Abstract
Survival of yeast during starvation has been shown to depend on the nature of the missing nutrient(s). In general, starvation for “natural” nutrients such as sources of carbon, phosphate, nitrogen, or sulfate results in low death rates, whereas starvation for amino acids or other metabolites in auxotrophic mutants results in rapid loss of viability. Here we characterized phenotype, gene expression, and metabolite abundance during starvation for methionine. Some methionine auxotrophs (those with blocks in the biosynthetic pathway) respond to methionine starvation like yeast starving for natural nutrients such as phosphate or sulfate: they undergo a uniform cell cycle arrest, conserve glucose, and survive. In contrast, methionine auxotrophs with defects in the transcription factors Met31p and Met32p respond poorly, like other auxotrophs. We combined physiological and gene expression data from a variety of nutrient starvations (in both respiratory competent and incompetent cells) to show that successful starvation response is correlated with expression of genes encoding oxidative stress response and nonrespiratory mitochondrial functions, but not respiration per se.
Keywords: mitochondrion, longevity, Warburg effect
Understanding global coordination of subcellular processes during adaptation to environmental change is a central challenge in systems biology. The ability of free-living organisms to adapt to changes in their nutritional environment is clearly one of the driving forces of their evolution. In natural environments, yeast are exposed to extreme variations in “natural nutrient” availability, particularly in their sources of carbon (and energy), phosphorus, sulfur, and nitrogen. Unlike wild type strains, auxotrophic mutant yeast strains unable to make an essential metabolite (e.g., leucine or uracil) can also be starved for the missing metabolite, but adaptation to this kind of “nonnatural” starvation has not been subject to evolutionary selection. Starvation of Saccharomyces cerevisiae for a single, growth-limiting nutrient offers the opportunity to study the coordination of nutrient sensing, metabolism, growth, and cell division. Proper coordination results in prolonged survival, concerted cell cycle arrest, and glucose conservation during starvation, and depends strongly on the specific nutrient being depleted. For instance, the survival of auxotrophic yeast starved for leucine, histidine, or uracil is substantially impaired (exhibiting a roughly 10-fold difference in half-life) relative to the same strain starved for the “natural” nutrients sulfate or phosphate (1). Starvation for sulfate or phosphate elicits rapid, nearly uniform G0/G1 cell cycle arrest and slows glucose consumption, whereas starvation for leucine, histidine, or uracil results in incomplete cell cycle arrest and markedly higher rates of glucose consumption.
We are interested in understanding what, if any, general principles determine starvation phenotype. Early work on nutrient starvation posited the existence of a starvation “signal” that promotes concerted cell cycle arrest and survival in response to nutritional scarcity (2). However, fundamental questions about the identity and effects of the starvation signal have never been addressed. The answers to these questions have implications for other areas of biology, because connections among the same processes that are influenced by nutrient starvation have been documented in cancer, aging, and the yeast metabolic cycle.
As a first step toward understanding general determinants of starvation phenotype, we sought to identify transcriptional correlates of “successful” or “survivable” starvation—that is, starvation that leads to concerted cell cycle arrest, glucose conservation, and, most importantly, survival. To this end, we compared the physiology and gene expression of S. cerevisiae in response to starvation for a variety of previously characterized nutrients, and for one additional nutrient, methionine. We chose methionine because previous studies hint that it coordinates diverse cellular functions, thereby acting as a regulatory hub. For example, methionine starvation of a methionine auxotroph causes rapid and uniform G0/G1 cell cycle arrest (2), unlike the result obtained with other auxotrophies (3). In addition, genome-wide analyses of periodic gene expression during the cell cycle showed that methionine is the only amino acid whose biosynthetic genes exhibit periodic expression (4). In the yeast metabolic cycle, the methionine-regulated transcription factors exhibit the most highly periodic transcriptional activity, implying a connection between the methionine regulon and the respiro-fermentative balance (5–7). This implied connection is strengthened by a study of glucose repletion in yeast, which shows that methionine biosynthetic genes are highly induced upon relief from glucose starvation (8). The methionine-regulated transcription factors are also well known to regulate the response to various toxins, including heavy metals and oxidizing agents (9, 10).
We began our comparison of multiple nutrient starvations by characterizing physiology, gene expression, and metabolite abundance during methionine starvation. We first created a panel of isogenic methionine auxotrophs. Two of these deletion mutants (met6Δ and met13Δ) lack genes encoding steps in methionine biosynthesis, creating a “metabolic” defect. The others lack genes encoding one or more methionine transcription factors, thereby creating “regulatory” defects that result in a methionine requirement. Phenotypic characterization of these strains confirmed that methionine starvation is indeed different from the previously studied auxotrophic starvations and results in a phenotype intermediate between that of the natural nutrient starvations and the other auxotrophic starvations. Methionine starvation of met6Δ or met13Δ mutants, like sulfate or phosphate starvation of a wild-type strain, results in uniform G0/G1 arrest, substantially increased survival relative to leucine or uracil starvation (assessed in mutants with analogous metabolic defects), and in conservation of residual glucose.
By comparing mRNA levels during methionine starvation in met6Δ and met13Δ with the previously published results of similar experiments during starvation for leucine, uracil, phosphate, and sulfate, we found a group of genes whose expression is strongly correlated with successful response to starvation. These nuclearly encoded genes are enriched for assorted mitochondrial functions, response to heat and oxidative stress, and regulation of the respiro-fermentative balance. We then narrowed down the role of these functions in survival: during starvation for various nutrients, we measured survival of ρ0 mutants and hydrogen peroxide-treated (ρ+) strains, which showed that oxidative phosphorylation is not required for survival but that survival half-life is correlated with ability to detoxify reactive oxygen species (ROS). This result is fortified by our observation that the short-lived regulatory methionine auxotroph, met31Δmet32Δ, differs from the long-lived metabolic methionine auxotrophs primarily in its reduced ability to combat oxidative stress during methionine starvation.
Our results speak against the intuitive and commonly held view that oxidative metabolism is deleterious per se. Instead, the data suggest that induction of the transcriptional programs associated with, but not dependent on, respiration exerts a protective effect on the cell during nutritional shortages, possibly because the many protective mechanisms against oxidative damage and related stresses become increasingly engaged as the cell shifts from fermentative to oxidative metabolism.
Results
Starvation for Methionine Resembles Starvation for Natural Nutrients.
The response of S. cerevisiae to starvation for phosphate, sulfate, ammonium, leucine, and uracil has been previously described (1, 3, 11). To determine where methionine starvation lies on the spectrum of starvation phenotypes, we measured survival, glucose consumption, and cell cycle arrest during methionine starvation of the metabolic methionine auxotrophs met6Δ and met13Δ. To calibrate our measurements against those in the literature, we also measured these parameters during starvation of a previously studied leucine auxotroph, leu2Δ. We compared all measurements with those made during phosphate starvation.
