Abstract
Protein allostery is based on the existence of multiple conformations in equilibrium linked to distinct functional properties. Although evidence of allosteric transitions is relatively easy to identify by functional studies, structural detection of a pre-existing equilibrium between alternative conformations remains challenging even for textbook examples of allosteric proteins. Kinetic studies show that the trypsin-like protease thrombin exists in equilibrium between two conformations where the active site is either collapsed (E*) or accessible to substrate (E). However, structural demonstration that the two conformations exist in the same enzyme construct free of ligands has remained elusive. Here we report the crystal structure of the thrombin mutant N143P in the E form, which complements the recently reported structure in the E* form, and both the E and E* forms of the thrombin mutant Y225P. The side chain of W215 moves 10.9 Å between the two forms, causing a displacement of 6.6 Å of the entire 215–217 segment into the active site that in turn opens or closes access to the primary specificity pocket. Rapid kinetic measurements of p-aminobenzamidine binding to the active site confirm the existence of the E*-E equilibrium in solution for wild-type and the mutants N143P and Y225P. These findings provide unequivocal proof of the allosteric nature of thrombin and lend strong support to the recent proposal that the E*-E equilibrium is a key property of the trypsin fold.
The hallmark of allosteric proteins is that they exist in multiple conformations in equilibrium (1, 2). When alternative conformations differ in their functional properties, linkage is established between structure and biological activity and allostery becomes the basis of the effects observed experimentally. The theoretical underpinnings of allosteric transitions have been defined for systems working at equilibrium (1) or under transient kinetics (3). Yet structural validation of a pre-existing equilibrium between alternative conformations remains a challenge even for textbook examples of allosteric proteins (2, 4).
Allostery is not an exclusive property of multimeric proteins. Indeed, the ability of monomeric enzymes to express complex behavior consistent with allosteric transitions has long been recognized (3, 5). Trypsin-like proteases are monomeric enzymes which constitute the largest and best studied group of homologous proteases in the human genome (6). They are phylogenetically grouped into six functional categories: digestion, coagulation and immunity, tryptase, matriptase, kallikrein and granzymes. Trypsin-like proteases share a common mechanism of catalysis that relies upon the coordinate action of three catalytic residues: H57, D102 and S195 (chymotrypsinogen numbering). In addition, they share a common mechanism of activation: an inactive zymogen precursor is proteolytically cut between residues 15 and 16 to generate a new N-terminus that ion-pairs with the highly conserved D194 next to the catalytic S195 and organizes both the oxyanion hole and primary specificity pocket (6–8). The irreversible zymogen→protease conversion affords a useful paradigm to explain the onset of catalytic activity as seen in the digestive system, blood coagulation or the complement system and is particularly useful to understand the initiation, progression and amplification of enzyme cascades, where each component acts as a substrate in the inactive zymogen form in one step and as an active enzyme in the subsequent step (9). However, considerable variation in catalytic activity is observed among members of the trypsin family following conversion from the inactive zymogen form. Digestive enzymes like trypsin, chymotrypsin and elastase, complement factors C1r and C1s, and coagulation factors like thrombin are highly active after the zymogen→protease conversion has taken place. On the other hand, complement factors B and C2 are mostly inactive until binding of complement factors C3b and C4b enable catalytic activity at the site where amplification of C3 activation leads to formation of the membrane attack complex (10–12). Coagulation factor VIIa circulates in the blood as a poorly active protease that acquires full catalytic competence only upon interaction with tissue factor that becomes exposed to the blood stream upon vascular injury (13, 14). Complement factor D assumes an inactive conformation with a distorted catalytic triad (15, 16) until binding to C3b and factor B promote substrate binding and catalytic activity (17, 18). The high catalytic activity of trypsin, C1r and thrombin and the ability of complement factor D or coagulation factor VIIa to remain in a zymogen-like form suggests that the trypsin fold may assume active and inactive conformations even after the zymogen→protease conversion has taken place. Existence of an allosteric equilibrium between active and inactive forms has been proposed for coagulation factor VIIa (14, 19). Rapid kinetics support a pre-existing equilibrium between active (E) and inactive (E*) forms for thrombin (20, 21), meizothrombin desF1 (22), factor Xa and activated protein C (23). Structures of thrombin in the free form reveal a conformation, E, with the active site open (24–26) and an alternative conformation, E*, with the active site blocked by repositioning of the 215–217 segment (27–30). However, no evidence currently exists that the same protease construct may assume alternative conformations that can be trapped by crystallographic analysis. This evidence is reported here for the first time.
