Abstract
Ribosomal (r-) RNA adopts a well-defined structure within the ribosome, but the role of r-proteins in stabilizing this structure is poorly understood. To address this issue, we use optical tweezers to unfold RNA fragments in the presence or absence of r-proteins. Here, we focus on Escherichia coli r-protein L20, whose globular C-terminal domain (L20C) recognizes an irregular stem in domain II of 23S rRNA. L20C also binds its own mRNA and represses its translation; binding occurs at two different sites—i.e., a pseudoknot and an irregular stem. We find that L20C makes rRNA and mRNA fragments encompassing its binding sites more resistant to mechanical unfolding. The regions of increased resistance correspond within two base pairs to the binding sites identified by conventional methods. While stabilizing specific RNA structures, L20C does not accelerate their formation from alternate conformations—i.e., it acts as a clamp but not as a chaperone. In the ribosome, L20C contacts only one side of its target stem but interacts with both strands, explaining its clamping effect. Other r-proteins bind rRNA similarly, suggesting that several rRNA structures are stabilized by “one-side” clamping.
Keywords: single molecule, optical trap, translation regulation, ribosome assembly
Whereas the structure of the bacterial ribosome has been solved in magnificent detail (1–3), far less is known about its mechanism of assembly (4, 5). In vitro, this assembly is highly cooperative (4), but how r-proteins affect the folding of rRNA remains obscure. A variety of ensemble techniques exist for comparing RNA structures in the presence or absence of proteins (see, e.g., refs. 6 and 7). Although these techniques can reveal net changes in rRNA structures following protein binding, they yield no information on the associated stability variations; moreover, they only reflect the average behavior of large populations of molecules. To bypass these limitations, we have used optical tweezers (8, 9) to stretch individual RNA fragments in the presence or absence of r-proteins. Force versus displacement curves reflect the nature and stability of the structural elements present within the RNA [(10); see ref. 11 for review]. This approach has been used to probe changes in RNA structure brought about by RNA helicases (12) or the ribosome itself (13), but the effect of proteins that bind specifically and statically to RNA, like r-proteins, has not been studied this way.
We focus here on the Escherichia coli r-protein L20 that binds rRNA early during in vitro assembly of the 50S subunit (4). It consists of two domains of similar size—i.e., an N-terminal alpha-helical domain that dives deeply into the 50S core and a globular C-terminal domain (L20C) that specifically binds the junction of helices (H) 40–41 (for numbering of rRNA helices and domains see ref. 1) at the exterior of the subunit (2) (Fig. S1). The rplT gene encoding L20 and the upstream rpmI gene encoding L35 are co-transcribed and translationally coupled. By binding to the mRNA at two nearby sites upstream of rpmI, L20 (and L20C) represses the translation of both genes (autogenous control) (14). There are three reasons for choosing L20, and more specifically its globular domain L20C, for this work (see L20 and L20C in SI Text for details). First, whereas many r-proteins (including the full-length L20; Fig. S1) bind different rRNA regions via multiple contacts whose contributions to the overall binding energy are undefined (2, 3), L20C binds a simple rRNA target with nanomolar affinity (14, 15). Second, L20 is dispensable for the function of the ribosome once assembled (4), and therefore the interactions of L20 (and L20C) with rRNA presumably only serves in rRNA folding. Third, the existence of well-characterized binding sites for L20C in both rRNA and mRNA allows comparing the effect of the same protein on the folding of two different RNA molecules.
Here, we analyze the effect of L20C on the unfolding of its rRNA and mRNA targets with a mechanical force. The protein dramatically affects the energetics of unfolding by stabilizing RNA structures that harbor its binding sites. Yet, it does not accelerate the formation of these structures from alternate conformations. The rationale for this “clamping” effect and its consequence for rRNA folding are discussed.
Results
RNA fragments of 0.15–0.2 kb in length carrying the known binding sites of L20C on rRNA and mRNA were designed and linked to two beads via 2.5 kb-long RNA/DNA handles (16) (Fig. 1A, Inset). After trapping the beads with double optical tweezers, we pull on the construct and record the force versus displacement curve (17).