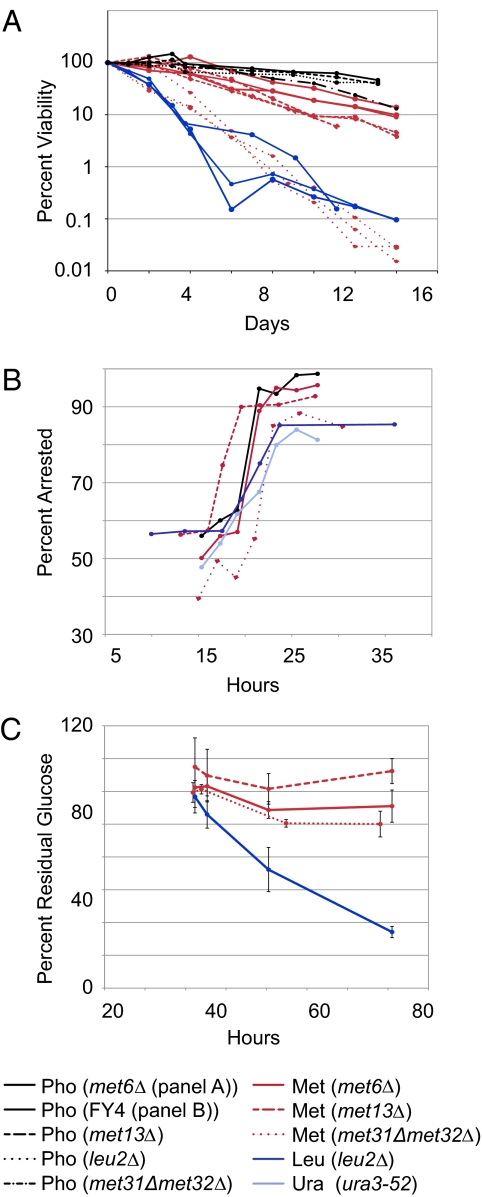
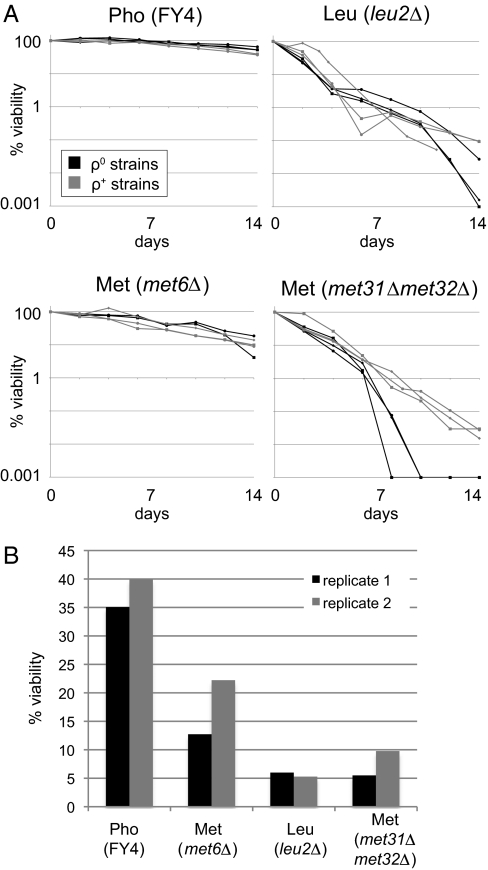
To measure the starvation survival of met6Δ, met13Δ, and leu2Δ, a single colony of each strain was grown in medium limited for a single nutrient, either phosphate or the required amino acid. Each strain was then diluted into medium lacking only that nutrient, and viability over time was measured (Materials and Methods). These methionine auxotrophs, when starved for methionine, survived longer than leucine or uracil auxotrophs starved for their respective requirements but not as long as the same strains starved for phosphate or sulfate (Fig. 1A and Table S1) (1). Fig. 1A also contains data for the regulatory methionine auxotroph met31Δmet32Δ, in which the methionine requirement is caused by the deletion of two genes, MET31 and MET32, which encode transcription factors; we address its properties below. We classified the starvations as long-lived [phosphate, sulfate, and methionine (metabolic; met6Δ or met13Δ)] or short-lived [(leucine, uracil, and methionine (regulatory; met31Δmet32Δ)] according to the data in Fig. 1A.
Fig. 1.
Physiological responses to nutrient starvation. (A) Survival over time during starvation for methionine (Met), phosphate (Pho), or Leucine (Leu) using the indicated strains. Survival was measured as the ability to form a colony on rich medium plates. (B) Cell cycle arrest, as measured by percentage of cells with no bud (assessed by light microscopy), during starvation for Met, Pho, Leu, or Uracil (Ura) using the indicated strains. Representative time courses are shown, with more comprehensive data provided in Table S1. (C) Glucose consumption (measured as described in Materials and Methods) after biomass (measured by optical density) has reached its plateau during starvation for Met or Leu. Error bars show SD of technical replicates.
Survival during starvation correlates strongly with uniformity of cell cycle arrest in stationary phase cultures (1, 3, 11). We measured bud index for three cultures each of met6Δ and met13Δ undergoing methionine starvation, and for one culture each of strains undergoing leucine (leu2Δ), uracil (ura3-52), and phosphate starvation (FY4) (Fig. 1B and Table S1). Combining our results with those in ref. 3, we found that methionine starvation is an outlier with respect to the other studied auxotrophic starvations: methionine starvation induces cell cycle arrest in G0/G1 to nearly the same extent as phosphate starvation (P = 0.07 by two-sided Student t test) but to a significantly greater extent than leucine and uracil starvation (P = 9.9 × 10−5). This, as expected, recapitulates the cell cycle arrest during methionine starvation that was initially observed decades ago (2).
In previously studied starvations, survival time and arrest phenotype correlated with the rate of glucose consumption after biomass accumulation stopped increasing (1, 12). We measured the concentration of residual, extracellular glucose in batch cultures of met6Δ, met13Δ, leu2Δ, and the exceptional met31Δmet32Δ undergoing starvation for the required amino acid. Like prototrophs starving for a natural nutrient, metabolic methionine auxotrophs starving for methionine stopped consuming glucose once biomass accumulation reached a plateau. In contrast, the leucine auxotroph continued consuming glucose, as reported previously (Fig. 1C) (1).
A diverse set of survival assays has been used to show that repression of glucose-activated signaling pathways, such as PKA and Tor, prolongs survival (1, 13, 14). To test the influence of glucose-activated signaling on survival during methionine starvation, we repeated the met6Δ and met13Δ survival measurements using a combination of glycerol and ethanol as the carbon source, which increased survival during methionine starvation almost twofold (Table S1).
Methionine Starvation Activates a Characteristic Gene Expression Program.
