Abstract
The tracking and projection of emerging epidemics is hindered by the disconnect between apparent epidemic dynamics, discernible from noisy and incomplete surveillance data, and the underlying, imperfectly observed, system. Behavior changes compound this, altering both true dynamics and reporting patterns, particularly for diseases with nonspecific symptoms, such as influenza. We disentangle these effects to unravel the hidden dynamics of the 2009 influenza A/H1N1pdm pandemic in London, where surveillance suggests an unusual dominant peak in the summer. We embed an age-structured model into a Bayesian synthesis of multiple evidence sources to reveal substantial changes in contact patterns and health-seeking behavior throughout the epidemic, uncovering two similar infection waves, despite large differences in the reported levels of disease. We show how this approach, which allows for real-time learning about model parameters as the epidemic progresses, is also able to provide a sequence of nested projections that are capable of accurately reflecting the epidemic evolution.
Keywords: Bayesian statistics, real-time modeling, general practice consultation data, infectious disease, seroepidemiology
An emerging epidemic engenders an increased demand upon health services. Resolving the extent to which this is due to high levels of disease transmission as opposed to a heightened public sensitivity is essential for determining the appropriate public health response.
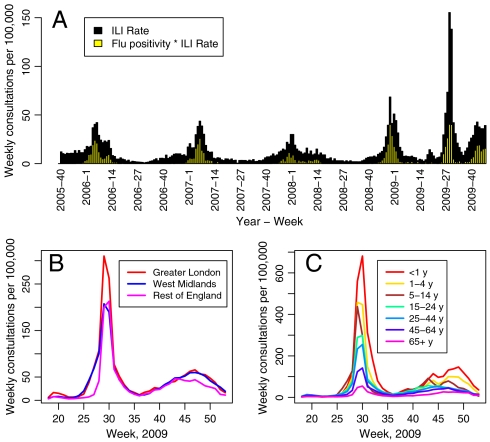
This was especially crucial when estimating the course of the 2009 influenza A/H1N1pdm outbreak in England, where, unusually, the pandemic resulted in a summer peak in rates of consultation at general practices (GPs) for influenza-like illness (ILI). This is clearly demonstrated by data from the return service of the Royal College of General Practitioners (RCGP) in Fig. 1A where weekly GP consultation rates per 100,000 population over the 2009 pandemic are compared with rates from the three previous years. Also shown is the proportion of swabbed individuals whose swabs tested positive for the presence of any flu virus (SI Data). Note that the GP consultation rate for 2009 is much higher than the usual seasonal rate, whereas the corresponding positivity is comparable to that observed in the preceding winters. This suggests that a substantial proportion of the peak in consultations was not directly attributable to A/H1N1pdm. Conversely, serological studies (3) have shown a marked increase in the prevalence of influenza antibodies among the population. Therefore, the degree to which the increased demand upon GPs is due to high levels of disease transmission as opposed to heightened public sensitivity remains unclear (4). Fig. 1 B and C show GP consultation rates by region and age group: consultations in Greater London and the West Midlands exhibit rapid early exponential growth, but the peak in London is much higher; rates appear to decrease markedly with age. Importantly, a first peak occurs immediately prior to the summer school holiday and the launch of the National Pandemic Flu Service (NPFS) phone line, ntroduced to relieve the pressure on GPs and expedite antiviral distribution (see SI Data); a second, much smaller peak, is observed in the autumn. This evidence, supported by the work of ref. 5, promotes the further hypothesis of a fluctuating propensity for individuals with symptoms of ILI to seek medical attention, perhaps induced by media coverage and changing governmental advice, as well as the social distancing effects of school holidays.
Fig. 1.
(A) Time series of GP consultation rate for ILI and virological positivity within the RCGP surveillance scheme in England and Wales (1), by week, from end-2005 to end-2009. The positivity is given by the proportion of each bar shaded yellow. (B) Observed weekly consultation rates per 100,000 within each group (see ref. 2 and SI Data, section 1.1) by region (Greater London, West Midlands, and Rest of England) and (C) by age group, over the weeks 18–53, 2009.
Traditionally, transmission modeling is used to investigate epidemic development. In the area of infectious respiratory diseases, many approaches have been proposed (6–13), including those that account for the effects of behavioral changes upon transmission (14), explicitly model the impact of school closure (15), and incorporate a temporally varying case-detection rate (16).
Here we model these aspects simultaneously while additionally accounting for the time-varying noise in the data due to consultations for non-A/H1N1pdm ILI. This is achieved by developing a model for integrating noisy GP consultation data, virological positivity data, virologically confirmed case data, and information from serological (seroprevalence) surveys (see SI Data). Each dataset is available and used at daily intervals. An age-structured transmission model is embedded within a Bayesian framework, allowing incorporation of any a priori information about model parameters from previous influenza strains via probability distributions. These prior distributions are then updated by available data to provide posterior statements about parameters of interest and their uncertainty, presented here in the form of 95% credible intervals (CrIs).
