Abstract
We report the transcriptional response of Escherichia coli MG1655 to damage induced by colicins E3 and E9, bacteriocins that kill cells through inactivation of the ribosome and degradation of chromosomal DNA, respectively. Colicin E9 strongly induced the LexA-regulated SOS response, while colicin E3 elicited a broad response that included the induction of cold shock genes, symptomatic of translational arrest. Colicin E3 also increased the transcription of cryptic prophage genes and other laterally acquired mobile elements. The transcriptional responses to both these toxins suggest mechanisms that may promote genetic diversity in E. coli populations, pointing to a more general role for colicins in adaptive bacterial physiology than has hitherto been realized.
Colicins are plasmid-encoded, multidomain bacteriocins produced by Escherichia coli during times of stress (12). Colicin E9 is a magnesium-dependent endonuclease that kills cells through cleavage of chromosomal DNA at its H-N-H active site (17, 22). The H-N-H motif is found in a diverse range of nucleases, including the caspase-activated DNases, responsible for the degradation of chromatin in eukaryotic apoptosis (27). Colicin E3 kills cells by inactivating the prokaryotic ribosome and abolishing protein synthesis. The ability of this enzyme to inhibit ribosome function is due to its cleavage of a single phosphodiester bond in the 16S rRNA between bases A1493 and G1494 at the ribosomal A site (2). In the present work, we show that the global transcriptional responses of E. coli to colicins E3 and E9 have distinct signatures that are consistent with their known cellular targets. Moreover, they indicate how bacterial cells respond to colicin intoxication and highlight molecular mechanisms by which microbial genetic diversity may be promoted.
Experimental design.
The effects of colicins E9 and E3 on E. coli were studied in type I microarray experiments (7). For each experiment, RNA was extracted in parallel from colicin-treated and control cultures of E. coli MG1655 grown at 37°C with shaking in 50 ml of Luria-Bertani broth, with the purified colicin E3-Im3 (28) or colicin E9-Im9 (29) complexes added (at an optical density at 600 nm of 0.6) to a final concentration of 5 μg/ml of culture. RNA samples were prepared from cultures taken at 0, 10, and, in the case of colicin E3, 20 min after the addition of the toxin. Each RNA sample was labeled in a reverse transcriptase reaction, and the cDNAs from the test and control cultures were combined and hybridized to E. coli MG1655 microarrays in quadruplicate (technical replicates). Each experiment was repeated with independently prepared RNA (biological replicates). Until about 20 min after the addition of colicin E9, the growth of the colicin-treated cells mirrored that of the control culture. For colicin E3, the arrest of growth was more rapid but did not cause any overall reduction in optical density after 10 min. The detailed protocols for RNA isolation, reverse transcription of RNA, and cDNA labeling with Cy3 and Cy5 are available at http://www.ifr.ac.uk/safety/microarrays/protocols.html. The E. coli K-12 microarrays consisted of 4,262 of the 4,279 protein-coding regions or open reading frames derived from the complete genome sequence (1). Entire coding sequences were amplified by using specific primer pairs (Sigma Genosys). DNA was spotted onto CMT-GAPS slides (Corning) by using a self-built “Stanford” arraying robot (23). Following the scanning of microarrays, spot and background fluorescence was quantified by using GenePix Pro software, version 3.0 (Axon Instruments, Inc.). Data centering was performed by bringing the median ln(red/green) to 0 for each group of spots printed by the same pin. Microarray data were filtered by using Significance Analysis of Microarrays (SAM) freeware (24). The raw data derived from colicin E3-treated cultures at 10 and 20 min were SAM filtered at delta values of 1.5 and 2.0, respectively, and the E9 data were filtered at a delta value of 2.0. The median coefficients of variation for the biological replicates were 11.2 and 6.5% for E9 at 0 and 10 min, respectively, and 10.8 and 29.8% for E3 at 0 and 20 min, respectively. The median coefficients of variation for the technical replicates were between 4.1 and 8%.
Colicin E9 induces the SOS response.