We followed gene expression during methionine starvation of the metabolic methionine auxotrophs met6Δ and met13Δ using a filter-based cell growth protocol that permits instantaneous removal of external methionine (Materials and Methods). This facilitates comparison of multiple strains because it allows the time courses to be aligned to the instant of methionine removal. As described in SI Materials and Methods, linear regression was used to select genes exhibiting (i) a statistically significant dependence on time and (ii) at least a twofold change in one or both time courses. In this regression analysis, the P value of the F statistic for the regression model represents the significance of the time dependence, and Q-VALUE software (15) was used to identify P values corresponding to a false discovery rate (FDR) of at most 0.01 (SI Materials and Methods). The resulting gene set shows that a wide variety of biological processes are perturbed during methionine starvation. We focused on identifying processes that are perturbed specifically during methionine starvation and might therefore provide insight into the unique role of methionine in the cell cycle, the metabolic cycle, and carbohydrate metabolism. To isolate such processes, we compared our methionine starvation gene expression data with previously published gene expression data collected during phosphate, sulfate, leucine, and uracil starvations, the “MegaCluster” in ref. 3.
We processed, combined, and filtered these data sets as described in SI Materials and Methods. To control for potential differences between our instantaneous starvation protocol and the gradual starvation protocols used for leucine, phosphate, sulfate, and uracil, we also collected expression data for met13Δ according to the protocol used for these other nutrients. A comparison of met13Δ expression data using the two alternate protocols showed essentially identical patterns of gene expression (Fig. S1 and Dataset S1). To identify genes whose differential expression is specific to methionine starvation, we clustered the combined expression data set into 40 distinct clusters using k-means clustering (SI Materials and Methods, Fig. S2A, and Dataset S1). We then focused on gene clusters displaying expression profiles specific to methionine starvation or to both methionine and sulfate starvation.
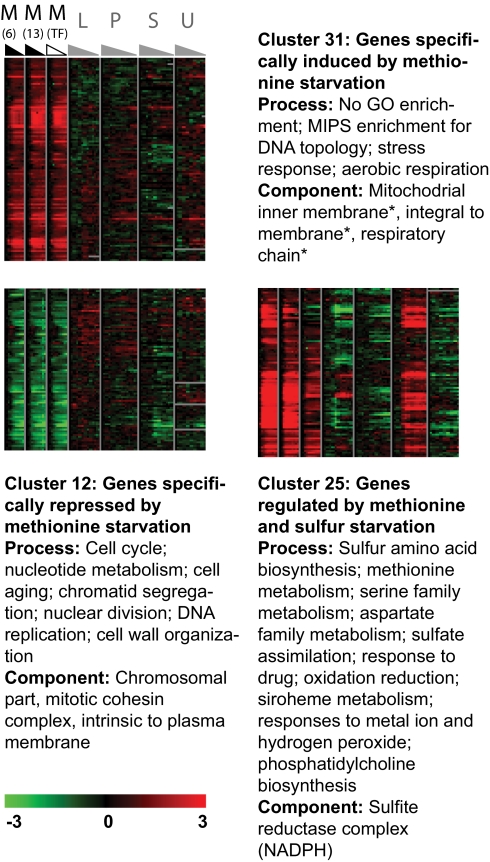
The k-means clustering yielded a cluster of genes (cluster 25) that are induced only in methionine or sulfate starvation (Fig. 2, Table S2, and Dataset S1). As expected, a significant number of these genes function in sulfur metabolism and methionine biosynthesis, including the entire methionine biosynthetic pathway. Remarkably, this cluster also contains a number of the NAD and NADP-binding enzymes on which methionine biosynthesis indirectly depends. Several other biological processes that depend heavily on methionine abundance are also enriched here, including the overlapping categories “oxidation reduction,” “oxidative stress response,” and “response to drug” (e.g., YAP1, GSH1, OPT1, GRX8, MXR1, and ZWF1) and “phosphatidylcholine biosynthesis” (OPI3, CKI1, SER33).
Fig. 2.
Expression profiles specific to methionine and sulfate starvation. K-means clusters characterized by gene expression specific to methionine starvation (cluster 31 and cluster 12) or methionine and sulfate starvation (cluster 25). Each heatmap shows microarray expression data for seven starvation time courses, separated by gray columns. The starvations are, from left to right, methionine using met6Δ [M(6)], methionine using met13Δ [M(13)], methionine using the Transcription factor mutant met31Δmet32Δ [M(T)], leucine using leu2Δ (L), phosphate using FY4 (P), sulfate using FY4 (S), and uracil using ura3-52 (U). The grayscale triangles above each time course show how the time courses were grouped for analysis and interpretation. Here, for example, methionine starvation in metabolic methionine auxotrophs (black triangles) was compared with phosphate, sulfur, leucine, and uracil starvation (gray triangles). met31Δmet32Δ expression data (white triangle) is displayed but was not used in the analysis. The functional enrichment (FDR ≤ 0.1, or, where indicated with an asterisk, between 0.1 and 0.2) of each gene set is summarized next to the corresponding heat map. Detailed enrichment data are provided in Table S2.
The substantial overlap between the transcriptional responses to sulfate and methionine starvation suggests that methionine might serve as an “indicator metabolite” for sulfate starvation. Despite this overlap, some genes are purely methionine starvation-specific. Of these, the repressed subset (cluster 12) is enriched for cell cycle regulation (e.g., TOR2, NDD1, PCL1, YOX1, RAD9, RAD53) and related processes, such as chromatid segregation, DNA replication, and nucleotide metabolism. It is also enriched for genes implicated in response to aging (SCH9, HDA2, LAC1, SGS1, and ACS2) and cell wall organization through the SHO1 osmosensing pathway.
Genes that are repressed in all starvations but methionine (cluster 27) are enriched for assorted mitochondrial functions, including the folate cycle (GCV1, GCV2, GCV3) and translation (e.g., AIM10, MST1, MSK1, MSD1, QCR6, MRPS17, PPA2, MRPL22, ALG6). This reflects the fact that the methionine biosynthetic pathway interacts directly with the folate pathway at its penultimate step, in which the methylenetetrahydrofolate reductase encoded by MET13 catalyzes the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Only a few terms are significantly overrepresented among the genes that are most strongly and specifically induced during methionine depletion (cluster 31) [e.g., the Munich Information Center for Protein Sequences (MIPS) categories related to DNA topology and aerobic respiration]. At a more lenient FDR, genes involved in ATP biosynthesis (QCR2, QCR8, and COX6) and metabolism of membrane phospholipids (e.g., IPT1, PBN1, PSD1, TGL2, and TLG2) are overrepresented.
Transcriptional Regulation of Metabolic Pathways and Isozymes During Methionine Starvation.
