Abstract
We identified an Arabidopsis thaliana mutant, clumped chloroplasts 1 (clmp1), in which disruption of a gene of unknown function causes chloroplasts to cluster instead of being distributed throughout the cytoplasm. The phenotype affects chloroplasts and nongreen plastids in multiple organs and cell types, but is detectable only at certain developmental stages. In young leaf petioles of clmp1, where clustering is prevalent, cells lacking chloroplasts are detected, suggesting impaired chloroplast partitioning during mitosis. Although organelle distribution and partitioning are actin-dependent in plants, the actin cytoskeleton in clmp1 is indistinguishable from that in WT, and peroxisomes and mitochondria are distributed normally. A CLMP1-YFP fusion protein that complements clmp1 localizes to discrete foci in the cytoplasm, most of which colocalize with the cell periphery or with chloroplasts. Ultrastructural analysis revealed that chloroplasts within clmp1 clusters are held together by membranous connections, including thin isthmi characteristic of late-stage chloroplast division. This finding suggests that constriction of dividing chloroplasts proceeds normally in clmp1, but separation is impaired. Consistently, chloroplast size and number, as well as positioning of the plastid division proteins FtsZ and ARC5/DRP5B, are unaffected in clmp1, indicating that loss of CLMP1-mediated chloroplast separation does not prevent otherwise normal division. CLMP1-like sequences are unique to green algae and land plants, and the CLMP1 sequence suggests that it functions through protein–protein interactions. Our studies identify a unique class of proteins required for plastid separation after the constriction stage of plastid division and indicate that CLMP1 activity is also required for plastid distribution and partitioning during cell division.
Keywords: mitochondrial distribution, organelle separation
Although plants are sessile organisms, plant cell organelles are motile. Their distribution and repositioning in the cell maximize their functions in response to internal and external stimuli and are essential for unbiased inheritance during cell division. In land plants, the positioning of chloroplasts, peroxisomes, and mitochondria is mediated mostly by the actin cytoskeleton (reviewed in ref. 1).
Chloroplasts are propagated by binary fission of preexisting organelles (2, 3). Constriction of chloroplasts is driven by several midplastid contractile rings, including the FtsZ ring, dynamin ring, and outer plastid-dividing (PD) ring (2–4). The FtsZ ring (Z ring), composed of the tubulin-like FtsZ protein, assembles inside the chloroplast, marks the division site, and constricts the inner membrane to initiate division (3, 5, 6). The dynamin ring, composed of the dynamin-related protein ARC5/DRP5B, and the outer PD ring, composed of bundled polyglucan fibrils, function outside the chloroplast, providing at least some of the force required for division (4, 7–10). Both the dynamin and outer PD rings function in constricting plastids through the late stages of division, but little is known about how final separation of the daughter plastids is achieved.
Here we describe the identification and functional analysis of a novel plant-specific protein, CLUMPED CHLOROPLASTS 1 (CLMP1). Chloroplasts constrict normally during division in Arabidopsis thaliana clmp1 mutants but are impaired in separation, resulting in chloroplast clustering. Loss of CLMP1 also results in clustering of other plastid types, indicating its function throughout the plant. Finally, the separation defect leads to impaired chloroplast partitioning during cell division, yielding some cells that lack chloroplasts. Thus, our results also uncover a connection between postconstriction plastid separation and plastid distribution and partitioning.
Results
Identification of the clumped chloroplasts 1 (clmp1) Mutant.
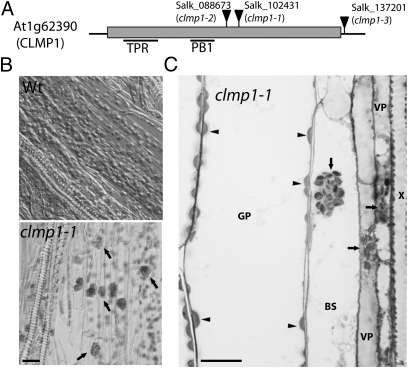
As part of the large-scale Arabidopsis Chloroplast 2010 project (http://www.plastid.msu.edu/), Arabidopsis mutants with T-DNA insertions in genes encoding predicted chloroplast proteins were screened for aberrant chloroplast morphology (11). Among the 5,200 homozygous lines screened, Salk_102431, bearing an insertion in At1g62390, displayed a unique phenotype; chloroplasts in some cells of this mutant appeared clustered (Fig. 1 A–C), whereas equivalent cells in the WT were dispersed normally (Fig. S1A). Two additional At1g62390 insertion mutants, Salk_088673 and Salk_137201, exhibited the same phenotype (Fig. S1B). All three alleles behaved as recessive mutations. We concluded that the chloroplast clustering phenotype resulted from loss of function of At1g62390, and named the gene CLUMPED CHLOROPLASTS 1 (CLMP1). Hereinafter, Salk_102431, Salk_088673, and Salk_137201 are referred to as clmp1-1, clmp1-2, and clmp1-3, respectively (Fig. 1A).
Fig. 1.