Colicin E9 treatment for 10 min led to the up-regulation of 30 genes by more than twofold. All but two of these genes can be identified as belonging to the LexA (SOS) regulon (6, 9, 26). In view of the nonspecific DNase activity of colicin E9, activation of the LexA regulon was not unexpected. It is, however, notable that few genes outside this response are induced, with only ppdC and b1012 not being readily identifiable as being LexA regulated. Five genes were down-regulated more than twofold on colicin E9 treatment (lpp, cspE, yeeD, cysH, and yhjR). Hence, the transcriptional changes that we observed are almost exclusively limited to the classical SOS response. Overall, the transcriptional response to colicin E9 treatment is similar to that reported by Courcelle et al. (6), who identified LexA-regulated genes in a broader microarray-based study in which DNA damage was induced through the exposure of wild-type and LexA-deficient cells to UV radiation. As in their study, sulA, recN, recA, umuC, and umuD were among the genes for which mRNA levels showed the greatest increase on induction by colicin E9, all showing >7-fold up-regulation. Additionally, similar transcriptional readthroughs from known LexA-regulated genes are apparent. For example, transcription of dinB leads to increased transcription of yafN, yafO, and yafP (see “Supplementary data” below).
The transcriptional response to colicin E3 shows similarities to the cold shock response.
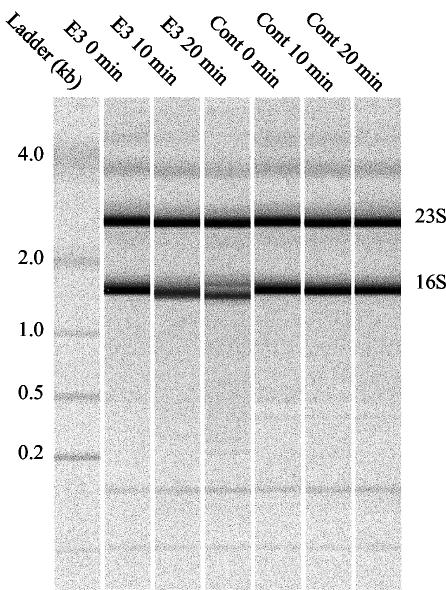
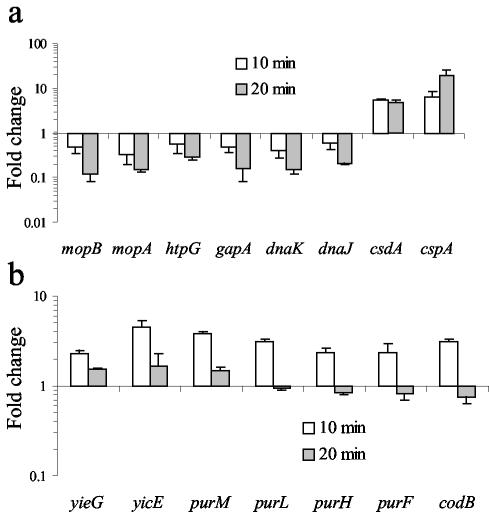
Colicin E3 selectively cleaves around 50 bases from the 5′ end of the 16S rRNA of the prokaryotic ribosome, leading to the inhibition of translation (2). This specific effect on the 16S rRNA could be observed directly in our experiments by analyzing the RNA samples isolated from colicin E3-treated cells, used subsequently for microarray analysis (Fig. 1); this indicated that there was almost complete cleavage of 16S rRNA in the culture after 20 min. In contrast to colicin E9 treatment, where changes in mRNA levels are almost exclusively limited to increased transcript levels of the LexA regulon, many genes show significantly decreased transcript levels after 10 min. It has previously been shown in protein expression studies that the response of E. coli to inhibitors of translation, such as the antibiotic chloramphenicol, is similar to that elicited by cold shock (25). Notable aspects of the cold shock response are increased expression of the major cold shock protein CspA and the cold shock-inducible DEAD box RNA helicase CsdA and decreased expression of heat shock-inducible proteins (14, 25). An increase in transcription of the cspA gene on treatment of E. coli with chloramphenicol has also been reported (13). In our study, both cspA and csdA showed increased transcript levels of six- and fivefold, respectively, on colicin E3 treatment at the 10-min time point and are among the most highly up-regulated genes (Fig. 2a). The σ32-regulated heat shock proteins encoded by dnaK, dnaJ, gapA, mopA, mopB, and htpG showed significantly decreased transcript levels of three- to eightfold at the 20-min time point (Fig. 2a). Down-regulation of the heat shock regulon is likely related to the reduced translational capacity of colicin E3-treated cells and the instability of the σ32 protein, which is rapidly degraded (half-life, <1 min) in growing MG1655 cells at 37°C (15). We also noted increased transcript levels for genes encoding the CspA homologues CspF and CspG (4- and 10-fold, respectively, at 20 min) and significant decreases in those encoding CspC and CspE (approximately 5-fold at 20 min). CspG is cold shock inducible, but little is known about CspF expression patterns (8). Both