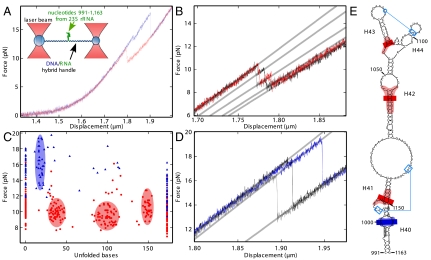
Fig. 1.
Stretching of the 991–1,163 rRNA fragment. (A) Force versus displacement (50 nm/s) curve for the rRNA fragment without (red) and with (blue) added L20C. The drops correspond to unfolding of the RNA structure. (Inset) Principle of the assay. (B) Three successive measurements (in red, black, and gray) on the same RNA molecule without L20C show reproducible intermediate states. (C) Distribution of the maximum force reached in a given state except for the fully unfolded state (173 bases) where the minimum force is given. The number of unfolded bases is deduced from the successive increase in length during the different substeps (SI Methods). This graph summarizes 38 and 87 individual measurements with (blue triangles) or without (red dots) L20C, respectively. (D) Same as B, except that L20C was present. The three successive measurements are in blue, black, and gray. (E) Positions of the intermediate states on the rRNA structure. Blue lines indicate tertiary interactions. Bars indicate the mean positions of unfolding intermediates and the colored regions represent the standard deviations around this mean. Intermediates with and without L20C are shown in blue and red, respectively. Nucleotides that contact L20 in the ribosome are in gray (2, 3).
L20C Specifically Stabilizes the Top of rRNA Helix 40.
First, we studied an RNA fragment (nt 991–1,163 of 23S rRNA) that encompasses H 40 to 44 (Fig. 1E), in the absence of L20C. When slowly stretched at constant speed (50 nm/s), the construct showed an elastic behavior, except for a force drop occurring in several substeps around 10 pN that corresponded to progressive unfolding of the RNA (Fig. 1 A and B). These substeps were reproducible when the molecule was allowed to refold and then stretched again (Fig. 1B). Each substep corresponds to an increase in length, from which the number of unfolded bases can be estimated (Fig. 1C and Fig. S2; see SI Materials and Methods). We observed three unfolding intermediates corresponding to 36.5 ± 5.3 (67 events), 100.0 ± 8.3 (66 events), and 149.6 ± 3.7 (39 events) unfolded bases. The standard deviations of these values exceeded the uncertainty of single measurements (ca three bases) and differed from state to state: Presumably, the boundaries of individual intermediates do not correspond to a particular base pair but to a small rRNA region, the size of which reflects the local curvature of the energy potential. Assuming sequential opening of the RNA, we placed these regions on the secondary/tertiary structure of the 991–1,163 subdomain as deduced from phylogeny (http://www.rna.ccbb.utexas.edu). Except for its branched top, this subdomain forms an irregular stem with two tertiary interactions (Fig. 1E). Interestingly, regions resisting unfolding (pink areas in Fig. 1E) matched stable helices of the structure. In particular, the first intermediate corresponded well to the unfolding of the stem up to the stable H 41 (36 unfolded bases). This result suggests that H 40–41, which constitutes the binding site of L20, preexists in the absence of L20, a situation that has been described for other r-protein binding sites as well (7). As for the second intermediate, it may be stabilized by the non Watson–Crick pairings that take place within the internal loop 1,044–1,050/1,109–1,111 (2, 3).
The data also yielded the energy changes associated with the unfolding process. In our experiment the system was close to equilibrium as shown by the fact that it flipped thermally between the fully folded and first intermediate states (10, 18, 19) (Fig. S3; see also The “close-to-equilibrium” hypothesis in SI Materials and Methods). Under these conditions, the free energy for unfolding equals the average difference in the elastic energy of the system before and after unfolding, which can be estimated from our data (SI Materials and Methods; Fig. S4A). The result (11.8 ± 2.5 kcal/mol) agreed well with the predicted energy (12.6 ± 1.3 kcal/mol) needed to unfold the secondary structure shown in Fig. 1E up to the bottom of H 41 (mfold software http://mfold.rna.albany.edu; see SI Materials and Methods). This suggested that in the absence of L20C the pseudoknot 1,005–1,006/1,138–1,137 contributes little to the overall stability of the structure.