To understand metabolic changes during methionine starvation from a transcriptional perspective, we mapped the most differentially regulated enzyme-encoding genes (≥32-fold change in met6Δ and/or met13Δ) onto the metabolic network (Fig. S3). Notable are the high induction of antioxidant biosynthetic genes (CTT1, GSH1, GTT1, GTT2, and GPX1); differential regulation of most pentose phosphate pathway genes; induction of the NAD metabolic genes BNA3 and PNC1; and repression of nucleotide biosynthesis and the folate cycle.
The set of differentially expressed genes contains several isozyme pairs that are particularly informative because they exhibit different specificities for anaerobic and aerobic conditions. In every pair, the isozyme specific to respiration is induced, whereas that specific to fermentation is repressed (cf. GLK1 and HXK1 vs. HXK2, PDC5 vs. PDC6, ALD4 vs. ALD6, GDH1 vs. GDH3, GND1 vs. GND2, and TKL1 vs. TKL2). This suggests that methionine starvation may cause the cell to rely increasingly on respiration for energy generation.
Direct Assessment of Metabolism During Methionine Starvation.
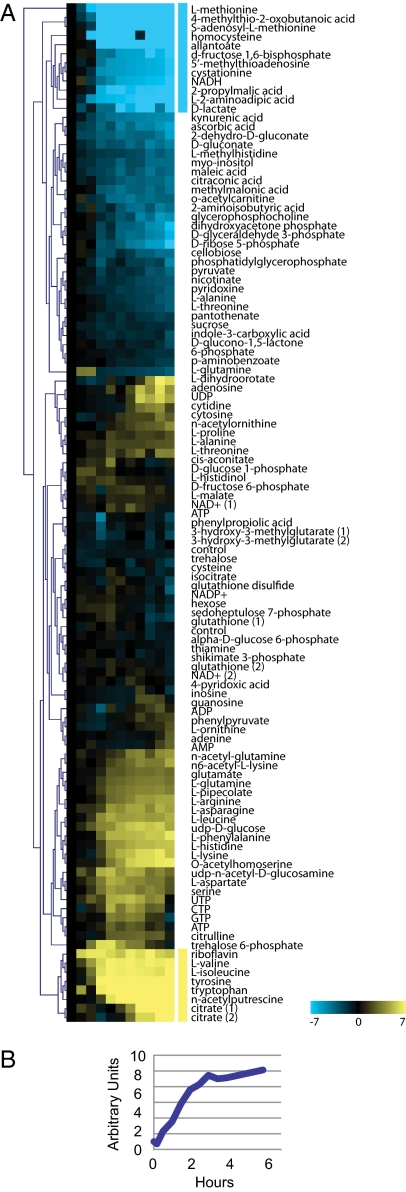
We measured intracellular metabolites for one methionine auxotroph, met13Δ, during starvation (Materials and Methods). Normalization and processing of these data are described in SI Materials and Methods. Hierarchical clustering was used to identify clusters of metabolites that exhibit coordinated changes in abundance (Fig. 3A and Dataset S1). As expected, the metabolites whose abundance decreases earliest and most dramatically (Fig. 3A, turquoise bar) are predominantly associated with methionine biosynthesis or salvage (e.g., methionine, 4-methylthio-2-oxobutanoic acid, S-adenosyl methionine, homocysteine, 5′-methylthioadensoine, and cysteine), reflecting the cell's need to replenish methionine. However, the set of rapidly depleted metabolites also includes fructose-1,6-bisphosphate and NADH, reflecting the broader metabolic reconfiguration that occurs during methionine starvation. Fructose-1,6-bisphosphate (F1,6BP) is produced from fructose-6-phosphate (F6P) by phosphofructokinase (PFK), the main regulator of glycolytic flux (16). Two observations suggest that PFK activity is reduced. First, F6P abundance increases concomitantly with the decrease in F1,6BP, implying inhibition of PFK. Second, citrate, a major inhibitor of PFK, is the metabolite whose abundance increases most. The decrease in NADH abundance causes an increase in the NAD:NADH ratio (Fig. 3B), which is also consistent with an increase in respiration. The NAD:NADH ratio is a potentially important clue to the mechanism by which methionine starvation prolongs survival. This ratio is widely thought to reflect the overall cellular energy balance, and an increase in NAD:NADH due to falling NADH has been explicitly implicated in longevity extension by dietary restriction (17). Taken together, the metabolite measurements support the inference made from gene expression patterns that methionine starvation tips the metabolic balance toward respiration.
Fig. 3.
Metabolite abundance during methionine starvation of met13Δ. (A) Metabolite abundance data in arbitrary units of fold-change relative to the zero time point. Colored bars indicate the metabolites exhibiting the greatest changes in abundance (range is ±128-fold). (B) NAD:NADH ratio during the time course.
There are two broad classes of metabolites whose abundance changes in a consistent and correlated fashion: amino acid levels increase throughout the time course, and the nucleotide triphosphates ATP, GTP, UTP, and CTP increase transiently. The increase in amino acid abundance may result from a combination of factors, including decreased amino acid consumption and degradation of ribosomes and other proteins.
Genes at the Intersection of Methionine Metabolism, Oxidative Stress Response, and Mitochondrial Function Are Required for Maximal Survival Under Multiple Starvations.
Methionine-specific gene expression depends largely on the activity of several transcription factors, Met4p, Met31p, Met32p, Met28p, and Cbf1p (18). We deleted these transcription factors individually and in pairwise combinations and found that the double mutant met31Δmet32Δ dies much faster than the met6Δ and met13Δ mutants during methionine starvation (Fig. 1A and Table S1). (met4Δ also dies much more quickly but was not further analyzed in this work.) The portion of methionine-specific gene expression regulated by Met31p and Met32p is therefore required for survival during methionine starvation. met31Δmet32Δ arrests in G0/G1 with slightly lower efficiency than the other (metabolic) methionine auxotrophs (Fig. 1B), but it does conserve glucose (Fig. 1C).
To test whether the MET31/MET32-dependent expression program is relevant to survival more generally, we measured the survival of the met31Δmet32Δ double mutant during phosphate starvation, which differs more from methionine starvation than the other starvations studied. Deleting MET31 and MET32 significantly decreases survival during phosphate starvation (Fig. 1A, P = 1 × 10−4), but the decrease is significantly smaller than the decrease observed under methionine starvation (P = 3 × 10−7) (SI Materials and Methods). The Met31p/Met32p regulon is therefore required for full survival during methionine and phosphate starvation.
To better understand how Met31p and Met32p contribute to survival, we measured gene expression during methionine starvation of met31Δmet32Δ. These data were combined with that from met6Δ and met13Δ, and multiple regression was used to identify genes whose expression is significantly different in met31Δmet32Δ compared with met6Δ and met13Δ (SI Materials and Methods). We then selected genes that rank higher than the 95th or 90th percentile with respect to the F statistic P value (P ≤ 9.19 × 10−6 or 2.07 × 10−4; FDR ≤ 0.01), depend statistically significantly on time (FDR ≤ 0.01), and change twofold or more in at least one time course. The resulting genes are strongly enriched for (i) ribosomal protein-encoding genes and (ii) genes that function at the intersection of methionine biosynthesis, regulation of cell division, and mitochondrial function, such as purine biosynthesis, folate metabolism, serine and glycine metabolism, iron metabolism, inositol metabolism, and aspartate metabolism (Table S3 and Dataset S1).