Materials and Methods
Thrombin mutants N143P and Y225P were expressed in mammalian cells or E. coli and purified to homogeneity as described previously (29, 31). Crystallization was achieved at 22 °C using the hanging drop vapor diffusion method, with each reservoir containing 500 μl of solution. Equal volumes (2 μl) of the protein sample (8–9 mg/ml) and reservoir solution (see Table 1) were mixed to prepare the hanging drops. Diffraction quality crystals grew in two weeks and were frozen using 15%–25% glycerol as cryoprotectant at 100 °K. X-ray diffraction data were collected to 1.9–2.1 Å with a home source (Rigaku 1.2 kW MMX007 generator with VHF optics) Rigaku Raxis IV++ detector and were indexed, integrated and scaled with the HKL2000 software package (32). Structures were solved by molecular replacement using MOLREP from the CCP4 suite (33) and using as search models PDB accession code 3JZ1 for N143P in the E form, 1SHH for Y225P in the E form and 3BEI for Y225P in the E* form. Refinement and electron density generation were performed with REFMAC (34) from the CCP4 suite and 5% of the reflections were randomly selected as a test set for cross-validation. Model building was performed in COOT (35). In the final stage of refinement, TLS tensors modeling rigid-body anisotropic temperature factors were calculated and applied to the model. Ramachandran plots were calculated using PROCHECK (36). Statistics for data collection and refinement are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the PDB as 3QGN (N143P E form), 3S7K (Y225P E form) and 3S7H (Y225P E* form).
Table 1.
Crystallographic data for thrombin mutants N143P and Y225P.
| N143P - E form | N143P - E* form | Y225P - E form | Y225P - E* form | |
|---|---|---|---|---|
| Buffer/salt | 0.2 M NH4I | 0.1 M imidazole | 0.2 M K formate | 0.1 M Tris, pH 8.0 |
| PEG | 3350 (20%) | 8000 (7%) | 3350 (20%) | 8000 (8%) |
| PDB ID | 3QGN | 3JZ2 | 3S7K | 3S7H |
|
| ||||
| Data collection: | Raxis IV++ | Mar345 | Raxis IV++ | Raxis IV++ |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 | 1.54 |
| Space group | C2 | P43 | P21212 | P43 |
| Unit cell dimensions (Å) | a=122.2 | a=57.9 | a=61.9 | a=57.6 |
| b=48.0 | b=57.9 | b=86.8 | b=57.6 | |
| c=52.0 | c=119.8 | c=101.0 | c=119.9 | |
| β=94.3° | ||||
| Molecules/asymmetric unit | 1 | 1 | 2 | 1 |
| Resolution range (Å) | 40-2.1 | 40-2.4 | 40-1.9 | 40-1.9 |
| Observations | 75156 | 83891 | 286602 | 185752 |
| Unique observations | 17583 | 14942 | 43419 | 30093 |
| Completeness (%) | 98.5 (97.3) | 96.7 (77.8) | 99.3 (98.2) | 97.8 (86.3) |
| Rsym (%) | 10.5 (16.0) | 7.6 (40.8) | 5.4 (30.5) | 7.3 (41.8) |
| I/σ(I) | 13.3 (6.6) | 19.0 (2.2) | 27.6 (4.0) | 20.7 (2.4) |
|
| ||||
| Refinement: | ||||
| Resolution (Å) | 40-2.1 | 40-2.4 | 40-1.9 | 40-1.9 |
| Rcryst, Rfree | 0.189, 0.239 | 0.189, 0.246 | 0.175, 0.216 | 0.175, 0.204 |
| Reflections (working/test) | 15588/892 | 14167/747 | 38897/2179 | 27031/1518 |
| Protein atoms | 2253 | 2242 | 4589 | 2273 |
| Solvent molecules | 150 | 116 | 382 | 191 |
| Rmsd bond lengthsa (Å) | 0.010 | 0.012 | 0.010 | 0.0080 |
| Rmsd anglesa (°) | 1.3 | 1.4 | 1.3 | 1.1 |
| Rmsd ΔB (Å2) (mm/ms/ss)b | 2.99/2.28/3.26 | 2.02/1.36/2.45 | 2.18/1.83/2.70 | 2.86/2.33/2.56 |
| <B> protein (Å2) | 41.8 | 34.1 | 35.4 | 39.0 |
| <B> solvent (Å 2) | 52.1 | 20.3 | 45.2 | 52.6 |
|
| ||||
| Ramachandran plot: | ||||
| Most favored (%) | 99.2 | 100 | 99.6 | 100 |
| Generously allowed (%) | 0.4 | 0 | 0.2 | 0 |
| Disallowed (%) | 0.4 | 0 | 0.2 | 0 |
Root-mean-squared deviation (Rmsd) from ideal bond lengths and angles and Rmsd in B-factors of bonded atoms.