L20C was then added in excess so that at equilibrium most RNA molecules were protein-bound (14, 15). Under these conditions, unfolding showed only one major substep that was different from those seen without protein. This substep occurred at forces above 13 pN and corresponded to 18.2 ± 3.2 unfolded bases (32 events) (Fig. 1 A, C, and D). The force needed to unfold the molecule beyond this point was so high (average 16 pN, or an extra 6 pN over the force needed to unfold the first 36 bases without L20C; Fig. 1C) that the remaining structure usually unfolded without further intermediates. That unfolding required higher forces in the presence of L20C than in its absence directly showed that the protein mechanically stabilizes the structure. From the number of base pairs unfolded in the intermediate state, we localized the region resisting unwinding within less than two base pairs in the H 40 intrahelical bulge—i.e., in the region contacted by L20 in the ribosome (2, 3) (Fig. 1E).
After complete unfolding, the molecule was allowed to refold by approaching the two traps at 50 nm/s. The corresponding force versus displacement curves lay well below the curve for unfolding, showing that the two processes are not the reverse of each other. Conceivably, refolding of the bottom of the stem requires the prior folding of the top, whereas during unfolding the bottom must obviously be opened first. Of note, no difference was seen whether L20C was present or not during refolding (Fig. S5).
Next, we attempted to quantify how strongly L20C stabilizes the rRNA structure. Upon going to the first intermediate (i.e., upon unfolding the first 18.2 ± 3.2 bases), the system again showed thermal flipping and thus was close to equilibrium (Fig. S6). Applying the same method as above, we found a free energy of 11.4 ± 2.4 kcal/mol for unfolding this region, which was again in good agreement with the free energy predicted for unfolding the secondary structure shown in Fig. 1E (12.1 ± 1.2 kcal/mol for the first 18 bases). This implies that L20C does not affect the stability of this region (essentially the bottom of H 40). The high force needed to unfold the RNA beyond this point made the rest of the process irreversible. Nevertheless, we could estimate the energy cost for extending unfolding from 18 to 36 bases (i.e., behind the L20C binding site) by extrapolating to higher forces the elasticity curve of the first intermediate in the absence of L20C (36 bases unwound) (SI Materials and Methods; Fig. S4B). Overall, the work necessary to unfold the first 36 bases in the presence of L20C was 23.7 ± 4.4 kcal/mol instead of 11.8 ± 2.5 kcal/mol in its absence. The difference, ca 12 kcal/mol, is the energy paid for removing L20C from its site.
Proper RNA Folding Is a Prerequisite for L20C Binding.
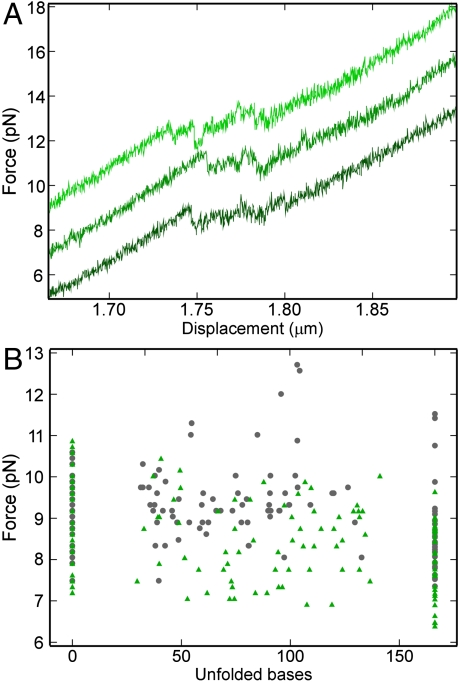
The above results suggested that H 40–41—i.e., the L20C binding site, preexists in solution in the absence of L20C. To test whether L20C can favor the formation of its binding site even from a different conformation, we used rRNA mutations. The double mutation A1009U, A1010U strongly reduces the interaction of L20C with the 991–1,163 rRNA fragment (14). Because in the ribosome structure L20 does not recognize specifically bases 1,009 and 1,010 (2, 3), this reduction presumably reflects a change in RNA conformation. Consistently, introduction of the mutations in the rRNA fragment dramatically modified the force versus displacement curve. Instead of four well-defined substeps (Fig. 1B and C), many small upward and downward substeps were apparent (Fig. 2A). Repetition of the experiment showed that, although the substeps occurred consistently within the same force range, they corresponded to the unfolding of a variable number of bases, from 15 to 85% of the total (Fig. 2B). The presence of L20C did not change this length distribution; simply, the drops occurred at slightly lower force. We concluded that the protein does not bind the mutated rRNA. The mfold predictions helped rationalize these results. The program suggested that the mutant adopts a branched structure, with an additional helix induced by pairing of the mutated nucleotides 1,009 and 1,010. The binding site for L20 was missing from this structure, hence the absence of binding. The branched structure presumably explains the continuous distribution of the substep length in the mutant, because unfolding of such structure can occur in either of two directions. Theory predicts that in this situation many intermediates close in energy should be observed (20).