Identification of CLMP1. (A) Gene structure and T-DNA insertion mutations. The box indicates the exon; the thin black lines represent UTRs. Positions of the predicted TPR and PB1 domains are indicated. (B) Chloroplast phenotype in WT and clmp1-1. Arrows indicate clumped chloroplasts. (C) Light micrograph of a petiole section from clmp1-1. The WT control is shown in Fig. S1A. Arrows indicate clumped chloroplasts; arrowheads, dispersed chloroplasts. X, xylem; VP, vascular parenchyma cell; BS, bundle sheath cell; GP, ground parenchyma cell. (Scale bars: 20 μm.)
CLMP1 encodes a unique protein of 751 amino acids containing predicted tetratricopeptide repeat (TPR) and octicosapeptide/Phox/Bem1p (PB1) domains (Fig. 1A). Both the TPR and PB1 domains function in protein–protein interactions (12, 13). CLMP1 is conserved in green algae and land plants, but CLMP1-like sequences have not been identified outside the green lineage. Three CLMP1 paralogs exist in A. thaliana: At2g25290, At4g32070, and At5g20360; however, these are in a separate clade that diverged from the CLMP1 clade before the divergence of monocots and dicots (Fig. S2). Neither homozygous T-DNA insertional mutants of At2g25290 or At4g32070 nor the double mutant generated from the two display the clumped-chloroplasts phenotype (Fig. S1 C–E), suggesting that they may have different functions from CLMP1. However, because no mutant was available for At5g20360, we cannot rule out the possibility that CLMP1 paralogs might have partial functional redundancy with CLMP1.
Clumped-Chloroplasts Phenotype Is Transient.
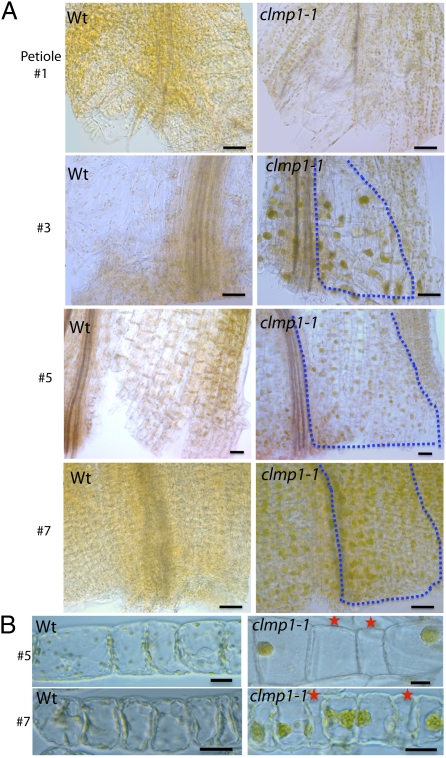
The mutant screen initially detected the chloroplast-clustering phenotype in vascular parenchyma (VP) and bundle sheath (BS) cells in petioles of clmp1-1 (Fig. 1 B and C). Further analysis showed that chloroplast distribution varied depending on the stage of leaf development. In juvenile leaves, clustered chloroplasts were observed in almost all cells of the petiole (Fig. 2A, leaf 7, dotted line; Fig. 2B, Bottom Right), including epidermal, ground parenchyma (GP), VP, and BS cells. In the basal half of the leaf blade, the phenotype became gradually restricted to the VP and BS cells of the midvein only. Clumping was not obvious in the rest of the leaf (Fig. S3A). The spatial distribution of cells with clumped chloroplasts in older leaves was similar to that in leaf 7, except fewer cells displayed the phenotype (Fig. 2A, leaves 5 and 3). In the oldest leaf (leaf 1), the phenotype was mostly absent but was observed infrequently in VP or BS cells. In WT, the chloroplasts were dispersed in all cell types irrespective of leaf developmental stage (Fig. 2A) and were most often situated around the cell periphery, as described previously (14).
Fig. 2.
More detailed characterization of clmp1. (A) Whole-mount differential interference contrast images of the petiole base from leaves 1, 3, 5, and 7 (old to young) of WT and clmp1-1. Dotted lines outline regions in which clumped chloroplasts are observed within half a petiole. (Scale bars: 50 μm.) (B) Ground parenchyma cells isolated from petioles of leaves 5 and 7 of WT and clmp1-1. Cells were from regions corresponding to outlined areas of leaves 5 and 7 in A. Red stars indicate cells lacking chloroplasts. (Scale bars: 20 μm.)
Disruption of CLMP1 Affects Multiple Plastid Types Throughout the Plant.
To evaluate whether the mutation in CLMP1 also affects nongreen plastids, we expressed a transgene encoding plastid-targeted YFP (RecA-YFP) (15) in clmp1-1 and WT plants and examined plastid distribution along the longitudinal axis of the root. In the root meristem, proplastids were not obviously clumped (Fig. S3B), although this examination was not conclusive because of the dense plastid packing in these small cells. In the elongation zone, where cells expand rapidly (16, 17), plastids were mostly scattered (Fig. S3B), except in a few cells in the vascular cylinder where plastids appeared clumped. In the maturation zone, where root hairs develop and xylem lignification is discernible, plastids were clumped in most cells, including those of the vascular cylinder (Fig. S3B, arrows), endodermis, and cortex and some epidermal cells (Fig. S3B, arrowheads). In the mature zone, clumped plastids were observed only in cells associated with the vascular cylinder (Fig. S3B, Top Right, arrows), at a lower frequency than seen in the maturation zone. Clumping of root plastids was not observed in WT (Fig. S3B).