mm, main chain-main chain; ms, main chain-side chain; ss, side chain-side chain. The structure of N143P in the E* form is from ref (29).
Stopped-flow fluorescence measurements of PABA binding to thrombin were performed over a time range of 100 ms using an Applied Photophysics SX20 spectrometer. An excitation wavelength of 330 nm and a cutoff filter of 375 nm were used for the experiments. All experiments were carried out in buffer containing 50 mM Tris, 0.1% PEG8000, pH 8.0 at 15 °C. Thrombin wild type or mutant at a final concentration of 1 μM was mixed 1:1 with increasing concentrations of PABA, having first established a baseline by mixing buffer and the same concentration of PABA. Applied Photophysics Pro-Data software was used to fit single exponential curves to the average of six or more kinetic traces for each PABA concentration. PABA was found to obey a two-step binding mechanism, with a fast phase too rapid to resolve and a slow, single exponential phase whose kobs decreases with increasing [PABA]. This dependence of the slow phase on ligand concentration is unequivocal proof of the kinetic mechanism of binding (20)
which is analogous to the binding mechanism of Na+. A pre-equilibrium between two forms, E* and E, precedes PABA (L) binding that can only occur to the active form E. The total fluorescence change observed upon PABA binding obeys the expression (20)
| (1) |
where F0 and F1 are the values of F at [PABA]=0 and [PABA]=∞, and
| (2) |
is the apparent PABA binding affinity that depends on the intrinsic PABA binding affinity and the equilibrium constant for the E*-E interconversion . Under conditions of separation of time scales, where PABA binding and dissociation are faster than the rates for the E*-E interconversion, the kobs for the evolution of the slow phase of fluorescence increase upon PABA binding obeys the expression
| (3) |
The value of kobs decreases hyperbolically with increasing [PABA] from kr+k−r ([PABA]=0) to kr ([PABA]=∞), thereby yielding kr, k−r and KA.
Results and Discussion
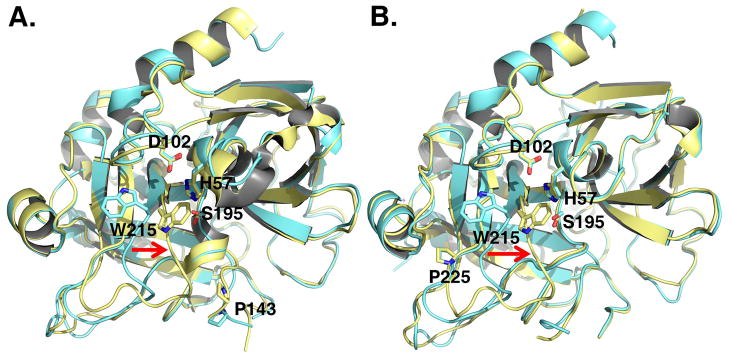
Structures of thrombin in the free form reveal a conformation with the active site fully accessible to substrate (24–26) and an alternative conformation with the active site blocked by repositioning of the W215 side chain and entire 215–217 segment (27–30). Although these conformations provide a structural basis for the allosteric E*-E equilibrium identified by kinetic studies (20, 21, 23), they have been documented with different protease constructs. The mutant N143P was engineered to abrogate the H-bond between the carbonyl O atom of E192 and the backbone N atom of N143, resulting in a flip of the E192-G193 peptide bond and disruption of the oxyanion hole formed by the backbone N atoms of G193 and S195 (29). A recently published structure of N143P in the free form reveals the predicted disruption of the oxyanion hole and a collapsed architecture of the active site (Figure 1) as observed in the E* form of the thrombin mutants D102N (28, 30) and Δ146-149e (27). The new structure of N143P in the free form reveals an open conformation of the active site (Figure 1), consistent with that observed in the E form of mutants R77aA (25) and C191A/C220A (24). The side chain of W215 moves back >10 Å toward F227 and relinquishes its interaction with the catalytic H57. The entire 215–217 segment moves >6 Å and opens up access to the active site. The mutant Y225P was engineered to abolish Na+ binding and constitutively stabilize thrombin in the Na+-free slow form (37, 38). Residue 225 has a dichotomous distribution in trypsin-like proteases: Pro is the preferred residue at this position, but Na+ binding requires the presence of Tyr or Phe as seen in clotting and complement proteases. The slow form is the form of thrombin free of any ligands and, based on the results of rapid kinetics, it is a mixture of the inactive E* and active E forms. The mutant was crystallized previously bound to an active site inhibitor (39). The new structures of Y225P in the free form reveal the two conformations, E and E*, analogous to those documented for N143P (Figure 1), thereby confirming that free thrombin (or slow thrombin) is a mixture of E* and E in allosteric equilibrium as predicted by kinetic studies on the wild-type (20),
Figure 1. X-ray crystal structures of the thrombin mutants N143P and Y225P in the E* and E forms.