CspE and CspC are not cold shock inducible and are in fact constitutively expressed at 37°C (21). In addition, transcript levels for rhlE, which encodes a DEAD box RNA helicase closely related to CsdA, was up-regulated by a factor of threefold after 10 min. Cold induction of a DEAD box RNA helicase has also been described for cyanobacteria (5) and archaea (19), and this appears to be a conserved feature of the response to cold shock and perhaps other forms of translational inhibition. DEAD box RNA helicases are ubiquitous; their importance to cold adaptation even in eukaryotes is highlighted by the mutation of the RNA helicase LOS4 of Arabidopsis thaliana which causes cold sensitivity (11).
FIG. 1.
Effect of colicin E3 on 16S rRNA from E. coli MG1655. Time-dependent cleavage of the 16S rRNA by colicin E3 monitored by size chromatography by using an Agilent 2000 bioanalyzer. Total RNA was isolated from colicin E3-treated cultures (E3) and compared to control cultures (Cont) of E. coli MG1655.
FIG. 2.
Time dependence of mRNA colicin/control ratios for selected genes. (a) Known cold shock- and heat shock-inducible genes. Colicin E3 treatment induces increased transcript levels for the cold shock-inducible genes cspA and csdA and decreased transcript levels for the σ32-regulated heat shock genes. (b) Transcription profiles for selected genes involved in de novo purine synthesis and nucleotide transport.
Functional classification and regulation of genes showing decreased mRNA levels.
In addition to the down-regulation of the σ32-dependent heat shock proteins, it is notable that a significant number of genes showing decreased transcript levels are in the σ38 regulon. Decreased transcript levels were also observed for the majority of genes coding for enzymes involved in central energy metabolism. In particular, those required for the conversion of fructose 1,6-biphosphate to pyruvate (fba, tpiA, gapA, pgk, gpmA, eno, and pykF [10]) were strongly affected and were down-regulated by three- to sixfold after 20 min. This metabolic pathway is highly conserved, and decreased transcript levels for some of these genes have been reported on treatment of Streptococcus pneumoniae with chloramphenicol, erythromycin, or tetracycline, but not puromycin (20). Genes encoding a number of tricarboxylic acid enzymes (gltA, icdA, acnA, fumA, and frdA) were also down-regulated on colicin E3 treatment.
Functional classification and regulation of genes showing increased mRNA levels.
Many genes showing increased transcript levels are of unknown function and in many cases are not conserved except in very close relatives of E. coli. A notable exception is provided by genes involved in de novo purine synthesis under the transcriptional control of the PurR repressor, which show significant up-regulation after 10 min (30). Indeed, all the enzymes required for the conversion of 5-phosphoribosyl-1-pyrophosphate to inosinate, i.e., the enzymes encoded by purFDNLMKCBH, showed a 1.5- to 4-fold increase in transcript levels after 10 min of colicin E3 treatment. However, after 20 min, these genes were down-regulated by around twofold relative to the 10-min time point (Fig. 2b). In S. pneumoniae, an increase in transcript levels for nearly all these genes was also observed 10 min after the addition of sublethal concentrations of antibiotics that inhibit translation (20). Although this aspect of the response to translational inhibition appears to be conserved between E. coli and S. pneumoniae, the transcriptional arrangement of the PurR regulon in the two organisms is very different. In S. pneumoniae, these genes form a single monocistronic cluster, whereas in E. coli they are distributed either singly or in pairs throughout the genome.