This gene set contains two particularly informative subsets (Fig. 4). The first, which depends on Met31p and Met32p for induction, is enriched for sulfur and methionine metabolism, oxidation-reduction, siroheme biosynthesis, glutathione biosynthesis, and response to oxidative stress (including key oxidative stress response genes such as GSH1, ZWF1, GTO1, OPT1, and MXR1). The second subset, which is repressed by met31Δmet32Δ, is strongly enriched for genes associated with purine, nucleotide, or glycine biosynthesis, or with iron homeostasis. The most striking examples are the purine biosynthetic genes SHM2, FCY2, ADE1, ADE2, ADE5,7, ADE13, and ADE17; the folate metabolic genes MTD1, GCV2, and GCV3; and several genes—FET3, FTR1, SIT1, and TIS11—that are induced when mitochondrial iron-sulfur cluster biogenesis is disrupted (19).
Fig. 4.
MET31/MET32-dependent genes and oxidative stress response. Genes whose expression depends most strongly on MET31 and MET32 as determined by multiple regression (Materials and Methods). Functional enrichment is indicated as in Fig. 2 and further described in Table S3.
Gene Expression Patterns Correlated with Starvation Phenotype Across Multiple Nutrient Starvations.
As a first step toward identifying and characterizing the starvation signal proposed in previous work (2), we compared gene expression data for methionine, phosphate, sulfate, leucine, and uracil starvations to find expression signatures that correlate with various starvation phenotypes. We analyzed the combined expression data for these nutrients with the hope of identifying groups of genes whose expression patterns are correlated with survival, arrest, and glucose conservation. To this end, we first filtered out expression patterns that are common to all starvations and expression patterns specific to a single starvation. We then asked three related questions: (i) Which genes are expressed differently in the “survivable” (i.e., glucose-conserving, cell cycle arresting, long-lived) starvations compared with the “unsurvivable” (i.e., glucose-wasting, shortest-lived) starvations? (ii) Which genes are expressed differently in all of the auxotrophs (including methionine) compared with the longest-lived, natural nutrient starvations? (iii) Which expression patterns best correlate with survival half-life?
Genes That Are Expressed Differently Between Survivable and Unsurvivable Starvations.
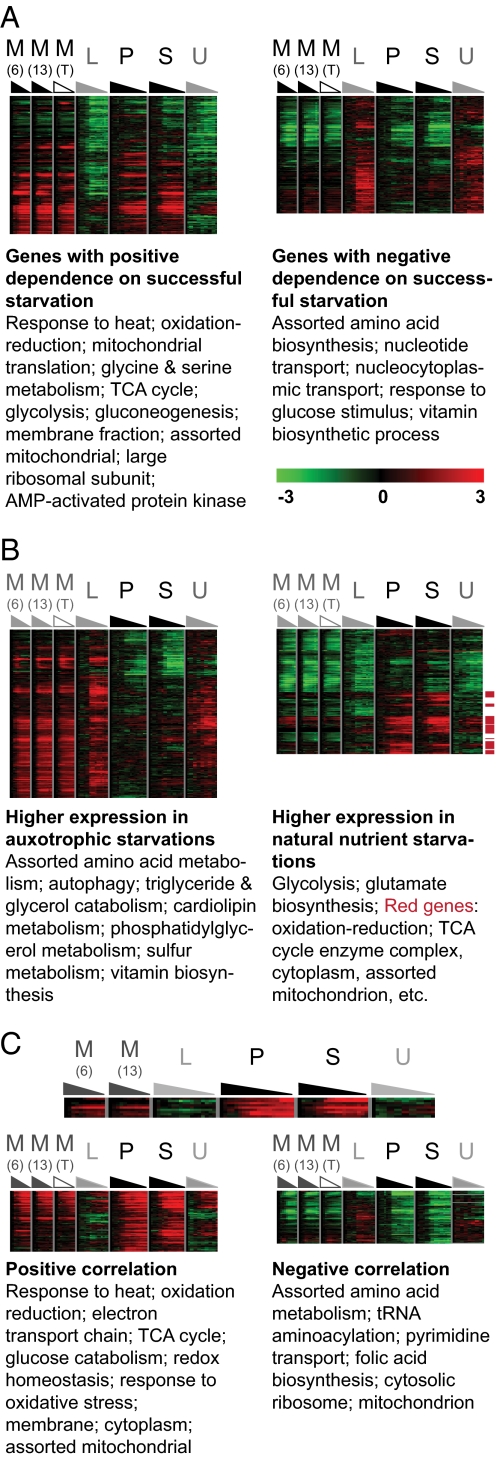
To identify genes that are expressed differently during the survivable starvations (methionine, phosphate, sulfate) than during the unsurvivable starvations (leucine, uracil), we used two independent methods, multiple regression (Fig. 5A) and k-means clustering (SI Materials and Methods and Fig. S2B), which yielded similar results. Multiple regression was used to extract a set of genes with a statistically significant dependence on the survivability of the starvation (SI Materials and Methods). As before, we selected genes that rank higher than the 95th or 90th percentile with respect to the F statistic P value (P ≤ 1.6 × 10−14 or 9.03 × 10−11; FDR ≤ 5.7 × 10−14 or 1.6 × 10−10), depend statistically significantly on time (FDR ≤ 0.01), and change twofold or more in at least one time course. Hierarchical clustering of these genes revealed two subclusters, one characterized by stronger induction in the successful starvations, the other by stronger repression (Fig. 5A and Dataset S1).
Fig. 5.
Genes that differ by starvation phenotype or nutrient class, as identified using multiple regression. Each panel shows genes ranking above the 90th percentile with respect to statistical significance for the given comparison. Functional enrichment and color coding are as specified in Fig. 2. These are independent analyses that address related questions (see text) and give consistent results; thus, the resulting gene sets (and their functional enrichments) necessarily overlap. (A) Expression profiles shared by successful starvations. Genes that are expressed differently in the survivable starvations (Met, Pho, and Sul) than in the unsurvivable starvations (Leu and Ura). Functional enrichment is further described in Table S4. (B) Expression profiles dependent on nutrient class. Genes that are expressed differently in the natural nutrient starvations (Pho and Sul) than in the auxtrophic starvations (Met, Leu, and Ura). Genes with particularly strong induction in Pho and Sul (red boxes) are strongly enriched for functions related to aerobic metabolism. Functional enrichment is further described in Table S5. (C) Expression profiles correlated with survival time. The gene expression template used for the Pavlidis template matching (SI Materials and Methods) is shown above genes whose expression profiles match the template mean with a Pearson correlation coefficient of at least 0.7 (“Positive correlation,” Left) or −0.7 (“Negative correlation,” Right). Functional enrichment is further described in Table S6.