Ribbon representation of the structure of the thrombin mutants N143P (A) and Y225P (B) in the E (cyan) and E* (gold) forms. In the E* form the side chain of W215 and the entire 215–217 segment collapse into the active site (red arrow). In the E form, W215 moves back 10.9 Å and the 215–217 segment moves 6.6 Å to make the active site accessible to substrate. The rmsd between the two forms is 0.328 Å (N143P) or 0.345 (Y225P). Relevant residues are labeled and rendered as sticks.
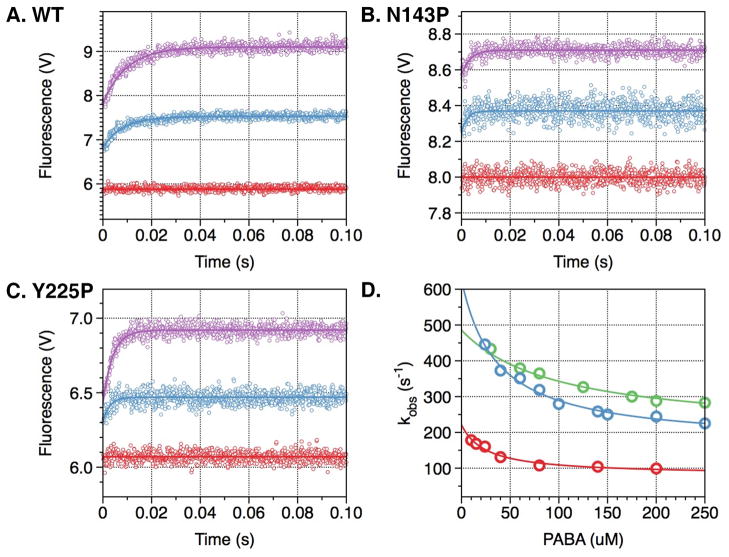
The E*-E equilibrium was originally discovered from rapid kinetic studies of Na+ binding to thrombin (20), recently extended to meizothrombin desF1 (22), activated protein C and factor Xa (23). Because Na+ cannot bind to the mutant Y225P due to the shift in the carbonyl O atom of K224 caused by the presence of P225 (37), an alternative probe of the E*-E equilibrium becomes necessary. Such probe could have wide applicability to trypsin-like proteases in general because Pro is the residue most represented at the 225 position in these enzymes (40) and Na+ activation is present only in a minority of enzymes in blood coagulation and the complement cascade (41). The active site inhibitor PABA has long been known as a useful reagent in fluorescence studies (42) and turned out to be an excellent probe of the accessibility of the active site in the E and E* forms (Figure 2). Rapid kinetics of PABA binding to wild-type thrombin reveals a two-step mechanism analogous to that observed for Na+ binding, with a fast phase completed within the dead time of the instrument and a slow phase of exponential increase in fluorescence whose kobs decreases hyperbolically as a function of [PABA] (Figure 2). This is an unequivocal signature of the existence of a pre-equilibrium that precedes PABA binding to the enzyme, and it likely reports the same E*-E equilibrium detected from Na+ binding measurements. Indeed, the rate constants for the E*-E interconversion derived from measurements of PABA binding (Table 2) are comparable to those derived from Na+ binding. Hence, the E*-E equilibrium in solution can be probed independently by binding to two different domains of the enzyme, the Na+ site or the active site. Rapid kinetic measurements of PABA binding to the mutants N143P and Y225P also reveal a slow phase with a kobs decreasing hyperbolically with increasing [PABA], thereby enabling detection of the E*-E equilibrium in systems where either Na+ cannot bind to its site (Y225P) or Na+ binding does not elicit a slow kinetic phase (N143P) (29). This offers a powerful new strategy to probe the E*-E equilibrium in other trypsin-like proteases.