Results
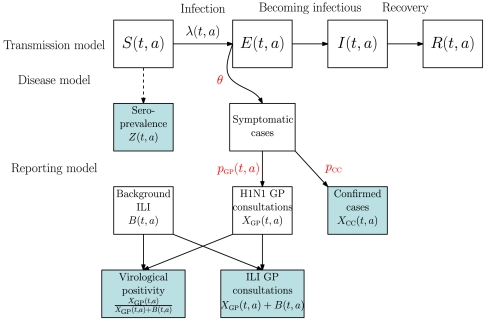
Fig. 2 is a schematic representation of the model used to describe the data-generating process. Three different components are knitted together: an age-structured transmission-governing component, a disease component, and a third component describing the mechanisms through which infected individuals report their symptoms to the health-care system. In the transmission component, susceptible individuals (S) become exposed (E) through an effective contact with infectious (I) individuals and become infective themselves after a short latent period, to be then removed (R) from the pool of infectious individuals after a further period. Transmission is governed both by a time- and age-varying force of infection λ(t,a), depending on the transmissibility of the virus and the mixing patterns in the population and by the transition rates among the S, E, I, and R states (see Material and Methods). Only a proportion, θ, of the newly exposed individuals develop febrile symptoms, from which further proportions pGP(t,a) and pCC consult their GP or have their illness virologically confirmed. Note that pGP(t,a) is calendar time and age-specific to accommodate potential fluctuations in consultation behavior. There are no direct data on transmission. However, serological surveys (see SI Data), carried out before and during the epidemic, provide data on indicators, Z(t,a) (see Material and Methods), informing the level of susceptibility within the population at the epidemic onset and over time. We make direct observation of XCC(t,a), the symptomatic cases that are virologically confirmed, though this is limited to the early stages of the epidemic (see SI Data). Only indirect information is available on the number of symptomatic cases consulting GPs, XGP(t,a), in the form of routine surveillance (see SI Data, section 1.1) counts of GP consultations for all ILI. This includes a background component, B(t,a), of non-A/H1N1pdm ILI. To identify these two components we use the total number of ILI consultations Y(t,a) = XGP(t,a) + B(t,a) and information on the virological positivity XGP(t,a)/(XGP(t,a) + B(t,a)) (see Material and Methods). By combining direct, indirect, and prior information, we produce posterior distributions for the process governing parameters (see Material and Methods) and other quantities of interest.
Fig. 2.
Model schematic diagram representing the data-generating process. The shaded boxes represent the quantities upon which we make observation.
Reconstructing the Epidemic.
Fig. 3A shows the posterior median and pointwise 95% CrI for the total number of weekly incident infections of A/H1N1pdm in Greater London, using 245 d of epidemic data covering May 1 to December 31 (i.e., from week 18 to week 53) of 2009. Additionally, Fig. 3A also shows the estimated age-specific incidences. Much like the GP consultation data, the epidemic occurs in two waves: a summer first wave (May to end-August) and an autumn second wave (September to December). The first wave rises sharply to a peak of 109,000 (81,000–146,000) new infections in the week immediately prior to the school holidays. The second wave has a smaller peak with posterior probability 0.885. Conversely, as can be seen from Table 1, which reports estimates of the infection attack rate (i.e., the cumulative incidence expressed as a proportion of the total population), there is slightly larger cumulative incidence in the second wave, a phenomenon not at all evident from the GP consultation data (Fig. 1 B and C). The discrepancy between the GP consultation data and the estimated infection pattern is clarified in Fig. 3B, which compares the cumulative consultations with the estimated cumulative infections, both calculated as proportions of their corresponding total. In this plot, a steep gradient identifies points in time during the pandemic when a relatively high density of GP consultations (or infections) occur. From the steep gradient in the GP consultations curve over weeks 27 and 28, it can be seen that the consultations were highly localized around this time. The separation between the two lines and the smaller gradient of the infection curve showed that, unlike the GP consultations, infection is shared out more evenly over the two waves.
Fig. 3.
(A) Estimated weekly infections for Greater London, spanning weeks 18 to 52 of 2009, as reconstructed from the model. The black line is the total incidence of infections over all ages and the dotted lines represent a 95% CrI. (B) The cumulative incidence of GP consultations (black line) versus the estimated (posterior median) cumulative incidence of infection (red line), both expressed as a fraction of the respective total incidence. (C) The estimated propensity for incident adult symptomatic cases to consult with a GP over time (black line). The red and blue lines present the propensity for ILI cases to call and visit (respectively) their GP from an Internet-based survey, FluSurvey (SI Materials and Methods).
Table 1.