Fig. 2.
Stretching of the rRNA fragment carrying the A1009U, A1010U mutations. (A) Three force versus displacement curves without L20C are shown. For clarity, the two upper curves have been shifted upward by 2 and 4 pN. (B) Same as Fig. 1C except that the mutated rRNA fragment was used. Gray dots: without L20C; green triangles: L20C added.
L20C Mechanically Stabilizes Two Sites on Its Own Messenger RNA.
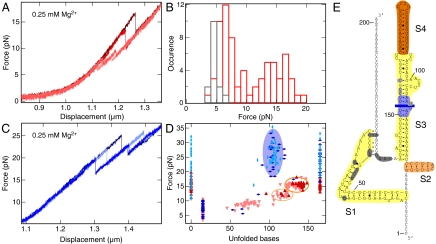
The region responsible for L20C binding (and for autogenous control) in the rpmI-rplT mRNA consists of two elements separated by a 155-nt nonessential linker (14). To probe the L20C–mRNA interactions with a mechanical force, we designed a 201-nt fragment carrying the two elements but lacking the linker. In solution, this RNA region folds into a branched structure closed by a pseudoknot (14). L20C binds to the pseudoknot and to the irregular stem S3 (Fig. 3E). In the absence of the protein, the force versus displacement curve again showed several substeps (Fig. 3A). Interestingly, the force corresponding to the first substep displayed a bimodal distribution centered at 6.6 and 14.9 pN (Fig. 3B). In the absence of Mg2+ ions, which are known to favor tertiary structures (21), only the low-force mode was observed (Fig. 3B). These results suggested that in the presence of Mg2+, the RNA exists as a mixture of two states with and without the pseudoknot, with the former state requiring a higher force to unfold. Depending upon the force needed to start unfolding (> 11.5 pN or < 11.5 pN), the initial state was assumed to bear or to lack the pseudoknot and thus was attributed either 0 or 15 unfolded bases, respectively (Fig. 3D). Subsequent unfolding events originating from these two initial situations are distinguished by different symbols on Fig. 3D. Beyond the pseudoknot, the three stems S1, S2, and S3–4 (Fig. 3E) can unfold in parallel. Consistently, the distribution of unfolded states from 50 to 80 unfolded bases showed a continuum like for the mutant rRNA fragment (Fig. 3D). Subsequently, two well-defined intermediate states were seen near the end of the unfolding process (ovals in Fig. 3D). Of note, when the pseudoknot was initially present, the continuum was never explored and the first intermediate was rarely seen (Fig. 3D), presumably due to the high force needed to start unfolding. As above, the intermediates were mapped on the structure based on the number of unfolded bases and the energy required to further unfold the molecule, assuming reversibility (Fig. S7). On this basis, the second intermediate (137.7 ± 4.4 unfolded bases; 21.8 ± 3.5 kcal/mole required for full unfolding) retains only the very stable stem S4 (137 unfolded bases; predicted unfolding energy 19 ± 1.9 kcal/mole), whereas the first intermediate presumably retains both S2 and S4 (119.7 ± 4.6 unfolded bases (predicted: 125) and 5.4 ± 2.6 kcal/mole for going to the second intermediate (predicted energy for melting S2: 4.5 ± 0.5 kcal/mole).
Fig. 3.