Figure 2. PABA binding to thrombin wild type and mutants N143P and Y225P.
Kinetic traces of PABA binding to thrombin wild type (A) and mutants N143P (B) and Y225P (C). In all cases, binding of PABA obeys a two-step mechanism, with a fast phase completed within the dead time (<0.5 ms) of the spectrometer, followed by a single-exponential slow phase. Traces were recorded in the presence of 40 μM (cyan) or 200 μM (purple) PABA. Controls with 200 μM PABA in buffer are shown in red. (D) The kobs for the slow phase decreases with increasing [PABA]. The values were obtained from analysis of the kinetic traces (A-C) and analyzed according to eq 3 in the text with best-fit parameter values reported in Table 2 for wild type (red), N143P (green) and Y225P (cyan). Experimental conditions are: 1 μM enzyme, 50 mM Tris, 0.1% PEG8000, pH 8.0, at 15 °C. The value of kobs features an inverse hyperbolic dependence on [PABA], thereby proving the existence of the E*-E equilibrium preceding PABA binding.
Table 2.
Parameters for PABA to thrombin wild type and mutants N143P and Y225P.
| Enzyme | F0 (V)a | F1 (V)a | ΔF/F0 (%)a | Kapp (mM−1)a | KA (mM−1)b | kr (s−1)b | k−r (s−1)b | rb |
|---|---|---|---|---|---|---|---|---|
| wt | 6.66±0.04 | 9.70±0.03 | 46 | 17±1 | 40±2 | 91±2 | 120±10 | 1.3±0.1 |
| N143P | 8.25±0.05 | 9.20±0.05 | 12 | 4.0±0.3 | 10±1 | 200±10 | 290±10 | 1.5±0.2 |
| Y225P | 5.96±0.03 | 7.20±0.04 | 21 | 7.6±0.5 | 28±2 | 170±10 | 460±20 | 2.7±0.3 |
From the data in Figure 3A–C using eq 1 in the text.
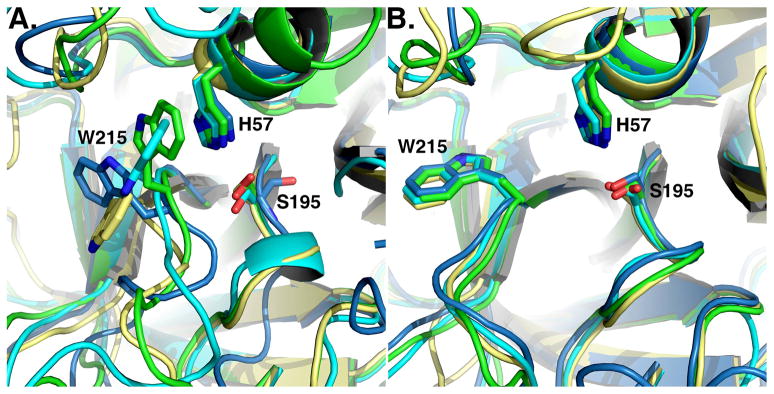
The relevance of these findings carries over to the entire family of trypsin-like proteases to which thrombin belongs. Indeed, a recent analysis of the entire structural database demonstrates that the E*-E equilibrium is a general feature of the trypsin fold, made possible by the conformational flexibility of W215 and the 215–217 segment (43). The E* form has been observed in the thrombin mutants D102N (28, 30), Δ146-149e (27), N143P (29) and Y225P reported here, complement factor D wild-type and mutants S915A, S215W and R218A (16–18), prostate specific antigen (44), tonin (45), prophenoloxidase activating factor II (46), hepatocyte growth factor activator (47) and prostasin (48, 49). The E form has been observed in the thrombin mutants R77aA (25, 26), C191A/C220A (24) and N143P and Y225P reported here, complement factors C1r (50) and C2a (51, 52), neuropsin (53) and trypsin (54). Mutant D216G of αI-tryptase crystallizes with the 215–217 segment in two conformations in a 3:1 occupancy ratio (55): one open and the other collapsed as in the wild-type (56). Relevant examples of these proteases in the E and E* forms are given in Figure 3. Detection of E* and E for the same protease construct remains challenging because it requires crystallization of the free form under conditions where either E* or E can be trapped. However, this difficulty can be overcome, as shown by the thrombin mutants N143P and Y225P reported here, and we expect more trypsin-like proteases to be crystallized in both the E* and E forms in the near future.