Posterior median and 95% CrI for cumulative incidence of infections and cases and age-specific attack rates
| End-August | September–December | |
| Infections | 672,362 | 784,934 |
| (544,466, 826,014) | (612,369, 981,410) | |
| Cases | 217,003 | 257,105 |
| (133,434, 324,027) | (160,860, 385,760) | |
| Attack rates, % | ||
| (Overall) | 8.9 | 10.4 |
| (7.2, 10.9) | (8.1, 13.0) | |
| < 1 y | 15.3 | 14.9 |
| (12.4, 18.9) | (12.1, 18.2) | |
| 1–4 y | 19.5 | 20.8 |
| (15.1, 26.1) | (16.9, 25.4) | |
| 5–14 y | 22.1 | 29.8 |
| (18.0, 26.7) | (24.1, 35.3) | |
| 15–24 y | 7.9 | 8.6 |
| (5.9, 10.5) | (6.3, 11.4) | |
| 25–44 y | 8.0 | 9.0 |
| (6.2, 10.4) | (6.7, 11.7) | |
| 45–64 y | 4.1 | 4.7 |
| (3.1, 5.5) | (3.4, 6.3) | |
| 65+ y | 2.5 | 3.0 |
| (1.9, 3.3) | (2.2, 4.0) |
Returning to Table 1, children, i.e., individuals younger than 15 y old, acquired the most disease: approximately 52% of 5–14, 40% of 1–4, and 30% of under 1 y-old children are estimated to have contracted the virus, substantially higher than the overall infection attack rate of 19%. Note that Fig. 1 shows that GP consultation rates decline with age. In contrast, the estimated attack rates peak in the 5–14 age group, indicating a greater component of background consultation in the < 5 s. The precipitous decline in the infections brought about by the school holidays (both peaks of Fig. 3A occur in the same week as the start of a school holiday) highlights the key role that children play as agents of transmission, as seen in estimates of scaling factors that modify contact rates (parameters mi in Table 2). Compared to school term time, we estimate a reduction in the rate of contact within the 5–14 age group of 72%(52%–97%) in the summer holiday (1 - m3) and of 48%(22%–72%) in the half-term school holidays (1 - m5). See Materials and Methods for further details. The data are, however, unable to identify a similar effect among the 1–4 y-olds (see the wide CrI attached to parameters m2 and m4). We further estimate that child-to-child infectious contacts are 2.13(1.86–2.47) (= 1/m1, Table 2) times as likely to result in transmission than those involving at least one adult. The effect of this estimated fall in contact rates in the summer holiday and the contribution of children to transmission translates into a reduction of 35.2%(30.2%–40.2%) in the effective reproductive number: the average number of secondary infections induced by a primary infection at a given time. In a fully susceptible population this reduction would be similar: 36.4%(30.9%–41.6%).
Table 2.
Posterior median and 95% CrI for key parameters
| Parameter | Estimate | (95% CrI) |
| R0 | 1.65 | (1.56, 1.75) |
| dI | 3.45 | (3.32, 3.57) |
| m1 | 0.469 | (0.405, 0.539) |
| m2 | 0.535 | (0.0395, 0.970) |
| m3 | 0.279 | (0.0323, 0.481) |
| m4 | 0.295 | (0.0115, 0.919) |
| m5 | 0.522 | (0.276, 0.784) |
| θ | 0.328 | (0.211, 0.468) |
Key parameters are R0, the basic reproductive number; dI, the average infectious period (in days); θ, the proportion symptomatic; mi, scaling factors applied to the mixing matrices to account for relative infectivity (m1) and school holiday effects
Also from Table 2 we can see that the proportion of infections that develop into symptomatic cases, θ, which has an informative prior (see Fig. 4B), is estimated to be 0.33(0.21–0.47). This corresponds to around 35,000 incident symptomatic cases at the peak of the first wave. The posterior median for the basic reproductive number, R0, is 1.65(1.56–1.75). As with θ, R0 shows considerable prior to posterior divergence, whereas the posterior for the mean infectious period, dI, is nearly identical to the prior (see Fig. 4B).