Clumping of chloroplasts was also observed in hypocotyls, petals, sepals, anther filaments, and papillae cells of the stigma, and of nongreen plastids in trichomes and root hairs (Fig. S3C). As in leaves and roots, in other organs the phenotype was detectable only at certain developmental stages, being generally more prominent in younger tissues than in more mature tissues (Fig. S3D).
To examine the spatial and temporal pattern of CLMP1 expression, we expressed a CLMP1::GUS transgene in WT Arabidopsis and performed histochemical staining on specimens of various plant organs at different developmental stages. We found that CLMP1 expression was developmentally regulated (Fig. S4), consistent with microarray data reported previously (17). The β-glucuronidase (GUS) staining pattern, particularly in petioles and roots, largely corresponded to the distribution of the plastid clumping phenotype in clmp1 (Fig. 2 and Fig. S3).
Despite the striking abnormality in the distribution of both chloroplasts and nongreen plastids, clmp1 exhibited overall normal growth and development, but flowered slightly earlier than WT. clmp1-1 and clmp1-2 flowered in 32.4 ± 2.5 d and 31.6 ± 2.1 d, respectively, whereas WT flowered in 34.4 ± 2.5 d (P < 0.02 and 0.001, respectively).
Chloroplast Partitioning Is Affected in clmp1.
Interestingly, we noticed that some petiole cells in clmp1 were devoid of chloroplasts. Fig. 2B shows a file of GP cells isolated from the basal region of the petiole in leaves 7 and 5, in which a few cells lack chloroplasts. Cells isolated from a similar region in WT plants had dispersed chloroplasts, and cells without chloroplasts were never observed (Fig. 2B, Left). Petiole cells in leaves at these developmental stages are still mitotic (18); our observations suggest that the improper distribution of chloroplasts in clmp1 also affects chloroplast partitioning to daughter cells during mitosis. We also looked for, but could not identify, cells lacking chloroplasts in more mature leaf tissues in which the phenotype is no longer detectable.
Actin Cytoskeleton Is Not Defective in clmp1.
A previous report that overexpression of CLMP1 in Schizosaccharomyces pombe produced elongated single-nucleate cells with a central bulge and diffuse actin staining promped speculation that the gene product might play a role in actin organization (19). Because chloroplast distribution and partitioning are actin-dependent (1, 20), we carried out a similar experiment by expressing either CLMP1 or a functional CLMP1-YFP fusion protein (see below) in S. pombe, but found no abnormal yeast cell morphology or growth (Fig. S5E). The difference in findings may be because the cDNA used in the earlier study contained an extra ∼300 nucleotides downstream of the stop codon that were not included in our construct, perhaps producing the abnormal morphology.
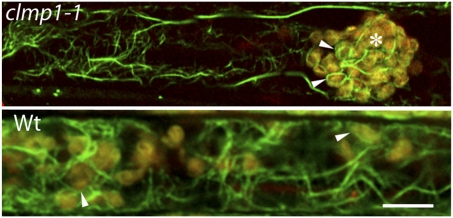
To investigate actin organization in clmp1 more directly, we stained petioles of clmp1 and WT plants with phalloidin (21). In mutant cells with clustered chloroplasts and in equivalent WT cells, we observed longitudinal arrays of thick actin cables and randomly oriented thin actin filaments (Fig. 3). Actin filaments were also wrapped around the chloroplasts in both mutant and WT cells (Fig. 3, arrowheads). These features are similar to those reported previously in WT Arabidopsis (22, 23), and there were no obvious differences between mutant and WT plants.
Fig. 3.
Alexa Fluor 488-phalloidin staining (green) of actin in VP cells in clmp1-1 and WT. Chlorophyll autofluorescence is indicated in red. (Scale bar: 10 μm.) Asterisk denotes clumped chloroplasts; arrowheads indicate chloroplast-associated actin filaments.
Mitochondria and Peroxisomes Are Distributed Normally in clmp1.