Figure 3. E* and E in the trypsin fold.
Active site conformations of relevant trypsin-like proteases in the E* form (A) or the E form (B). Note how the active site is fully open in the E form (B), but occluded to various degrees in the E* form (A). Structures are colored as follows: (A) prostasin (3DFJ, yellow), thrombin mutant N143P (3JZ2, cyan), complement factor D mutant S215W (1DST, green), prophenoloxidase activating factor II (2B9L, blue), (B) complement factor C1r (1MD8, yellow), thrombin mutant N143P (cyan, 3QGN), neuropsin (1NPM, green), trypsin (2G51, blue).
The physiological relevance of the E*-E equilibrium deserves attention. When E* is stabilized, the protease possesses low activity and acts as a switch that can be turned on by binding of specific cofactors or allosteric activators that facilitate conversion to the active E form. Relevant examples are complement factor D that shifts from the E* to the E form upon binding to the complex of C3b and factor B (17) and coagulation factor VIIa that makes a similar transition upon binding to tissue factor (14, 19). Stabilization of E produces a highly active enzyme upon conversion from the zymogen form without requiring macromolecular cofactors, as seen in trypsin, complement factor C1r and thrombin. Therefore, the E*-E equilibrium provides a reversible mechanism of regulation of enzyme activity following the irreversible transition from the zymogen form. The repertoire of activities available to the protease is greatly expanded in ways that fit the diverse requirements of enzymes acting in different biological environments or at different levels of cascades like blood coagulation and the complement.
Acknowledgments
We are grateful to Ms. Tracey Baird for her help with illustrations.
Abbreviations used
- PABA
p-aminobenzamidine
- PDB
Protein Data Bank
- PEG
polyethyleneglycol
Footnotes
This work was supported in part by the National Institutes of Health Research Grants HL49413, HL58141 HL73813 and HL95315 (to E.D.C.) and a PostDoctoral Research Fellowship from the American Heart Association (to W.N.).
References
- 1.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Perutz MF. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 3.Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970;245:5788–5799. [PubMed] [Google Scholar]
- 4.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botts J, Morales M. Analytical description of the effects of modifiers and of multivalency upon the steady state catalyzed reaction rate. Trans Faraday Soc. 1953;49:696–707. [Google Scholar]
- 6.Page MJ, Di Cera E. Serine peptidases: classification, structure and function. Cell Mol Life Sci. 2008;65:1220–1236. doi: 10.1007/s00018-008-7565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 8.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 10.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 11.Arlaud GJ, Barlow PN, Gaboriaud C, Gros P, Narayana SV. Deciphering complement mechanisms: the contributions of structural biology. Mol Immunol. 2007;44:3809–3822. doi: 10.1016/j.molimm.2007.06.147. [DOI] [PubMed] [Google Scholar]
- 12.Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 13.Banner DW, D’Arcy A, Chene C, Winkler FK, Guha A, Konigsberg WH, Nemerson Y, Kirchhofer D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 14.Eigenbrot C, Kirchhofer D, Dennis MS, Santell L, Lazarus RA, Stamos J, Ultsch MH. The factor VII zymogen structure reveals reregistration of beta strands during activation. Structure. 2001;9:627–636. doi: 10.1016/s0969-2126(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 15.Jing H, Macon KJ, Moore D, DeLucas LJ, Volanakis JE, Narayana SV. Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. Embo J. 1999;18:804–814. doi: 10.1093/emboj/18.4.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayana SV, Carson M, el-Kabbani O, Kilpatrick JM, Moore D, Chen X, Bugg CE, Volanakis JE, DeLucas LJ. Structure of human factor D. A complement system protein at 2.0 A resolution. J Mol Biol. 1994;235:695–708. doi: 10.1006/jmbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 17.Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing H, Babu YS, Moore D, Kilpatrick JM, Liu XY, Volanakis JE, Narayana SV. Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. J Mol Biol. 1998;282:1061–1081. doi: 10.1006/jmbi.1998.2089. [DOI] [PubMed] [Google Scholar]