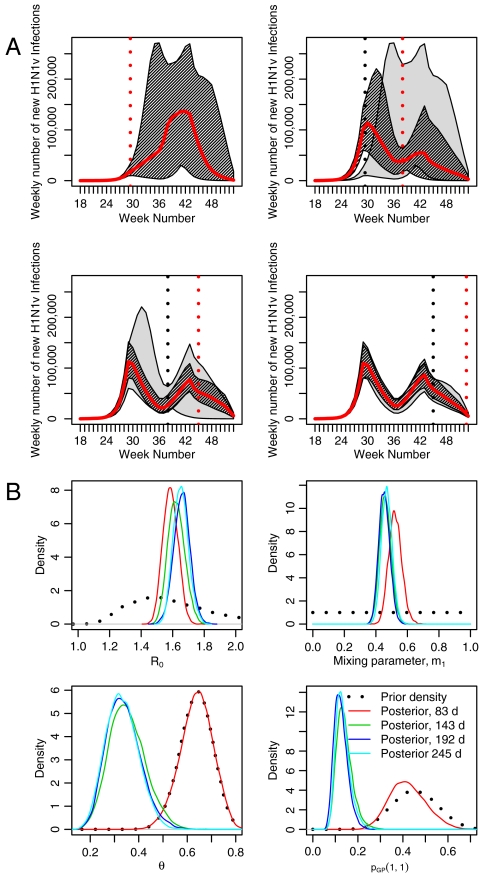
Fig. 4.
(A) Sequential epidemic reconstructions/projections based on 83, 143, 192, and 245 d of surveillance data. The gray shaded area shows the 95% CrI for the epidemic construction from the temporally previous analysis, with the darker shaded area showing the “current” analysis. The solid red line indicates the posterior median number of infections, the dotted red vertical line shows the time at which the surveillance data ends, and the dotted gray vertical line shows the time at which the previous batch of surveillance data ended. (B) Sequentially obtained posterior density estimates, plotted alongside corresponding prior distributions for the parameters R0, m1, θ, and pGP(1,1).
At NPFS launch, the propensity for adults to consult is estimated to fall from 16% to 1.8% (Fig. 3C). Only a small increase follows at a second breakpoint in early September, but by a third breakpoint, in late October, the propensity returns to a value close to 10%. Similar results are obtained for this parameter in children. These estimates are similar to values expected during seasonal influenza epidemics (17, 18), but lower than estimates from the Internet-based Flusurvey (see Fig. 3C and SI Data), possibly reflecting biases in the population captured by the survey.
Predicting the Epidemic.
The above results are related to an epidemic that is now over. A crucial question is whether the model can be used as a tool for inferences and predictions while an epidemic is ongoing. To assess this, further analyses were conducted based on 83, 143, and 192 d of epidemic surveillance data. The 83-d analysis contains no serological data except those used to inform the baseline prevalence of antibodies, whereas the 143- and 192-d analyses incorporate serological data collected during the epidemic (see ref. 3 and SI Data).
Fig. 4A illustrates how the predictions evolve as data accumulate. Fig. 4B shows how the estimated posterior densities for the parameters R0, m1, θ, and pGP(1,1), the consultation propensity in children in the first 83 d (see SI Materials and Methods), evolve over time, starting with their prior distributions. From the 143-d analysis onward, credible intervals for the future number of infections appear to enclose the estimated numbers in the subsequent analysis. However, this is not so in moving from the 83- to the 143-d analysis. This is due to the lack of serological data in the 83-d analysis. Given the large degree of dispersion, the GP consultation data are too weakly informative to overcome the informative priors placed, partly for the sake of identifiability, upon parameters such as θ and pGP(·,·) (see SI Materials and Methods). The densities of Fig. 4B show that, in the earliest analysis with no serological data, the posterior distributions for these parameters are near identical to the priors, centered on values far larger than the posteriors obtained from the subsequent analyses. The inclusion of the serological data in the 143-d analysis provides a clear indication of the level of cumulative incidence, which is higher than the 83-d results might suggest. With stronger information on the incidence, the data become sufficiently informative to overcome the prior distributions for θ and pGP, and this is reflected in a shifting of the posterior distributions to values that remain consistent across the 143-, 192-, and 245-d analyses.
Discussion
Our approach allows reconstruction and projection of the trajectory of an epidemic by disentangling epidemic and behavioral dynamics. Combining data from different sources is crucial, as each plays an important role: virological data partition consultations between A/H1N1pdm and other ILI, GP consultations determine the temporal trend, and the serological data give the scale of the epidemic.
Our estimates of attack rates are lower than has been obtained elsewhere (19), importantly providing improved understanding of how a third wave of A/H1N1pdm infection occurred in late 2010. Although our estimate of R0 is consistent with that obtained by others (13, 16), the estimate that only a third of infections are symptomatic is much lower than corresponding estimates from Mexico and Hong Kong [0.86 and 0.64, (13, 20)] but comparable to estimates from New Zealand [0.45 (21)] and France [0.20 (22)] and in broad agreement with the systematic review of ref. 23. Our estimate of 0.47( = 1/2.13) for the relative risk of transmission in infectious contacts involving at least one adult is in direct agreement with a previous estimate of 0.485 (95% CrI 0.302–0.625) (13). By combining our estimated θ and attack rates, we obtain a number of symptomatic cases, which is a fourfold increase on the central and a twofold increase on the upper bound of the official estimates for the two waves (24). Previous work (10) uses these central estimates as data, multiplying them by a factor of 10 in order to achieve a good model fit. This factor can be interpreted as a product of two components: one that accounts for the asymptomatic infections (1/θ) and one that accounts for underascertainment in the symptomatic case number estimates. Here, these two components multiply to give a factor of approximately 12.