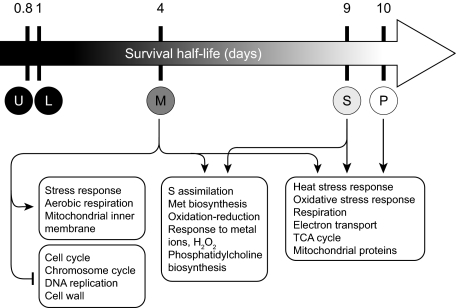
Overall, the functional enrichment of these clusters implies that successful starvation is correlated with expression of genes that support stress response and several functions executed largely within the mitochondria, such as aerobic metabolism, oxidative stress response, and redox homeostasis (Fig. 5A and Table S4). More specifically, genes expressed more highly in the successful starvations are enriched for heat stress, mitochondrial translation, and energy generation through respiration, including the Gene Ontology (GO) categories “oxidation reduction” and “generation of precursor metabolites and energy.” (In GO, the descendents of the “oxidation reduction” term are involved in mitochondrial respiration and its constituent functions, such as electron transport and ATP synthesis. “Generation of precursor metabolites and energy” contains TCA cycle components and enzymes that transfer electrons from metabolites to the electron transport chain.) Specifically, this gene set contains several indicators of aerobic metabolism (e.g., HXK1, ENO1, PGK1, and GPM1), which we also identified using k-means. With respect to oxidative stress response, it contains key regulators of oxidative stress, including the superoxide dismutase SOD1, the thioredoxin TRX3, and the glutathione synthetase GTT1, as well as TDH1, BNA2, RCK2, PST2 (a Yap1p target), OYE3, and the pentose phosphate pathway components TAL1 and GND2.
This gene set is also enriched for the mitochondrial folate cycle [including glycine and serine metabolism (e.g., SER33)] (Table S4) and for several documented starvation-response genes (SVF1, SSA4, GIS1, SIP2, and PNC1) that were also identified using k-means. Although not statistically enriched as a GO class, we also identified in this cluster a number of phospholipid biosynthetic genes, including OPI1 (a PKA-activated transcriptional repressor of phospholipid biosynthetic genes), OPI3, CHO1, CHO2, GPI6, and SER33. This gene set is enriched for genes whose products are localized to the mitochondrion, cytoplasm, and cell membrane.
Genes That Are Expressed Differently During Starvation for Natural Nutrients vs. Starvation for Auxotrophic Requirements.
Presumably, the auxotrophic starvations—including methionine—share some physiological features that limit survival relative to the natural nutrient starvations. We used multiple regression to identify genes with a statistically significant dependence on nutrient class (either auxotrophic or “natural”), selecting genes that rank higher than the 95th or 90th percentile as above (P ≤ 1.2 × 10−13 or 1.5 × 10−9; FDR ≤ 5.4 × 10−13 or 3.3 × 10−9). Hierarchical clustering of the significant genes yielded two main subclusters, one containing genes that are more highly induced during phosphate or sulfate starvation than in methionine, leucine, or uracil starvation, and a second with the opposite expression pattern (Fig. 5B, Table S5, and Dataset S1). [Results obtained using k-means clustering (Fig. S2C) are discussed in SI Results.]
The first cluster, containing genes that are more highly expressed in the natural nutrient starvations, is enriched for oxidative stress response (e.g., BNA2, GTO3, GRX2, TRX2, OYE3, and HYR1). Using FunSpec (20) to identify enrichment for MIPS functional categories (21), we found enrichment for genes involved in glutamate biosynthesis and genes involved in generating input to the citric acid cycle and thence to the electron transport chain (e.g., IDH1, IDH2, ACO1, and MEU1). [This cluster is also enriched for genes annotated to the GO glucose metabolism category (YOR283W, PGK1, HXK2, TDH3, ENO2, and TDH1); because most of those genes function in both glycolysis and gluconeogenesis, the meaning of this enrichment is ambiguous.]
The second cluster, characterized by greater induction during the auxotrophic starvations, is strongly enriched for amino acid biosynthesis (as expected), autophagy, metabolism of glycerolipids, cardiolipin, phosphatidylglycerol, and related compounds. This cluster is also enriched for the MIPS categories related to morphogenesis and cell wall integrity (e.g., MKK2, SLT2, RLM1, SOG2, and SOG2) and contains PPM1, whose deletion dramatically increases the survival during leucine starvation (1, 22, 23).
Gene Expression Program Correlated with Survival Time over All Types of Starvations.
The preceding analyses began with classifications based on either (i) the observation that met6Δ and met13Δ auxotrophs survive longer during starvation than leu2Δ or ura3Δ auxotrophs, or (ii) the fact that methionine-requiring mutants are auxotrophs. To avoid potential bias introduced by prior classification, we used Pavlidis template matching (24) to identify genes that are highly correlated with survival time per se (91 genes with correlation at least ±0.7; Fig. 5C, Table S6, and Dataset S1). These genes are significantly enriched for the same heat and oxidative stress, redox, citric acid cycle, and respiration-related processes previously identified, with many genes having annotations in several of these categories.
Overlap Between Met31p/Met32p-Dependent Gene Expression and Survival-Correlated Gene Expression.
Deletion of MET31 and MET32 significantly decreases survival during methionine and phosphate starvation. To determine whether Met31p and Met32p promote survival by the same means as the survival-correlated gene expression signature discussed above, we examined the intersection between Met31p/Met32p-dependent genes and survival-correlated genes. Nine genes are shared between these gene sets. They are involved in methylmethionine import and metabolism (MMP1 and MHT1), methionine import (MUP1), glutathione transport (OPT1), purine biosynthesis (SHM2 and ADE5,7), the folate cycle (MTD1 and GCV2), and glycine/serine metabolism (SER33). From this we conclude that, through Met31p and Met32p, these methionine-regulated processes must be properly regulated during starvation for a variety of nutrients, not just methionine.
Cells Undergoing Survivable Starvations Consume More Oxygen, but an Intact Electron Chain Is Not Required for Survival.
The analysis of gene expression patterns directly correlated with survival implicates processes associated with mitochondria and processes associated with oxidative stress response. We carried out two experiments to better characterize the involvement of these processes. In one, we studied oxygen consumption during starvation, and in the other, we studied survival in strains lacking mitochondrial DNA (and therefore lacking an electron transport chain). Fig. S4 shows that there are indeed differences in oxygen consumption for different starvation regimes and that the cultures with poorer survival consume less oxygen. By the second day of starvation, methionine-starved (met6Δ) cells consume slightly more oxygen than leucine-starved or methionine-starved (met31Δmet32Δ) cells, but the difference is not statistically significant. Strikingly, cells starved for phosphate (FY4 or met6Δ) consume by far the most oxygen (P = 4.3 × 10−7), paralleling their extraordinarily prolonged survival.