- 19.Dickinson CD, Kelly CR, Ruf W. Identification of surface residues mediating tissue factor binding and catalytic function of the serine protease factor VIIa. Proc Natl Acad Sci U S A. 1996;93:14379–14384. doi: 10.1073/pnas.93.25.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bah A, Garvey LC, Ge J, Di Cera E. Rapid kinetics of Na+ binding to thrombin. J Biol Chem. 2006;281:40049–40056. doi: 10.1074/jbc.M608600200. [DOI] [PubMed] [Google Scholar]
- 21.Di Cera E. Thrombin. Mol Aspects Med. 2008;29:203–254. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaconstantinou ME, Gandhi PS, Chen Z, Bah A, Di Cera E. Na(+) binding to meizothrombin desF1. Cell Mol Life Sci. 2008;65:3688–3697. doi: 10.1007/s00018-008-8502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogt AD, Bah A, Di Cera E. Evidence of the E*-E equilibrium from rapid kinetics of Na(+) binding to activated protein C and factor Xa. J Phys Chem B. 2010;114:16125–16130. doi: 10.1021/jp105502c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bush-Pelc LA, Marino F, Chen Z, Pineda AO, Mathews FS, Di Cera E. Important role of the Cys-191: Cys-220 disulfide bond in thrombin function and allostery. J Biol Chem. 2007;282:27165–27170. doi: 10.1074/jbc.M703202200. [DOI] [PubMed] [Google Scholar]
- 25.Pineda AO, Carrell CJ, Bush LA, Prasad S, Caccia S, Chen ZW, Mathews FS, Di Cera E. Molecular dissection of Na+ binding to thrombin. J Biol Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 26.Pineda AO, Savvides SN, Waksman G, Di Cera E. Crystal structure of the anticoagulant slow form of thrombin. J Biol Chem. 2002;277:40177–40180. doi: 10.1074/jbc.C200465200. [DOI] [PubMed] [Google Scholar]
- 27.Bah A, Carrell CJ, Chen Z, Gandhi PS, Di Cera E. Stabilization of the E* form turns thrombin into an anticoagulant. J Biol Chem. 2009;284:20034–20040. doi: 10.1074/jbc.M109.012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi PS, Chen Z, Mathews FS, Di Cera E. Structural identification of the pathway of long-range communication in an allosteric enzyme. Proc Natl Acad Sci USA. 2008;105:1832–1837. doi: 10.1073/pnas.0710894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu W, Chen Z, Bush-Pelc LA, Bah A, Gandhi PS, Di Cera E. Mutant N143P reveals how Na+ activates thrombin. J Biol Chem. 2009;284:36175–36185. doi: 10.1074/jbc.M109.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pineda AO, Chen ZW, Bah A, Garvey LC, Mathews FS, Di Cera E. Crystal structure of thrombin in a self-inhibited conformation. J Biol Chem. 2006;281:32922–32928. doi: 10.1074/jbc.M605530200. [DOI] [PubMed] [Google Scholar]
- 31.Marino F, Pelc LA, Vogt A, Gandhi PS, Di Cera E. Engineering thrombin for selective specificity toward protein C and PAR1. J Biol Chem. 2010;285:19145–19152. doi: 10.1074/jbc.M110.119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Bailey S. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 37.Dang QD, Di Cera E. Residue 225 determines the Na(+)-induced allosteric regulation of catalytic activity in serine proteases. Proc Natl Acad Sci U S A. 1996;93:10653–10656. doi: 10.1073/pnas.93.20.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang QD, Guinto ER, Di Cera E. Rational engineering of activity and specificity in a serine protease. Nat Biotechnol. 1997;15:146–149. doi: 10.1038/nbt0297-146. [DOI] [PubMed] [Google Scholar]
- 39.Guinto ER, Caccia S, Rose T, Futterer K, Waksman G, Di Cera E. Unexpected crucial role of residue 225 in serine proteases. Proc Natl Acad Sci U S A. 1999;96:1852–1857. doi: 10.1073/pnas.96.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krem MM, Di Cera E. Molecular markers of serine protease evolution. Embo J. 2001;20:3036–3045. doi: 10.1093/emboj/20.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Cera E. A structural perspective on enzymes activated by monovalent cations. J Biol Chem. 2006;281:1305–1308. doi: 10.1074/jbc.R500023200. [DOI] [PubMed] [Google Scholar]