In its transmission component, the model is similar to that used elsewhere (16), where data on laboratory-confirmed cases, modeled using a time-varying reporting rate, have been considered. Here, in addition to some lab-confirmed cases, other sources of information are used, notably noisy GP consultation data. This enables us to advance earlier efforts to model the epidemic in England. Using a hybrid of estimation approaches, ref. 10 treats estimated weekly incident cases of A/H1N1pdm (24) as data, with no propagation of the error inherent in the estimation process. We employ a more rigorous, statistical approach, which utilizes a richer array of raw data. For the same pandemic in Singapore, ref. 25 implements an algorithm for online updating of estimates arising from an S, E, I, R transmission model fitted to GP consultation data alone. This approach, which makes no stratification by age, suffers similarly from a lack of nesting in the early stages and masks the interage group transmission dynamics. In the same vein, ref. 18 develops a methodology for real-time inference, but this offers little opportunity to make any learning about many key model parameters. Our model also allows for the quantification of the impact of school holidays, the level of non-A/H1N1pdm consultation, and obtains estimates for the propensity for patients affected by the A/H1N1pdm virus to seek consultation.
Assumptions.
One key strength of any model used to predict or assess epidemic impact is a robustness to (often unavoidable) modeling assumptions. In SI Sensitivity Analyses we investigate the impact of dropping, or changing, a number of the assumptions made in producing the epidemic estimates obtained here. Our results are generally robust to reasonable deviations from modeling assumptions, yet also suggest avenues for further investigation. Specifically, we examine three modeling components: the performance of the virological testing procedure, the assumed contact patterns, and the assumed functional form of the propensity to consult, pGP(t,a).
Test Sensitivity.
Thus far, it has been assumed that the virological testing procedure has a sensitivity of 1; i.e., there are no false negatives. If we relax this assumption to reasonable values for the test sensitivity, say 0.8 and 0.9, the key results change very little. As expected, there is a small increase in the estimated symptomatic attack rates, as a lower sensitivity allows for more A/H1N1pdm cases among the GP consultations, but there is negligible impact on the total infection attack rates. Of the values investigated, a test sensitivity of 0.8 had larger values for the likelihood in its posterior distribution.
Mixing Matrices.
The mixing matrices used to describe rates of contact between the different age groups are based upon United Kingdom (UK) data from the POLYMOD (Improving Public Health Policy in Europe through Modeling and Economic Evaluation of Interventions for the Control of Infectious Diseases) study (see ref. 26). Results are robust to small changes in both parameterization of the mixing matrices (see Materials and Methods) and the rates of contact themselves. More interestingly, a preliminary attempt to estimate the entire mixing matrices (SI Sensitivity Analyses), using the POLYMOD data only as prior information, indicates that contacts among the 5–14 age group are particularly important, while also suggesting that contacts between this age group and the 15–24 age group may be more influential than previously thought. Adopting these estimates results in a slight fall in the estimated R0, while increasing the total infection attack rate. This is due to a shift in the infection profile from small children to the bigger pool of susceptibles in the 15–24 age group. Developing a more rigorous approach to estimation of contact patterns, and in particular the choices of the informative priors, constitutes a promising avenue for future work.
Propensity to Consult.
A piecewise linear parameterization for pGP(t,a) in the post-NPFS era gave results not materially different from those featured here. Attempts to adopt this piecewise linear parameterization over the entire epidemic period resulted in lack of identifiability and undue influence of prior distributions (see SI Sensitivity Analyses).
Further Applicability.
Our modeling approach has focused on the reconstruction of the epidemic in a globally prominent metropolitan region. However, the general methodology presented here is highly applicable to any influenza epidemic within any similar health-care network. Although it is unreasonable to carry out this modeling exercise on England as a whole, due to the unrepresentativeness of the pooled virological and serological data, we have repeated the analyses in three other disjoint regions, which, together with London cover the whole of England (see SI Further Applicability). London and West Midlands experience very similar epidemics, whereas the remainder of the country is split into two regions, North and South, neither of which had a substantial first wave of infection. There is mostly nonsignificant variation in the estimated parameters across the four regions, with the exception of R0, which suggests a possible link between the reproductive number of the epidemic and the population density within the affected region (London and West Midlands are England’s two most densely populated areas).
Serological Data.