Stretching of the mRNA fragment. (A) Four successive stretching measurements (shown with different red tones) on the same RNA molecule without L20C. (B) Distributions of the force required to start unfolding without L20C, in the presence (red) or absence (dark gray) of Mg2+ ions (see text). (C) Same as A but with L20C (three curves are shown with different blue tones). (D) Same as Fig. 1C except that the mRNA fragment was used instead of the rRNA fragment. The fully unfolded state corresponds to 165 bases. Initial states that carried or lacked the pseudoknot were assigned 0 (fully folded) or 15 unfolded bases, respectively (E). In the absence of L20C, the unfolding intermediates corresponding to these two initial situations are distinguished with upward and downward red triangles, respectively. In the presence of L20C, the corresponding symbols are vertical and horizontal rhombuses. The graph represents 62 and 72 individual measurements with and without L20C, respectively. (E) Structure of the mRNA fragment. Stems S1–S4 are highlighted in yellow, sand, and orange. S2 and S4 are the last two structures to unfold in the absence of L20C and their unfolding correspond to the ovals with the same colors in D. A blue bar indicates the mean position of the intermediate state observed in the presence of L20C, with the light blue region representing the corresponding standard deviation. Bases from the pseudoknot and from S3 that contact L20C are shown in dark and light gray, respectively (ref. 14 and references therein).
When L20C was present, the forces needed to unfold the mRNA fragment were much higher than without the protein, and the unfolding patterns were very different (Fig. 3 C and D). Two states were especially stabilized: the fully folded one and an intermediate state centered at 105 ± 4.9 unfolded bases. These states corresponded well to the position of the two protein binding sites—i.e., the pseudoknot and the middle of stem S3 (Fig. 3E). Interestingly, while strongly hampering the unfolding of the mRNA fragment, L20C had little effect on its refolding, as with the rRNA fragment (Fig. S8).
Discussion
The interaction of the globular C-terminal domain of L20 (L20C) with its rRNA and mRNA targets has been extensively characterized in vivo and in vitro. Despite their different nature, L20C recognizes its three binding sites (an irregular stem around H 40–41 on rRNA, a pseudoknot and an irregular stem on mRNA) with similar affinities in the 10 nM range (∆G° ca 10 kcal/mol) (14, 15). The ribosome structure reveals the details of this recognition: In the H 40–41 region, L20C contacts only the RNA backbone and not individual bases (2, 3). Using this well-characterized L20C–RNA interaction as an example, we show here that the stretching of single RNA fragments can tell us where, and how strongly, these fragments interact with specific RNA-binding proteins. To our knowledge, no other single technique can yield this dual information. The measurements show that our double tweezers setup, combining high stiffness (total trap stiffness of 0.1 pN/nm) and force resolution (about 100 fN at 130 Hz), allows us to dissect the secondary structure of RNA molecules with close to single base resolution.
Our results are consistent with more conventional methods for the location of the binding sites and the associated binding energies. An important finding of this work is that, while L20C binds tightly and stabilizes RNA structures that carry its binding sites, it cannot use its binding energy for inducing the formation of these structures from alternate conformations. In short, it acts as a clamp resisting unfolding and not as a chaperone facilitating folding. These points, and their bearing for ribosome assembly, are discussed successively.
Binding Sites and Energies.
The presence of L20C makes RNA fragments encompassing the L20C binding sites more resistant to unfolding. We could localize the outer edge of the L20C binding site from the number of bases that have unfolded at the onset of this resistance. For the binding site on H 40–41 of rRNA (Fig. 1E) or on stem 3 of mRNA (Fig. 3E), this edge lies within 2–3 bases of the nucleotides in contact with L20C as deduced from footprinting and mutational analyses and from the ribosome structure (2, 3, 14). The localization of the binding site on the mRNA pseudoknot is less precise because in this case the entire tertiary structure resists unfolding (Fig. 3E). Altogether, the L20C binding sites identified here agree well with those deduced from other methods.
The energy required to remove L20C from its rRNA binding site (ca 12 kcal/mol) has also been estimated from our data; its ensemble equivalent would be the free energy of activation for L20C dissociation. Incidentally, this value compares well with the standard free energy for L20C binding to the same fragment according to fluorescence anisotropy measurements (Kd = 13 nM, or ΔG0 = -10.6 kcal/mol) (14). This result suggests that the reverse reaction, the binding of L20C to its specific site, has negligible activation energy—i.e., that it is diffusion-controlled or nearly so. This inference is reasonable because this reaction does not involve large conformational changes: The structure of L20C does not change upon RNA binding (22), and the L20 binding site on rRNA seems to exist prior to this binding (see below).