These results led us to test whether respiration is required for starvation survival. We used ethidium bromide to create ρ0 (cytoplasmic petite) derivatives of the strains tested above (Materials and Methods) and measured starvation survival as described above (Fig. 6A). The ρ0 derivatives survive nearly perfectly in phosphate starvation, almost as well in methionine starvation of the metabolic methionine auxotroph, and very poorly in leucine starvation and methionine starvation of the regulatory methionine auxotroph. Loss of the electron transport chain (and all of the other functions encoded in the mitochondrial DNA) results in only marginal impairment of survival of the methionine regulatory mutant and the leucine mutant. Because the presence of an electron transport chain had no consistent effect on survival, we conclude that survival does not depend on respiration per se (although respiration may contribute to survival in certain cases). Of course, this experiment does not rule out the possibility that the efficiency of respiration in yeast with an intact electron transport chain also influences survival.
Fig. 6.
(A) Survival of ρ0 derivatives during starvation for phosphate, methionine, and leucine. Survival of ρ0 derivatives of FY4, met6Δ, leu2Δ, and met31Δmet32Δ (black lines) was measured as the ability to form colonies on rich medium plates (Materials and Methods). Survival of the ρ+ parent strains is shown for comparison (gray lines). (B) Viability after hydrogen peroxide treatment. For duplicate cultures limited for phosphate (FY4), methionine (met6Δ or met31Δmet32Δ), or leucine (leu2Δ), colony-forming units on rich medium plates after treatment with 20 mM hydrogen peroxide, a source of oxidative stress, is reported as a percentage of colony-forming units after treatment with water. Black and gray bars indicate biological replicates.
Survival Is Correlated with Ability to Detoxify ROS.
If survival is not affected by the respiration genes we identified in the expression analysis, it may be affected by the other respiration-associated genes we identified. In our data, oxidative stress response is one of the most prominent respiration-associated functions. Moreover, the data in Fig. 4 suggest that impairing the oxidative stress response, such as by deleting MET31 and MET32, impairs survival in multiple nutrient starvations. To test the idea that starvation survival correlates more generally with the ability to survive oxidative stress, we measured the survival of hydrogen peroxide-treated cultures limited for methionine (met6Δ and met31Δmet32Δ), leucine (leu2Δ), or phosphate (FY4) (Materials and Methods). Fig. 6B shows that starvation survival is indeed highly correlated (Pearson correlation, 0.997) with the robustness of the oxidative stress response.
Discussion
Previous work suggested the existence of a starvation signal-and-response network that is differently activated in different nutritional environments, thereby leading to different phenotypes (2). Under this interpretation, the signal-and-response network is triggered during starvation for naturally required nutrients (e.g., sulfate, phosphate, nitrogen, and carbon), resulting in cell cycle arrest, glucose conservation, and survival, but is not triggered during starvation for auxotrophic requirements. We sought to better characterize the function of this signal by studying starvation for diverse nutrients. We have shown that cells undergoing survivable starvations (i.e., those in which cells survive for many days or weeks) induce expression of genes involved in mitochondrial function and/or stress response. They also survive externally applied oxidative stress far better than those undergoing unsurvivable starvations. In addition, we have shown that cells undergoing survivable starvations consume more oxygen than cells undergoing unsurvivable starvations, but that respiration per se is not required for prolonged starvation survival. We conclude that the differences between the survivable and unsurvivable starvations represent differential activation of the signal-and-response network, and that this network controls functions that support stress response and nonrespiratory mitochondrial functions.
Role of Mitochondrial Function and Stress Response in Starvation Survival.
According to our gene expression analyses, “successful” response to starvation is associated with induction of genes that can be broadly characterized as supporting stress response and/or mitochondrial function. When we identified genes whose expression is correlated with survival time during starvation, we found that many of them are involved in three basic cellular functions: (i) electron transport, aerobic respiration, and general mitochondrial activity (such as mitochondrial translation), (ii) acetyl-CoA and storage carbohydrate metabolism, and (iii) response to heat and oxidative stress. Some of the genes involved in the latter are mitochondrial and help maintain mitochondrial integrity and respiratory efficiency—for example, through synthesis of antioxidants such as superoxide dismutase, thioredoxin, peroxiredoxin, glutaredoxin, and glutathione, which neutralize ROS generated by electron transport (25). Additionally, aerobic-specific isozymes are preferentially induced relative to their fermentation-specific counterparts, particularly in methionine starvation. The fact that many genes typically associated with the canonical MSN2/MSN4-mediated stress response cocluster with genes involved in aerobic metabolism suggests that stress response and aerobic metabolism are transcriptionally coordinated.
Physiological measurements suggest that the relationship between survival and mitochondrial function may well be causative rather than merely correlative: we and others have shown that forcing the cell to respire during starvation (for example, by providing only a nonfermentable carbon source), increases survival time. The case for causality is supported by a recent study that measured survival of the prototrophic yeast deletion collection during phosphate and leucine starvation (26): genes whose deletion decreased survival in both starvations were more highly enriched for localization in the mitochondrion than any other cellular component.
Determining more precisely whether, how, and why mitochondria influence starvation survival is a complex task. The various processes that occur in the mitochondrion might influence starvation survival in different, even opposing, ways. For instance, electron transport generates ROS that might shorten survival, but compensatory stress response mechanisms might ultimately lengthen survival. Indeed, the literature is rife with conflicting reports on the role of mitochondrial function in chronological aging (e.g., refs. 14 and 27). Our results concerning survival of mutants lacking mitochondrial DNA show that respiration per se is not the major determinant of survival.
If survival does not depend on respiration, then it may depend on the stress response pathways that seem to be coupled to respiratory growth. This dependency is supported by our results with the short-lived met31Δmet32Δ mutant, which cannot mount a response to oxidative stress, and our results showing that ability to detoxify hydrogen peroxide is correlated with starvation survival across a variety of starvations. Previous work suggested that glucose signaling during leucine and uracil starvation activates the Tor pathway, leading to an inappropriate progrowth signal that results in cell death (1). Our data suggest that the survival-correlated stress response pathways are the same ones that are repressed by the Tor pathway [many of which are regulated by MSN2/MSN4 (23)]. Thus, survivable starvation conditions may repress the Tor pathway, thereby derepressing the Tor-regulated stress response pathways.