As epidemic surveillance data accumulate over time, the model is capable of producing sequential epidemic estimates that converge (in the sense that successive credible intervals are nested) and could be used for real-time modeling and prediction. However, we have shown surveillance data alone are insufficient unless or until the epidemic is very far progressed. In general, to generate reliable projections early in the epidemic, the timely availability of relevant data on cumulative incidence and/or the proportion of infections reported in surveillance (a role originally envisaged for the Flusurvey during England’s 2009 A/H1N1pdm outbreak; see SI Data) is required. Serological data, in particular, are shown in this paper to be vital to ensure convergence of sequentially obtained estimates, given our choice of priors. Analysis conducted in the absence of serological data (see SI Serological Studies) shows that with surveillance data alone, a realistic epidemic reconstruction is still impossible at 192 d, i.e., after the peak of the second wave. As a result, any online inference is rendered highly infeasible. This highlights the critical importance of the timely availability of serological information in an emergent epidemic, when information on key parameters may be lacking and/or priors may be misspecified, as here. This is clearly a challenge, given current limitations on test developments, facilities, and recruitment of appropriately representative populations (3, 19), but one that is very important to meet.
Materials and Methods
Our approach integrates data from a number of sources, combining information from GP surveillance networks with epidemic specific data. The SI Data section provides an in-depth description of the available data on GP consultations for ILI, virological positivity, virological confirmed cases, and serological surveys.
An Integrated Model.
The proposed model in Fig. 2 comprises a transmission model and a disease and reporting model. The model dynamics are deterministic and discrete. We model from the period May 1 to December 31, 2009, and for the following age groups: < 1 y, 1–4, 5–14, 15–24, 25–44, 45–64, and 65+ y. The epidemic is initiated with a small number of infectious individuals and a pool of susceptible individuals. At each subsequent time point the transmission model generates a number of newly infected individuals, which enter the disease and reporting model, while the pool of susceptible individuals diminishes. The disease and reporting models then govern the proportion of these incident infections that appear in the GP consultation and confirmed case datasets and the delay inherent in doing so.
In the age-structured S, E, I, R model, transmission is dictated by a time- and age-varying force of infection λ(t,a) and transition rates σ and γ, which describe the rates of transition between states E → I and I → R, respectively. These rates are functions of the mean latent period, dL, and the mean infectious period, dI, the expected times spent in states E and I, respectively. The force of infection depends on two key quantities: the basic reproduction number of the virus, R0, and the relative rates of contact between the different age groups, introduced through the time-varying matrix, M(t). Details of how these quantities combine to give the incident number of infections can be found in SI Materials and Methods, Eq. 7. A proportion, θ, of the exposed individuals become clinical cases, with further fractions pGP(t,a) and pCC of these symptomatic individuals consulting their GP or being virologically confirmed, respectively. Typically, there will also be a time lag from infection to either of these events. This delay is assumed to be distributed as a gamma random variable and arises from three independent processes: the incubation time until symptom onset, the delay in reporting the GP consultation or having illness virologically ascertained, and the subsequent reporting delay. These component delays are assumed to have known mean and variances, which are summed to give the mean and variance of the distribution governing the overall time from infection to each event. This is discussed in more detail in SI Materials and Methods. The size of the initially susceptible population within an age group, S(0,a), is informed by baseline serological data from 2008 (3). Subsequently, for serological data taken at time t, the expected seropositivity is given by 1 - (S(t,a)/Na), where Na is the size of the population in age group a.
Modeling Challenges.
Consultation Behavior.
The propensity of individuals to consult with their GP given symptomatic ILI varied significantly over the course of the study period. Initially, this propensity was high, as seen from the marked increase in the consultation rates during the first wave, with only a modest increase in the accompanying virological positivity. However, government advice that patients were to consult through the NPFS, rather than their GP, drastically reduced this propensity. The model has to be sufficiently flexible to account for this, as well as permitting some temporal variation in the levels of adherence to the governmental guidelines over time. This impacts upon our model in two ways: (i) through the propensity to consult with a GP conditional upon symptomatic infection with A/H1N1pdm, pGP(t,a); this is modeled as a piecewise function over time, with differing rates for children and adults, the details of which can be found in SI Materials and Methods; (ii) through the “background” consultation of non-A/H1N1pdm patients with ILI symptoms. The background component of the consultation, B(t,a), is parameterized as a piecewise constant function over time, with varying rates for each age group, thus allowing for temporal fluctuation in the behavior and prevalence of individuals with non-A/H1N1pdm ILI. See SI Materials and Methods for further details of the model and the estimation of these background rates of consultation, using informative priors derived on the basis of pandemic data heralding from other regions of England.
Mixing Rates and School Holidays.