Structural Basis for L20C-Mediated Stabilization of RNA Structures.
Certain DNA-binding proteins, e.g., the restriction enzymes EcoRV, XhoI or BsoBI (23–25), protect their double-stranded targets against unzipping by an external force in a manner similar to L20C. However, these enzymes differ from L20C in the way they “clamp” their substrate. When bound to their recognition sequence, restriction enzymes largely or even completely encircle the DNA double helix (26) (Fig. S9C). In contrast, L20C binds only one side of the irregular stem H 40–41, but the contacts involve both helix strands, which explains the helix stabilization (Fig. S9 A, B, and D). Logically, this mode of clamping is less efficient than helix encirclement. At a loading rate of 2.3 pN/s, pulling L20C out of its rRNA binding site requires an extra force of ca 6 pN compared to unfolding without L20C (Fig. 1C) whereas dislodging BsoBI or XhoI with the same loading rate requires extra forces of 15 pN or more (cf. figure 4 in ref. 25).
Is single-sided clamping unique to L20C? The crystal structure of the ribosome shows that L13, which interacts with the same H 40–41 region as L20C (Fig. S1), also binds one side of this stem and contacts both strands, although at nucleotides different from L20C. Presumably, L13 acts as another clamp for this stem. Similarly, the 30S proteins S4, S7, and S8 mainly bind to one side of a helix but contact both strands (2, 3). Presumably, single-sided clamping is widespread among r-proteins.
L20C Helps Resisting Unfolding But Cannot Assist Folding.
There is evidence that the binding sites for L20C in our rRNA and mRNA fragments (Figs. 1E and 3E) preexist in the absence of L20C. With the rRNA fragment, the number of bases unfolded during the first substep (36.5 ± 5) corresponds well to the unfolding of H 40, up to the stable H 41 (36 bases) and the associated free energy change fits the calculated free energy for the secondary structure of this region. Similarly, our data support the existence of the pseudoknot and of stems S2 and S4 for the mRNA fragment. This result, together with chemical probing data (14), suggests that the structure of Fig. 3E, including the two L20C binding sites, preexists in solution. Thus, L20C appears to stabilize existing structures. This stabilization is quite strong, as judged from the energies involved (10–12 kcal/mol; see above).
Yet, our data show that this large binding energy cannot be used to drive the formation of L20C binding site if it is not initially present in the rRNA and mRNA fragments. The refolding process is not affected by L20C when the unfolded molecule is slowly relaxed, in spite of the stabilization of the final folded state. Another illustration is given by the rRNA fragment carrying the double mutation A1009U–A1010U for which the force versus displacement curve reveals a new, presumably branched, conformation. Although the energy provided by the binding of L20C to its binding site is arguably higher than the energy difference between the wild-type and mutated structures, our data (Fig. 2) and spectroscopic measurements (14) show that L20C does not bind the mutated fragment. Thus L20C, while strongly stabilizing structures that carry its binding site, cannot induce their formation from alternate conformations. In short, it cannot act as an RNA chaperone (27). Chaperones catalyze the interconversion of RNA structures by stabilizing intermediary states. L20C may be unable to play this role because of the high specificity with which it recognizes RNA. Interestingly, the chaperone activities of all Escherichia coli r-proteins have been compared using a trans splicing assay. Although this assay is unrelated to the folding of rRNA, it is noteworthy that the full-length L20 was found to lack chaperone activity in contrast with many other r-proteins (28).
Concluding Remarks
The “clamping” of the bottom of subdomain 991–1,163 of 23S rRNA by L20C, as documented here, is presumably highly significant. L20 is required for the recruitment of L13, L21, and L22 (4), all of which bind to the same subdomain or, for L22, in proximity of it (2, 3) (see Fig. S1). Rather than a direct interaction of L20 with all these proteins, this requirement presumably reflects the need for a correctly and stably folded subdomain that is mediated by L20C. As discussed in Subdomain 991–1,163 and Ribosome Assembly in SI Text, this folding seems to occur early during the 50S assembly and it may be essential to this process.