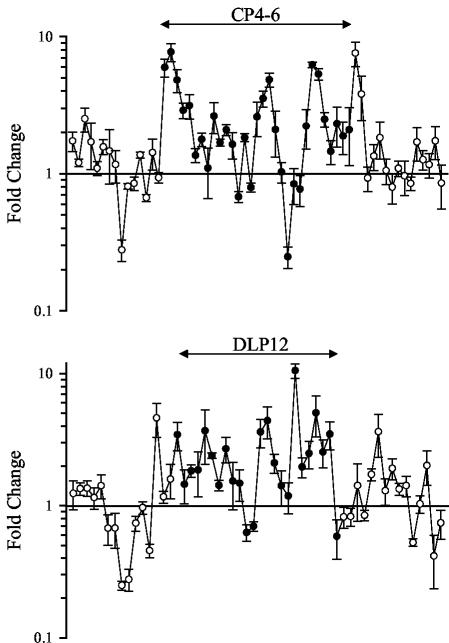
A surprising observation stemming from our array data for colicin E3-treated cells is that a number of genes showing increased transcript levels were located in phage-derived regions of the genome. E. coli K-12 harbors at least seven cryptic prophages, CP4-6, DLP12, e14, Rac, Qin, CP4-44, and CP4-57, the locations of which have been either precisely or approximately delineated (1, 3). At the 20-min time point, around 15 of the 100 genes showing the greatest increases in transcript levels are located within prophage regions. Figure 3 shows the changes (n-fold) for those genes lying within and bordering prophages CP4-6 and DLP12 (1, 3). Genes in the cryptic prophage regions showing increased expression include predicted DNA integrases, invertases, and recombinases (e.g., ybcK, b1345, b1374, and b1545). It is notable that other laterally acquired elements such as the insertion sequence IS1 genes, insA and insB, and genes of the Rhs elements, rhsD and rhsE, also show large increases in transcript levels (four- to sixfold after 20 min).
FIG. 3.
Colicin E3 up-regulates transcription of cryptic prophage genes. Effect of colicin E3 on genes of the cryptic prophages CP4-6 and DLP12 after 20 min. Filled circles indicate genes of the prophage, and open circles represent genes bordering the prophage.
Conclusion.
Although the cellular targets of colicins E9 and E3 are quite different, the present study suggests that both may play a role in the adaptive physiology of a bacterium. In the case of colicin E9, this may be due to the activation of SOS-inducible error-prone DNA polymerases and perhaps other SOS-induced functions (16, 18). For colicin E3, increased mRNA levels for mobile genetic elements, which include DNA recombinases of phage and insertion sequence origin, may provide another route to the generation of genetic diversity. It is interesting to speculate that the administration of sublethal quantities of translation inhibitors to a bacterium (in the present case, this administration pertains to colicins but could apply equally to small-molecule antibiotics) may promote lateral gene transfer and/or chromosomal rearrangements and that these changes may lead to the accelerated acquisition and spread of antibiotic resistance. Indeed, the relationship between stress and the mobility of transposable elements in prokaryotes and eukaryotes has been well documented (reviewed in reference 4). Colicin E9 does not produce the same global effect, and so this response cannot simply be due to colicin-induced cell death but must be linked to the translational arrest caused by colicin E3. The mechanism by which this arrest signals the elevated transcription of mobile elements in the genome is currently unknown. Moreover, we are unaware of a similar phenomenon in biology, where the cellular response to untimely cell death is akin to “rats leaving a sinking ship.”
Supplementary data.
The SAM-filtered data for genes showing a >2-fold increase in transcript levels on colicin E9 treatment are listed in Table S1 found at http://www.biolws1.york.ac.uk/echobase/colicinarrays/. Filtered data sets for genes showing increased or decreased transcript levels on colicin E3 treatment after 10 min are listed in Table S2 and Table S3, and those after 20 min of treatment are listed in Table S4 and Table S5.