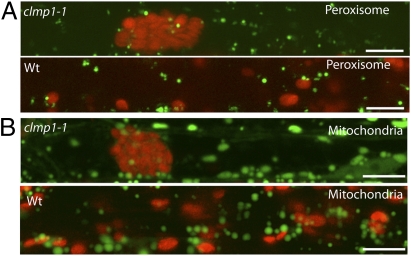
Mitochondrial and peroxisome distribution and partitioning are also actin-dependent (1, 20). To assess whether disruption of CLMP1 affects these organelles, we examined their distribution in clmp1-1 mutants expressing either the peroxisomal marker protein YFP-PTS1 (24) or the mitochondrial marker protein mt-YK (25). Unlike chloroplasts, both peroxisomes and mitochondria appeared to be distributed normally in all mutant lines (Fig. 4). To assess this finding quantitatively, we compared the average distances between peroxisomes or mitochondria in petiole cells of clmp1-1 containing clustered chloroplasts, as well as in equivalent cells from WT (Fig. 4). The average distances between peroxisomes were 32.9 ± 23.7 μm in WT and 32.0 ± 21.9 μm in clmp1-1, a statistically insignificant difference (P = 0.57, Student t test). Similar results were obtained for mitochondria, with average distances of 36.4 ± 25.2 μm in WT and 36.0 ± 25.1 μm in mutant (P = 0.41). The distribution of distances between peroxisomes and mitochondria was also similar in clmp1-1 and WT (Fig. S6). Based on the normal distribution of mitochondria and peroxisomes, normal actin organization, and lack of abnormal cell morphology in S. pombe cells expressing CLMP1-YFP, we conclude that general defects in the actin cytoskeleton do not account for the chloroplast clumping in clmp1.
Fig. 4.
Distribution of peroxisomes (A) and mitochondria (B) in the VP cells of clmp1-1 and WT. Peroxisomes and mitochondria are in green; chlorophyll autofluorescence is in red. Images were superimposed from three Z-section images. (Scale bars: 10 μm.)
CLMP1 Is Localized to Foci in the Cytoplasm.
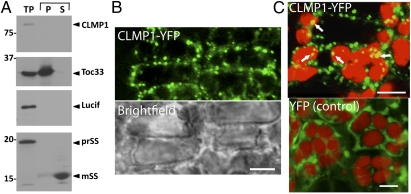
CLMP1 has a weakly predicted transit peptide based on TargetP (26), accounting for its inclusion in the Chloroplast 2010 gene list (http://www.plastid.msu.edu/about/gene_list.html). However, TargetP localization predictions for CLMP1 orthologs (Fig. S2, clade 1) were variable. To assess the subcellular localization of CLMP1, we performed an in vitro chloroplast import assay using radiolabeled CLMP1 with the chloroplast membrane protein Toc33, stroma-localized small subunit of RUBISCO (SS), and cytosolic luciferase (lucif) as markers. All proteins migrated at their predicted sizes on SDS gels (TP in Fig. 5A). As expected, Toc33 and SS were associated with the membrane and soluble fractions, respectively. Furthermore, the transit peptide of SS was cleaved on import, producing a molecular weight shift (prSS to mSS in Fig. 5A). In contrast, luciferase was absent from both the membrane and soluble fractions, as expected for a cytosolic protein. CLMP1 behaved like luciferase, suggesting that it is not imported into the chloroplast.
Fig. 5.
CLMP1 localization. (A) Chloroplast import assay of CLMP1 and controls. TP, translation product; P, chloroplast membrane fraction; S, chloroplast soluble fraction; PrSS, SS precursor; mSS, mature SS. Positions of molecular mass markers (in kDa) are shown at the left. (B) CLMP1-YFP localizes to foci near the cell periphery in complemented clmp1-1 plants. A brightfield image of the same sample is shown below the fluorescence image. (C) Some CLMP1-YFP foci colocalize with chloroplasts (arrows). YFP localization in WT is shown as a control. YFP fluorescence is indicated in green; chlorophyll autofluorescence is in red. (Scale bars: 5 μm.)
To further define the localization of CLMP1, we generated a transgene encoding full-length CLMP1 fused to YFP (35S:CLMP1-YFP) and expressed it in clmp1-1. The transgene complemented the mutant phenotype (Fig. S5A), indicating that it encodes a functional protein. CLMP1-YFP was localized to distinct foci in the cytosol, which frequently colocalized with the cell periphery and with chloroplasts (Fig. 5 B and C). Punctate localization also was observed in anther filaments and roots (Fig. S5 B and C) and in S. pombe cells (Fig. S5D). Taken together, these data indicate that CLMP1 is not imported to the plastid, but is localized to distinct foci in the cytoplasm.
Chloroplasts in clmp1 Are Interconnected.
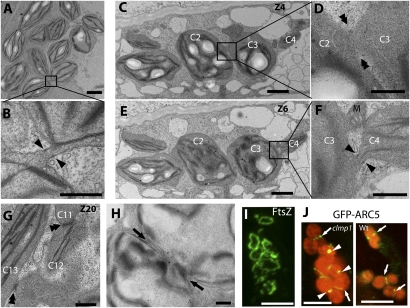
To understand the basis of chloroplast clustering in clmp1, we examined petiole tissue in clmp1-1 using transmission electron microscopy (TEM). Chloroplasts within clumps were ultrastructurally similar to those in WT (Fig. S7 B and C), with one major exception: Clustered chloroplasts in the mutant were often connected. The appearance of the connections between chloroplasts varied. Some were narrow stroma-filled isthmi in which the double membranes were evident (Fig. 6 B and F). In such connections, small electron-dense deposits, probably representing PD rings (4, 27), were often visible at the edges (Fig. 6 B and F, arrowheads). To the best of our knowledge, PD rings have not been previously reported in Arabidopsis. These morphological features suggest that the narrow isthmi represent chloroplasts at a very late stage of division (27). We also observed some chloroplasts at earlier stages of division in clmp1 (Fig. 6H), which we also found in the WT (Fig. 6J, Right). Other chloroplast connections in clmp1-1 were less well defined in appearance. Some were wider, darker, and less organized than the narrow isthmi (Fig. 6G, double arrowheads, and Fig. S7M), whereas others appeared even more disorganized (Fig. S7D). Neither thin isthmi nor disorganized chloroplast connections were observed in the equivalent tissues from WT (Fig. S7A and Fig. S8A).