Materials and Methods
The double optical tweezers setup, including a frequency shifting procedure for increased precision, has been described (17). The extremities of the molecular constructions are linked to two different beads—i.e., an antidigoxygenin coated silica bead (diameter 0.97 μm) and a streptavidin coated polystyrene bead (diameter 1.87 μm), prepared from carboxylated beads (Bangs Laboratories Inc.). The RNA/DNA hybrid constructions were prepared as in ref. 16 and attached to the beads by mixing all components together for 1–2 h at room temperature. L20C, corresponding to amino acids 59–118 of L20, was prepared as in ref. 29. To ensure saturation of RNA with protein, L20C (3 μM) was present at more than 200-fold excess over Kd. Experiments on the rRNA fragment were performed at 29 °C in 20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2. For the mRNA fragment, we used 50 mM HEPES-K pH 7.5 with or without 0.25 mM MgCl2. Data were acquired at a sampling rate of 300 Hz with an antialiasing filter at 132 Hz.
Supplementary Material
Acknowledgments.
We thank Julien Riposo and Romain Dubreuil for help with experiments, Dr. F. Allemand for gift of L20C, Drs. K. Nierhaus and Y. Timsitt for fruitful discussions, and Dr. N.K. Tanner for looking over the manuscript. This work was funded by the Centre National de la Recherche Scientifique and Agence Nationale pour la Recherche [grants 07-blan-0351-03 (STIR) to M.S., NT05-1_44659 (CARMa) to M.D. and U.B., and 2010 BLAN 1503 01 and 1503 02 (HelicaRN) to M.D. and U.B.].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107121108/-/DCSupplemental.
References
- 1.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 2.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 3.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 4.Nierhaus K. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 5.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 6.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: Folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 7.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Bockelmann U. Single-molecule manipulation of nucleic acids. Curr Opin Struct Biol. 2004;14:368–373. doi: 10.1016/j.sbi.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang X. Single-molecule RNA science. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 12.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillier M, et al. Double molecular mimicry in Escherichia coli: Binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol Microbiol. 2005;56:1441–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- 15.Allemand F, Haentjens J, Chiaruttini C, Royer C, Springer M. Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007;35:3016–3031. doi: 10.1093/nar/gkm197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangeol P, Cote D, Bizebard T, Legrand O, Bockelmann U. Probing DNA and RNA single molecules with a double optical tweezer. Eur Phys J E Soft Matter. 2006;19:311–317. doi: 10.1140/epje/i2005-10060-4. [DOI] [PubMed] [Google Scholar]
- 17.Mangeol P, Bockelmann U. Interference and crosstalk in double optical tweezers using a single laser source. Rev Sci Instrum. 2008;79:083103. doi: 10.1063/1.2957652. [DOI] [PubMed] [Google Scholar]
- 18.Bockelmann U, Thomen P, Essevaz-Roulet B, Viasnoff V, Heslot F. Unzipping DNA with optical tweezers: High sequence sensitivity and force flips. Biophys J. 2002;82:1537–1553. doi: 10.1016/S0006-3495(02)75506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerland U, Bundschuh R, Hwa T. Force-induced denaturation of RNA. Biophys J. 2001;81:1324–1332. doi: 10.1016/S0006-3495(01)75789-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 22.Raibaud S, et al. NMR structure of bacterial ribosomal protein l20: Implications for ribosome assembly and translational control. J Mol Biol. 2002;323:143–151. doi: 10.1016/s0022-2836(02)00921-x. [DOI] [PubMed] [Google Scholar]
- 23.Bockelmann U, Thomen P, Heslot F. Dynamics of the DNA duplex formation studied by single molecule force measurements. Biophys J. 2004;87:3388–3396. doi: 10.1529/biophysj.104.039776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys J. 2002;83:1098–1105. doi: 10.1016/S0006-3495(02)75233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch SJ, Wang MD. Dynamic force spectroscopy of protein-DNA interactions by unzipping DNA. Phys Rev Lett. 2003;91:028103. doi: 10.1103/PhysRevLett.91.028103. [DOI] [PubMed] [Google Scholar]
- 26.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 28.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haentjens-Sitri J, Allemand F, Springer M, Chiaruttini C. A competition mechanism regulates the translation of the Escherichia coli operon encoding ribosomal proteins L35 and L20. J Mol Biol. 2008;375:612–625. doi: 10.1016/j.jmb.2007.10.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.