Estimated contact rates based on UK weekday data from within the POLYMOD study (26) formed the basis of the contact matrices M(t) used in our analysis. In school term times, these POLYMOD matrices were modified through the introduction of a scaling factor, m1, applied to all matrix elements representing a contact rate involving adults. This confers an interpretation upon m1 of a relative infectivity of adult infectious contacts in comparison to those solely involving children. Effects of school holidays upon disease transmission were accounted for by introducing further factors, m2 and m3, which describe the proportionate reduction in rates of contact among 1–4 and 5–14 y-olds, respectively, during the summer holiday. During the shorter half-term holidays, additional multipliers, m4 and m5, were applied to the same contact rates to permit differing effects of social distancing brought about by the two types of holiday. See SI Materials and Methods for further details.
Inference.
Parameters.
Inference is carried out within the Bayesian framework, based upon the posterior distributions of parameters and derived quantities of interest, obtained through the combination of the prior distributions and the likelihood function. We estimate posterior distributions for parameters R0, dI, mi (i = 1,…,5), and the size of the initial spark of infection. Conversely, the mean latent period dL is assumed to be known. Preliminary attempts to estimate both dL and dI highlighted that only their sum, not the individual components, is easily identified and these findings have been formalized elsewhere (27). For the disease and reporting models, we estimate θ, pCC, and the parameters describing pGP(t,a). Furthermore, we estimate the nuisance parameters used to model B(t,a).
Likelihood.
If we denote the collection of all model parameters by the vector φ, and
wta is a realization of W(t,a), the virological positivity at time t in age group a, based on a sample of size
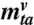 .
.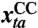 is a realization of XCC(t,a), the number of lab-confirmed cases at time t in age group a.
is a realization of XCC(t,a), the number of lab-confirmed cases at time t in age group a.yta is a realization of Y(t,a), the number of GP consultations at time t in age group a.
zta is a realization of Z(t,a), the seropositivity at time t in age group a, based on a sample of size
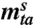 .
.
Then, treating the above as independent data, the likelihood is given by
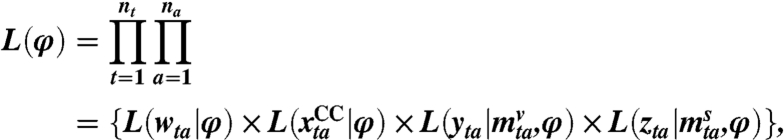 |
where nt and na are the number of time points (245) and age groups (7), respectively. The third term in the product gives the likelihood of the GP consultation data. This is modeled through a negative binomial distribution to account for the overdispersion in the count data. This overdispersion is in part due to the within-week pattern of consultation characterized by very few consultations on weekends or bank holidays and a higher rate of reported consultations on Mondays, gradually declining through the week until a small increase on Fridays. The negative binomial distribution is parameterized in terms of the mean number of consultations (as found in SI Materials and Methods, Eq. 8) and a piecewise constant dispersion parameter, with one breakpoint at the time of NPFS launch. Otherwise, the confirmed cases, xCC, are modeled as Poisson count data, and the positivity and serological data are both treated as realizations of binomial random variables with known denominators.
Priors.
A list of the model parameters comprising φ can be found in SI Materials and Methods, section 2.3.2. Where possible, parameters have been included as stochastic quantities; i.e., we have placed a prior upon them, so that we can learn about them through the data and so that the modeling procedure incorporates as much a priori knowledge/uncertainty as possible. Some parameters, due to reasons of identifiability, are held to fixed values. Fixed values and the majority of prior distributions are taken from the literature (see SI Materials and Methods for details). Such information is deemed to be unknown or unavailable for the parameters of the mixing matrices, mi, and the overdispersion parameters, and so we place priors that are reasonably uninformative upon them.
Implementation.
Posterior distributions for the unknown parameters are evaluated through Markov chain Monte Carlo methods, using a random walk Metropolis algorithm (28, 29). The algorithm was implemented using a bespoke C++ code specifically generated for this class of models. Two separate chains, each consisting of 450,000 iterations, were run in parallel, with the results presented based on a thinned subsample of the final 250,000 iterations from the two chains.
Supplementary Material
Acknowledgments.
The authors thank the Health Protection Agency Pandemic Influenza team for the timely availability of data; Professor E. Miller for providing serological data; the RCGP Research and Surveillance Centre; the University of Nottingham, Egton Medical Information Systems (EMIS), and EMIS practices contributing to the QSurveillance database. P.J.B., A.M.P., and D.D.A. were funded by the UK Medical Research Council (Grant G0600675). D.D.A. was funded also by the UK Health Protection Agency, as were B.S.C., R.J.H., A.C., X.S.Z., P.J.W., and R.G.P. G.K. was supported by the Commission of the European Community under the Sixth Framework Program Specific Targeted Research Project, SARS (Severe Acute Respiratory Syndrome) Control “Effective and Acceptable Strategies for the Control of SARS and new emerging infections in China and Europe” (Contract SP22-CT-2004-003824). B.S.C. acknowledges support by the Oak Foundation. P.J.W. thanks the Medical Research Council Centre for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103002108/-/DCSupplemental.