Fig. 6.
Structure of clumped chloroplasts in clmp1. (A–H) Transmission electron micrographs of chloroplasts within clumps. C–G are selected images from serial sections of a single chloroplast cluster shown more completely in Figs. S7 and S9. Chloroplasts with the same number in C–G and Fig. S7 (arbitrarily numbered C1–C13) represent the same chloroplast viewed in different serial sections. A, B, and H are from a different cluster. B, D, and F are magnified views of the boxes in A, C, and E, respectively. Black arrowheads indicate presumed PD rings at the edges of isthmi. Double arrowheads indicate chloroplast connections that are disorganized in appearance. Black arrows indicate chloroplast constriction sites. M, mitochondrion. (Scale bars: 2 μm in A, C, and E; 500 nm in B, D, F, G, and H.) (I) FtsZ immunolocalization in clmp1. (Scale bar: 5 μm.) (J) GFP-ARC5 localization in clmp1 and WT. (Scale bar: 5 μm.)
Serial sectioning through clusters confirmed that most of the structures described above, regardless of appearance, represent connections between chloroplasts and not different regions of the same chloroplast. For example, the connection between the chloroplasts labeled C2 and C3 in Fig. 6 C and D is not visible in sections above or below them (Fig. S7 E–G). In addition, the serial sections showed that one chloroplast was commonly adjoined to two or more chloroplasts within a clump; for example, chloroplast C3 was connected not only to chloroplast C2 (Fig. 6 C and D), but also to chloroplast C4 in a different plane (Fig. 6 E and F and Fig. S7 E–J). The extent of connectedness between chloroplasts varied. In some clusters the chloroplasts were extensively interconnected, whereas in others fewer connections were present. However, among the three clusters examined by serial sectioning from three individual clmp1-1 mutants, multiple connections within a cluster were always observed. This variability may reflect the transience of the phenotype; clusters with fewer connections are probably from slightly older cells in which the phenotype is beginning to disappear.
In contrast to chloroplasts, the fine structures of mitochondria and peroxisomes in clmp1 were indistinguishable from those in WT (Fig. S8 A–D). This is consistent with our finding that the distribution of these organelles is not altered in clmp1-1 (Fig. 4 and Fig. S6).
Localization of Plastid Division Proteins in clmp1.
The persistent thin connections between chloroplasts within clmp1 clusters prompted us to investigate whether the chloroplast contractile machinery might be affected. We examined the localization of two division proteins, FtsZ and the dynamin-related protein ARC5/DRP5B (5, 7). Single FtsZ rings were consistently observed in clustered chloroplasts of clmp1, as in WT (Fig. 6I). Similarly, ARC5/DRP5B localized to mid-plastid rings and to bright spots between chloroplasts (Fig. 6J, white arrows and arrowheads, respectively). The latter may represent the persistence of ARC5/DRP5B at the pole of one daughter plastid late in division, as occurs in WT (28). The midplastid localization of FtsZ and ARC5/DRP5B in clmp1 clusters, coupled with the presence of deep constrictions between chloroplasts, suggests that division of clustered chloroplasts in clmp1 is sufficient to allow proper repositioning of the contractile machinery and effect subsequent divisions, even though chloroplasts remain connected. These findings are consistent with the data showing that CLMP1 does not localize to the midplastid (Fig. 5) as do FtsZ, ARC5/DRP5B, and other components of the contractile machinery (2, 5, 7), and indicate a role for CLMP1 specifically in separation, but not constriction, of plastids.
Discussion
We have identified a cytosolic protein, CLMP1, that is required for normal plastid distribution in Arabidopsis. In the absence of CLMP1, both chloroplasts and nongreen plastids are clustered, whereas mitochondria and peroxisomes are distributed normally. TEM showed that chloroplasts in clmp1 are physically interconnected, explaining the clustering phenotype. Although chloroplast clustering has been reported in several previous studies, physical connections have not been observed (29–31), indicating that clustering itself does not induce the formation of such connections. Thus, clmp1 represents a unique phenotype and defines a specific class of proteins required for plastid separation.
