2002. Fission yeast Rad26 is a regulatory subunit of the Rad3 checkpoint kinase. Mol. Biol. Cell 13:480-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolkow, T. D., and T. Enoch. 2003. Fission yeast Rad26 responds to DNA damage independently of Rad3. BMC Genetics 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood, R. D. 1997. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272:23465-23468. [DOI] [PubMed] [Google Scholar]
- 40.Wright, J. A., K. S. Keegan, D. R. Herendeen, N. J. Bentley, A. M. Carr, M. F. Hoekstra, and P. Concannon. 1998. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc. Natl. Acad. Sci. USA 95:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You, Z., L. Kong, and J. Newport. 2002. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277:27088-27093. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 43.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542-1548. [DOI] [PubMed] [Google Scholar]