References
- 1.Fleming DM. Weekly Returns Service of the Royal College of General Practitioners. Commun Dis Public Health. 1999;2:96–100. [PubMed] [Google Scholar]
- 2.Harcourt SE, et al. Use of a large general practice syndromic surveillance system to monitor the progress of the influenza A(H1N1) pandemic 2009 in the UK. Epidemiol Infect. 2011 doi: 10.1017/S095026881100046X. 10.1017/S095026881100046X. [DOI] [PubMed] [Google Scholar]
- 3.Miller E, et al. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet. 2010;375:1100–1108. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 4.Fleming DM. Influenza surveillance, the swine-flu pandemic, and the importance of virology. Clin Evid. 2009 Nov 16; [Google Scholar]
- 5.Rubin GJ, Potts HW, Michie S. The impact of communications about swine flu (influenza A H1N1v) on public responses to the outbreak: Results from 36 national telephone surveys in the UK. Health Technol Assess (Rocky) 2010;14:183–266. doi: 10.3310/hta14340-03. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson NM, et al. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper BS, Pitman RJ, Edmunds WJ, Gay NJ. Delaying the international speed of pandemic influenza. PLoS Med. 2006;3:e212. doi: 10.1371/journal.pmed.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowell G, Ammon CE, Hengartner NW, Hyman JM. Transmission dynamics of the great influenza pandemic of 1918 in Geneva, Switzerland: Assessing the effects of hypothetical interventions. J Theor Biol. 2006;241:193–204. doi: 10.1016/j.jtbi.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: Impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 10.Baguelin M, Van Hoek AJ, Flasche S, White PJ, Edmunds WJ. Vaccination against pandemic influenza A/H1N1v in England: A real-time economic evaluation. Vaccine. 2010;28:2370–2384. doi: 10.1016/j.vaccine.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Bettencourt LMA, Ribeiro RM. Real time Bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS One. 2008;3:e2185. doi: 10.1371/journal.pone.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boëlle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat Med. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- 13.Fraser C, et al. Pandemic potential of a strain of influenza A(H1N1): Early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bootsma MC, Ferguson NM. The effect of public health measures on the 1918 influenza pandemic in U.S. cities. Proc Natl Acad Sci USA. 2007;104:7588–7593. doi: 10.1073/pnas.0611071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cauchemez S, Valleron A.-J, Boëlle P-Y, Flahault A, Ferguson NM. Estimating the impact of school closure on influenza transmission from Sentinel data. Nature. 2008;452:750–754. doi: 10.1038/nature06732. [DOI] [PubMed] [Google Scholar]
- 16.Wu JT, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infect Dis. 2010;16:538–541. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming DM, Zambon M, Bartelds AI. Population estimates of persons presenting to general practitioners with influenza-like illness, 1987–96: A study of the demography of influenza-like illness in sentinel practice networks in England and Wales, and in The Netherlands. Epidemiol Infect. 2000;124:245–253. doi: 10.1017/s0950268899003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall IM, Gani R, Hughes HE, Leach S. Real-time epidemic forecasting for pandemic influenza. Epidemiol Infect. 2007;135:372–385. doi: 10.1017/S0950268806007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardelid P, et al. Assessment of baseline age-specific antibody prevalence and incidence of infection to novel influenza AH1N1 2009. Health Technol Assess (Rocky) 2010;14:115–192. doi: 10.3310/hta14550-03. [DOI] [PubMed] [Google Scholar]
- 20.Cowling BJ, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010;362:2175–2184. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandanarayake D, Bissielo A, Huang S, Wood T. Wellington: New Zealand Ministry of Health; 2010. Seroprevalence of the 2009 influenza A(H1N1) pandemic in New Zealand. Technical report. [Google Scholar]
- 22.Flahault A, de Lamballerie X, Hanslik T, Salez N. Symptomatic infections less frequent with H1N1pdm than with seasonal strains. Plos Curr. 2009;RRN1140 doi: 10.1371/currents.RRN1140. 10.1371/currents.RRN1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrat F, et al. Time lines of infection and disease in human influenza: A review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 24.Evans B, et al. Has estimation of numbers of cases of pandemic influenza H1N1 in England in 2009 provided a useful measure of the occurrence of disease? Influenza Other Respi Viruses. 2011;5 doi: 10.1111/j.1750-2659.2011.00259.x. 10.1111/j.1750-2659.2011.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong JBS, et al. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PloS One. 2010;5:e10036. doi: 10.1371/journal.pone.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossong J, et al. Social contacts and mixing patterns relevant to the spread of infectious disease. PLoS Med. 2008;5:e74. doi: 10.1371/journal.pmed.0050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc R Soc Lond B Biol Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equations of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1091. [Google Scholar]
- 29.Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.