Although chloroplast clustering in clmp1 occurs in many cell types, CLMP1 expression is developmentally regulated, and clustering is observed in a developmental pattern largely paralleling CLMP1 promoter activity. The presence of cells lacking chloroplasts in petioles of young leaves implies that CLMP1 activity contributes to proper chloroplast partitioning during cell division in some mitotically active tissues. How plastid partitioning is regulated in dividing cells remains unclear, but previous work in both green algae and land plants suggests an active partitioning mechanism that distributes chloroplasts to daughter cells fairly equally and prevents the formation of aplastidic cells (20, 32). If cells lacking chloroplasts in clmp1 petioles also lacked nongreen plastids, then they presumably would die during leaf growth, because plastids are assumed to be essential to the viability of most cells. This presumption is consistent with our finding that no cells lack chloroplasts in older leaves, although one study reported that cells lacking detectable plastids were viable (33). The transient nature of the clustering phenotype suggests that plastid separation may be delayed in clmp1 or that another mechanism of separation becomes active as cells mature. Alternatively, CLMP1 paralogs, which have different expression patterns from that of CLMP1 (Arabidopsis eFP browser) (34), could supplant CLMP1 function at other stages of development.
Although clustering prevented accurate measurement, chloroplast size and number appeared normal in clmp1 clusters, indicating that plastids continue to divide (i.e., constrict) even though they do not separate. This is in contrast with mutants defective in late-acting chloroplast division proteins, such as arc5 and pdv2, in which chloroplasts are enlarged and dumbbell-shaped. In these mutants, multiple adjacent Z rings or spirals occur at constriction sites (28), suggesting that division does not progress beyond a point at which the Z ring becomes repositioned to the middle of the two new daughter plastids. In contrast, single Z rings, as well as midplastid ARC5/DRP5B rings, were present in clmp1 clusters, suggesting proper repositioning of these molecular components of the division machinery. Furthermore, the clustered chloroplasts frequently exhibited thin isthmi and presumed PD rings, typical of the late stages of division in various plants (27, 35). We did not observe such structures in WT, probably because plastids separate rapidly once they reach the final stages of constriction (27, 36). Taken together, these findings indicate that the latest stage of constriction is prolonged in the mutant, and that CLMP1 functions after constriction specifically to promote plastid separation. However, delayed separation in the mutant does not appear to interfere with the otherwise normal operation of the chloroplast contractile machinery.
How chloroplasts separate in the final stage of division is not known, but envelope membrane remodeling must be involved. Some chloroplast connections in clmp1 appear more disorganized than the narrow isthmi and may represent disordered PD rings or other components of the contractile machinery. These connections might also represent a normally short-lived phase of membrane remodeling that is too transient to have been observed in WT. Alternatively, the persistence of isthmi or PD rings between chloroplasts in the mutant could induce the formation of some disorganized membrane connections. Thus, it is possible that CLMP1 could function in a membrane remodeling process that promotes plastid separation after the latest stages of constriction, or in disassembly or removal of the PD ring and/or other late-acting cytosolic division components. The localization of CLMP1 near the cell periphery suggests that such a function might involve removal or recycling of such components to the plasma membrane or a nearby compartment.
The normal actin filament morphology and distribution of peroxisomes and mitochondria in clmp1 argue against a role for CLMP1 in the organization of cytoplasmic actin cables. However, short chloroplast-associated actin filaments anchor chloroplasts to the plasma membrane and have been suggested to provide motive force for blue light-mediated chloroplast movement (23), possibly through interaction with a protein localized partially in the plasma membrane (37). The localization of CLMP1 to both the cell periphery and chloroplasts (Fig. 5) raises the alternative possibility that CLMP1 could play a role in anchoring chloroplasts to the plasma membrane, perhaps through interaction with chloroplast-associated actin. If such CLMP1-mediated anchoring occurred, it could implicate chloroplast movement in plastid separation, perhaps via a pulling mechanism.
Although the biochemical role of CLMP1 has yet to be established, the presence of predicted TPR and PB1 domains, both of which mediate protein–protein interactions (12, 13), suggests that CLMP1 function likely involves attachment to other proteins. A recent in silico analysis suggested a potential interaction with the Hsp90/Hsp70 chaperone system (38). Intriguingly, disruption of a conserved cytosolic protein, CluA, impairs separation of mitochondria in Dictyostelium and other eukaryotes, resulting in mitochondrial clustering (39–41). Like CLMP1, CluA bears a predicted TPR domain, and its localization in Drosophila resembles that of CLMP1 (40). How CluA and its orthologs effect mitochondrial separation is not known, but they have been postulated to promote outer membrane scission at a late stage of division (42) and to facilitate separation via movement on the cytoskeleton (40, 41, 43). Thus, CluA and CLMP1 may have analogous functions in the separation and distribution of mitochondria and chloroplasts. Examining the interactions of CLMP1 with other plant proteins may shed light on the mechanisms underlying final separation of both organelles.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana ecotype Columbia (Col-0) was used in all experiments. Plant material, growth conditions, and generation of transgenic plants are detailed in SI Materials and Methods.
Brightfield and Fluorescence Microscopy.
Brightfield microscopy was performed with differential interference contrast on a Leica inverted microscope system (11) or Leica DMRA2 microscope (7), as indicated. Epifluorescence microscopy was performed with a Leica DMRA2 or an Olympus Fluoview 1000 laser scanning confocal microscope. Alexa Fluor 488 (Invitrogen), YFP, and chlorophyll were excited with 488-nm, 543-nm, and 543-nm lasers, respectively, and emissions were collected with BA505-525, BA535-565, and BA650IF filters, respectively.