- 42.Evans SA, Olson ST, Shore JD. p-Aminobenzamidine as a fluorescent probe for the active site of serine proteases. J Biol Chem. 1982;257:3014–3017. [PubMed] [Google Scholar]
- 43.Gohara DW, Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011 doi: 10.1016/j.tibtech.2011.06.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho AL, Sanz L, Barettino D, Romero A, Calvete JJ, Romao MJ. Crystal structure of a prostate kallikrein isolated from stallion seminal plasma: a homologue of human PSA. J Mol Biol. 2002;322:325–337. doi: 10.1016/s0022-2836(02)00705-2. [DOI] [PubMed] [Google Scholar]
- 45.Fujinaga M, James MN. Rat submaxillary gland serine protease, tonin. Structure solution and refinement at 1.8 A resolution. J Mol Biol. 1987;195:373–396. doi: 10.1016/0022-2836(87)90658-9. [DOI] [PubMed] [Google Scholar]
- 46.Piao S, Song YL, Kim JH, Park SY, Park JW, Lee BL, Oh BH, Ha NC. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. Embo J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shia S, Stamos J, Kirchhofer D, Fan B, Wu J, Corpuz RT, Santell L, Lazarus RA, Eigenbrot C. Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B. J Mol Biol. 2005;346:1335–1349. doi: 10.1016/j.jmb.2004.12.048. [DOI] [PubMed] [Google Scholar]
- 48.Rickert KW, Kelley P, Byrne NJ, Diehl RE, Hall DL, Montalvo AM, Reid JC, Shipman JM, Thomas BW, Munshi SK, Darke PL, Su HP. Structure of human prostasin, a target for the regulation of hypertension. J Biol Chem. 2008;283:34864–34872. doi: 10.1074/jbc.M805262200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spraggon G, Hornsby M, Shipway A, Tully DC, Bursulaya B, Danahay H, Harris JL, Lesley SA. Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations. Protein Sci. 2009;18:1081–1094. doi: 10.1002/pro.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budayova-Spano M, Grabarse W, Thielens NM, Hillen H, Lacroix M, Schmidt M, Fontecilla-Camps JC, Arlaud GJ, Gaboriaud C. Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism. Structure. 2002;10:1509–1519. doi: 10.1016/s0969-2126(02)00881-x. [DOI] [PubMed] [Google Scholar]
- 51.Krishnan V, Xu Y, Macon K, Volanakis JE, Narayana SVL. The crystal structure of C2a, the catalytic fragment of classical pathway C3 and C5 convertase of human complement. J Mol Biol. 2007;367:224–233. doi: 10.1016/j.jmb.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milder FJ, Raaijmakers HCA, Vandeputte MDAA, Schouten A, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P. Structure of complement component C2A: implications for convertase formation and substrate binding. Structure. 2006;14:1587–1597. doi: 10.1016/j.str.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Kishi T, Kato M, Shimizu T, Kato K, Matsumoto K, Yoshida S, Shiosaka S, Hakoshima T. Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis. J Biol Chem. 1999;274:4220–4224. doi: 10.1074/jbc.274.7.4220. [DOI] [PubMed] [Google Scholar]
- 54.Mueller-Dieckmann C, Panjikar S, Schmidt A, Mueller S, Kuper J, Geerlof A, Wilmanns M, Singh RK, Tucker PA, Weiss MS. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Crystallogr D Biol Crystallogr. 2007;63:366–380. doi: 10.1107/S0907444906055624. [DOI] [PubMed] [Google Scholar]
- 55.Rohr KB, Selwood T, Marquardt U, Huber R, Schechter NM, Bode W, Than ME. X-ray structures of free and leupeptin-complexed human alphaI-tryptase mutants: indication for an alpha-->beta-tryptase transition. J Mol Biol. 2006;357:195–209. doi: 10.1016/j.jmb.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 56.Marquardt U, Zettl F, Huber R, Bode W, Sommerhoff C. The crystal structure of human alpha1-tryptase reveals a blocked substrate-binding region. J Mol Biol. 2002;321:491–502. doi: 10.1016/s0022-2836(02)00625-3. [DOI] [PubMed] [Google Scholar]