Acknowledgments
This work was funded by The Wellcome Trust and the BBSRC.
We thank Peter Young (York, United Kingdom) and Mark Buttner (Norwich, United Kingdom) for helpful discussions and Geraint Barton (York) for additional statistical analysis.
REFERENCES
- 1.Blattner, F. R., G. Plunket III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatric, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, C. M., J. E. Dahlberg, T. Ikemura, J. Konisky, and M. Nomura. 1971. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc. Natl. Acad. Sci. USA 68:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, A. M. 1996. Cryptic prophages, p. 2041-2046. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 4.Capy, P., G. Gasper, C. Biémont, and C. Bazin. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85:101-106. [DOI] [PubMed] [Google Scholar]
- 5.Chamot, D., W. C. Magee, E. Yu, and G. W. Owttrim. 1999. A cold shock-induced cyanobacterial RNA helicase. J. Bacteriol. 181:1728-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcelle, J., A. Khodursky, B. Peter, P. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 8.Etchegaray, J.-P., and M. Inouye. 1999. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 181:1827-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 10.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 11.Gong, Z., H. Lee, L. Xiong, A. Jagendorf, B. Stevenson, and J.-K. Zhu. 2002. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99:11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James, R., C. N. Penfold, G. R. Moore, and C. Kleanthous. 2002. Killing of E. coli cells by E group nuclease colicins. Biochimie 84:381-389. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, W., P. Jones, and M. Inouye. 1993. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J. Bacteriol. 175:5824-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, P. G., M. Mitta, Y. Kim, W. Jiang, and M. Inouye. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanemori, M., H. Yanagi, and T. Yura. 1999. Marked instability of the σ32 heat shock transcription factor at high temperature. J. Biol. Chem. 274:22002-22007. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S.-R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleanthous, C., U. C. Kuhlmann, A. J. Pommer, N. Ferguson, S. E. Radford, G. R. Moore, R. James, and A. M. Hemmings. 1999. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6:243-251. [DOI] [PubMed] [Google Scholar]
- 18.Kleanthous, C., and D. Walker. 2001. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26:624-631. [DOI] [PubMed] [Google Scholar]
- 19.Lim, J., T. Thomas, and R. Cavicchioli. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J. Mol. Biol. 297:553-567. [DOI] [PubMed] [Google Scholar]
- 20.Ng, W.-L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommer, A. J., S. Cal, A. H. Keeble, D. Walker, S. J. Evans, U. C. Kuhlmann, A. Cooper, B. A. Connolly, A. M. Hemmings, G. R. Moore, R. James, and C. Kleanthous. 2001. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314:735-749. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, A., S. Lucchini, and J. C. D. Hinton. 2001. It's easy to build your own microarrayer. Trends Microbiol. 9:154-156. [DOI] [PubMed] [Google Scholar]
- 24.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 27.Walker, D. C., T. Georgiou, A. J. Pommer, D. Walker, G. R. Moore, C. Kleanthous, and R. James. 2002. Mutagenic scan of the H-N-H motif of colicin E9: implications for the mechanistic enzymology of colicins, homing enzymes and apoptotic endonucleases. Nucleic Acids Res. 30:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker, D., G. R. Moore, R. James, and C. Kleanthous. 2003. Thermodynamic consequences of bipartite immunity protein binding to the ribosomal ribonuclease colicin E3. Biochemistry 42:4161-4171. [DOI] [PubMed] [Google Scholar]
- 29.Wallis, R., A. Reilly, A. Rowe, G. R. Moore, R. James, and C. Kleanthous. 1992. In vivo and in vitro characterization of overproduced colicin E9 immunity protein. Eur. J. Biochem. 207:687-695. [DOI] [PubMed] [Google Scholar]
- 30.Zalkin, H., and P. Nygaard. 1996. Biosynthesis of purine nucleotides, p. 561-579. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.