Analysis of Plastid Morphology.
Leaves were detached from the base of a 2.5-wk-old plant and measured. Leaves were named consecutively, with leaf 1 the oldest and leaf 7 the youngest. Petiole length and total leaf length were 2 mm and 5 mm, respectively, for leaf 1, 4 mm and 8 mm for leaf 3, 2.5 mm and 7 mm for leaf 5, and 0.5 mm and 3 mm for leaf 7 (Fig. 3). Leaves were fixed (44) and placed on a microscope slide under a coverslip for whole-mount imaging. To image single cells, cells were further separated by gently tapping the coverslip. Samples were viewed on the Leica inverted microscope (11). Chloroplast morphologies in anther filaments and other flower parts were imaged similarly but without fixation. Morphology of nongreen plastids was evaluated by introducing RecA-YFP (15) into clmp1-1 or WT plants and then imaging YFP fluorescence by epifluorescence or confocal microscopy.
Phalloidin Staining.
Juvenile leaves with ∼2-mm-long petioles from WT and clmp1-1 plants were attached to a slide with double-sided tape. Thin slivers were peeled from the petiole, placed on a slide in 50 mM PIPES, 10 mM EGTA, 5 mM MgSO4, 0.01% Nonidet P-40, 5% DMSO, and 0.1 M mannitol containing 0.1 μM Alexa Fluor 488-phalloidin (Invitrogen), incubated for 5 min in darkness, and imaged by confocal microscopy.
Analysis of Mitochondrial and Peroxisome Distribution.
The peroxisomal and mitochondrial marker proteins YFP-PTS1 and mt-YK (24, 25) were transformed into clmp1-1 or WT and imaged by confocal microscopy. Quantitative analysis of organelle distribution is detailed in SI Materials and Methods.
Analysis of Subcellular Localization.
Generation of plants expressing 35S:CLMP1–YFP is described in SI Materials and Methods. YFP was detected using confocal microscopy.
Ultrastructural Analysis.
Petioles (1–3 mm long) were prepared for TEM (45) using microwave-assisted processing and embedded in Poly/Bed 810 (Polysciences). Qualitative observations were made on serial 70-nm-thick sections through a minimum of 10 cells per petiole per plant for each of three clmp1-1 and WT plants. Images were captured with a Phillips 201 transmission electron microscope equipped with an Advantage HR camera system (Advanced Microscopy Techniques).
See SI Materials and Methods, Table S1, and Dataset S1 for details on promoter:GUS analysis, phylogenetic analysis, chloroplast import assays, and analysis of FtsZ and ARC5/DRP5B localization.
Supplementary Material
Acknowledgments
We thank Kathy Sault, Alicia Pastor, Rebecca Roston, Giovanni Stefano, Melinda Frame, and Wei Ma for technical support; Linda Savage, Jessica Reif, and Robert Last for mutant screening; Susan Forsburg for reagents; and Aaron Schmitz for comments on the manuscript. This work was supported by the U.S. National Science Foundation (Grant 0519740, to K.W.O.), funding from the International Rice Research Institute C4 Rice Program (to T.L.S.), an American Society of Plant Biologists Summer Undergraduate Research Fellowship (to Y.L.), and the U.S. Department of Energy, Office of Basic Energy Sciences, Office of Science, Chemical Sciences, Geosciences, and Biosciences Division, (Grants DE-FG02-10ER15808, to K.W.O., and DE-FG02-91ER20021, supporting J.E.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106706108/-/DCSupplemental.
References
- 1.Wada M, Suetsugu N. Plant organelle positioning. Curr Opin Plant Biol. 2004;7:626–631. doi: 10.1016/j.pbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Miyagishima SY, Kabeya Y. Chloroplast division: Squeezing the photosynthetic captive. Curr Opin Microbiol. 2010;13:738–746. doi: 10.1016/j.mib.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Osteryoung KW, Nunnari J. The division of endosymbiotic organelles. Science. 2003;302:1698–1704. doi: 10.1126/science.1082192. [DOI] [PubMed] [Google Scholar]
- 4.Kuroiwa T, et al. Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol. 2008;271:97–152. doi: 10.1016/S1937-6448(08)01203-3. [DOI] [PubMed] [Google Scholar]
- 5.Vitha S, McAndrew RS, Osteryoung KW. FtsZ ring formation at the chloroplast division site in plants. J Cell Biol. 2001;153:111–120. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA. 2003;100:4328–4333. doi: 10.1073/pnas.0530206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyagishima SY, et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell. 2003;15:655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida Y, et al. Isolated chloroplast division machinery can actively constrict after stretching. Science. 2006;313:1435–1438. doi: 10.1126/science.1129689. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida Y, et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science. 2010;329:949–953. doi: 10.1126/science.1190791. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, et al. New connections across pathways and cellular processes: Industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol. 2008;146:1482–1500. doi: 10.1104/pp.107.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Andrea LD, Regan L. TPR proteins: The versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007:re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 14.Kinsman EA, Pyke KA. Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development. 1998;125:1815–1822. doi: 10.1242/dev.125.10.1815. [DOI] [PubMed] [Google Scholar]
- 15.Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. Exchange of protein molecules through connections between higher plant plastids. Science. 1997;276:2039–2042. doi: 10.1126/science.276.5321.2039. [DOI] [PubMed] [Google Scholar]
- 16.Aeschbacher RA, Schiefelbein JW, Benfey PN. The genetic and molecular basis of root development. Annu Rev Plant Physiol. 1994;45:25–45. [Google Scholar]
- 17.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 19.Xia G, et al. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 1996;10:761–769. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]
- 20.Sheahan MB, Rose RJ, McCurdy DW. Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 2004;37:379–390. doi: 10.1046/j.1365-313x.2003.01967.x. [DOI] [PubMed] [Google Scholar]
- 21.Krzeszowiec W, Rajwa B, Dobrucki J, Gabryś H. Actin cytoskeleton in Arabidopsis thaliana under blue and red light. Biol Cell. 2007;99:251–260. doi: 10.1042/BC20060077. [DOI] [PubMed] [Google Scholar]
- 22.Kandasamy MK, Meagher RB. Actin–organelle interaction: Association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton. 1999;44:110–118. doi: 10.1002/(SICI)1097-0169(199910)44:2<110::AID-CM3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Kadota A, et al. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:13106–13111. doi: 10.1073/pnas.0906250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan JL, et al. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139:231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 27.Leech RM, Thomson WW, Platt-Aloia KA. Observations on the mechanism of chloroplast division in higher plants. New Phytol. 1981;87:1–9. [Google Scholar]
- 28.Miyagishima SY, Froehlich JE, Osteryoung KW. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell. 2006;18:2517–2530. doi: 10.1105/tpc.106.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo A, Kaikawa J, Funaguma T, Ueno O. Clumping and dispersal of chloroplasts in succulent plants. Planta. 2004;219:500–506. doi: 10.1007/s00425-004-1252-3. [DOI] [PubMed] [Google Scholar]
- 30.Chalcroft J, Matthews RE. Cytological changes induced by turnip yellow mosaic virus in Chinese cabbage leaves. Virology. 1966;28:555–562. doi: 10.1016/0042-6822(66)90240-6. [DOI] [PubMed] [Google Scholar]
- 31.Vaughn KC, Hickok LG, Warne TR, Farrow AC. Structural analysis and inheritance of a clumped-chloroplast mutant in the fern Ceratopteris. J Hered. 1990;81:146–151. [Google Scholar]
- 32.Hennis AS, Birky CW., Jr Stochastic partitioning of chloroplasts at cell division in the alga Olisthodiscus, and compensating control of chloroplast replication. J Cell Sci. 1984;70:1–15. doi: 10.1242/jcs.70.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, et al. Plant cells without detectable plastids are generated in the crumpled leaf mutant of Arabidopsis thaliana. Plant Cell Physiol. 2009;50:956–969. doi: 10.1093/pcp/pcp047. [DOI] [PubMed] [Google Scholar]
- 34.Winter D, et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modrusan Z, Wrischer M. Studies on chloroplast division in young leaf tissues of some higher plants. Protoplasma. 1990;154:1–7. [Google Scholar]
- 36.Possingham JV, Lawrence ME. Controls to plastid division. Int Rev Cytol. 1983;84:1–56. [Google Scholar]
- 37.Whippo CW, et al. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr Biol. 2011;21:59–64. doi: 10.1016/j.cub.2010.11.059. [DOI] [PubMed] [Google Scholar]
- 38.Prasad BD, Goel S, Krishna P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS ONE. 2010;5:e12761. doi: 10.1371/journal.pone.0012761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu QL, Hulen D, Liu TY, Clarke M. The cluA- mutant of Dictyostelium identifies a novel class of proteins required for dispersion of mitochondria. Proc Natl Acad Sci USA. 1997;94:7308–7313. doi: 10.1073/pnas.94.14.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox RT, Spradling AC. Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis Model Mech. 2009;2:490–499. doi: 10.1242/dmm.002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logan DC, Scott I, Tobin AK. The genetic control of plant mitochondrial morphology and dynamics. Plant J. 2003;36:500–509. doi: 10.1046/j.1365-313x.2003.01894.x. [DOI] [PubMed] [Google Scholar]
- 42.Fields SD, Arana QY, Heuser J, Clarke M. Mitochondrial membrane dynamics are altered in cluA− mutants of Dictyostelium. J Muscle Res Cell Motil. 2002;23:829–838. doi: 10.1023/a:1024492031696. [DOI] [PubMed] [Google Scholar]
- 43.Logan DC. Mitochondrial fusion, division and positioning in plants. Biochem Soc Trans. 2010;38:789–795. doi: 10.1042/BST0380789. [DOI] [PubMed] [Google Scholar]
- 44.Pyke KA, Leech RM. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1991;96:1193–1195. doi: 10.1104/pp.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Fan J, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009;21:3885–3901. doi: 10.1105/tpc.109.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.