Other recent systems-level gene expression analyses have also found connections between mitochondrial functions and stress response pathways. It may be that over evolutionary time, induction of oxidative metabolism became associated with induction of protective mechanisms (e.g., thioredoxins, glutathione, superoxide dismutases, etc.) that provide recourse against respiration-generated ROS. Consequently, inducing oxidative metabolism may increase the abundance of antioxidants, whose protective effects may benefit the entire cell. This interpretation suggests that oxidative metabolism indirectly exerts a protective effect on the cell, in direct contrast with the standard view that increased oxidative metabolism is necessarily associated with increased abundance of damaging ROS.
Methionine Regulates Cellular Redox Homeostasis and Oxidative Stress Response, Which Contributes to Longevity During Methionine Starvation.
Our results beg the question of why starvation for methionine (in met6Δ or met13Δ mutants) is more like starvation for a natural nutrient. Our interpretation, based on the similarity of gene expression during methionine starvation and sulfur starvation, is that methionine is the “indicator metabolite” for sulfate availability. Our data show that regulation of methionine metabolism, iron and sulfur metabolism, oxidative metabolism, antioxidant biosynthesis, and one-carbon metabolism is highly interdependent. Thus, methionine auxotrophs ultimately up-regulate some or all of the protective functions normally associated with increased oxidative metabolism. It is therefore not surprising that deleting MET31 and MET32, which curtails survival during methionine starvation, phosphate starvation, and hydrogen peroxide exposure, severely affects the expression of oxidative stress response genes.
We conclude that proper regulation of these interdependent processes, particularly induction of the oxidative stress response genes, contributes to longevity in multiple starvations. This supports previous evidence for the role of oxidative stress in aging (28–32) and contributes to a growing body of evidence suggesting that methionine abundance regulates the pathways that protect the cell from the ravages of oxidative metabolism. In higher organisms, methionine restriction has been found to increase longevity and reduce oxidative damage to proteins and mitochondrial DNA and to decrease ROS generated in the mitochondria (32).
Nutrient Starvation in Yeast as a Model for the Warburg Effect in Cancer Cells.
A major metabolic hallmark of cancer is rapid glucose consumption even in the presence of oxygen, known as the Warburg effect. Although the uncontrolled glucose consumption of yeast starving for leucine or uracil is superficially analogous, there was no previous evidence that the two phenomena have a common cause. The Warburg effect was long thought ancillary to more fundamental molecular changes in cancer, but recent work suggests that it results directly from oncogene activation. Mounting evidence suggests that oncogene activation may cause mitochondrial dysfunction, which increases the cell's dependence on glycolysis (33) and the abundance of ROS (25, 34).
Our gene expression data suggest that glucose wasting in yeast may also be caused by failure to induce a variety of mitochondrial functions. In our data, a major difference between glucose-conserving and glucose-wasting starvations is greater induction of the mitochondrial genes in the glucose-conserving starvations. This suggests that glucose wasting during certain yeast starvations may be a useful model for the Warburg effect.
Nutrient Starvation in Yeast as a Model for Aging.
A variety of studies on chronological and replicative aging show that a metabolic regime favoring respiration instead of glycolysis increases yeast lifespan (1, 14, 35–38), but there is conflicting evidence for the role of respiration itself (27, 39, 40). Our results suggest that the situation is more nuanced: survival during starvation, often called “longevity,” is correlated with the induction of a nonrespiratory subset of mitochondrial functions, particularly the pathways that alleviate heat and oxidative stress. Protective mechanisms may be coinduced with oxidative metabolism, such that metabolic perturbations that favor respiration over fermentation may indeed increase longevity. We believe that the essential connections between longevity, metabolism, and stress response may well be conserved in whole or in part between yeast and higher organisms, and that data from yeast may well lead to a better understanding of the interplay between metabolism, stress response, cell growth control, and longevity.
Materials and Methods
Media compositions are listed in Table S7. Strain descriptions are listed in Table S8.
Establishing the Range of Linear Dependence on Methionine Concentration.
The range of linear dependence was determined as in ref. 3, using 0, 5, 10, 20, 40 or 200 mg/L [i.e., 0, 33.5, 67, 134, 268.1, or 1340.4 μM (μmoles/L)] methionine. A methionine concentration of 7.5 mg/L (i.e., 50.3 μM) lies in the center of the linear range.
Methionine Dependence and Growth on Different Carbon Sources.
A single colony of each strain was grown to steady state in rich medium and spotted in threefold serial dilutions on YNB agar plates containing 20 g/L glucose or 12.6 g/L glycerol and 8.1 g/L ethanol as the carbon source, with or without 20 mg/L methionine.
Cell Cycle Arrest.
A single colony of each strain was grown for 24 h in 5–7 mL of methionine-limited (7.5 mg/L) minimal medium, then diluted to a density around 1 × 105 to 5 × 105 cells/mL in fresh methionine-limited medium. Beginning at the end of lag phase (12–15 h after dilution), turbidity (Klett units), cell density (Coulter Count), and bud index were measured periodically. Bud index, the fraction of unbudded cells in a stationary phase culture (Table S1 lists strain-specific stationary phase Klett values), was measured by manual scoring of a sonicated culture under a microscope.
Starvation Survival Assays.
Survival assays on various carbon sources were performed as described in ref. 1. Here, nutrient-limited medium contained 7.5 mg/L methionine, 40 mg/L leucine, or 13.3 mg/L phosphate plus 200 mg/L methionine or 240 mg/L leucine. ρ0 mutants were generated using ethidium bromide as described in ref. 41.
Hydrogen Peroxide Survival Assays.
A single colony was inoculated into nutrient-limited medium and grown for 24 h with shaking at 30 °C. The culture was diluted into fresh nutrient-limited medium and grown with shaking at 30 °C until midexponential phase (the length of time required is strain-specific). An aliquot containing 1 × 108 cells was resuspended in 10 mL of fresh nutrient-limited medium, treated with 0 mM or 20 mM hydrogen peroxide for 20 min, washed twice with water, and plated for viable cells. Viability in 20 mM hydrogen peroxide is reported as a percentage of the viable cells counted after treatment in 0 mM hydrogen peroxide.
Residual Glucose Measurements.
A single colony was inoculated into nutrient-limited medium (7.5 mg/L methionine or 40 mg/L leucine) and grown for 24 h with shaking at 30 °C. Each culture was diluted into fresh nutrient-limited medium to a density between 1 × 105 and 4 × 105 cells/mL and grown with shaking at 30 °C. Starting 12–15 h after dilution, turbidity was measured, and 1-mL samples were frozen periodically over the course of 3 d. The glucose concentration in each thawed sample was measured in triplicate using the Boehringer-Mannheim d-glucose UV test.
Methionine-Starvation Filter-Switching Time Courses for met6Δ, met13Δ, and met31Δmet32Δ.
A single colony of each strain was inoculated into methionine-limited minimal medium and grown with shaking at 30 °C. After 24 h, each culture was diluted to low density (1 × 105 to 1 × 106 cells/mL) in 250–300 mL of fresh methionine-limited minimal medium and grown with shaking for 12–15 h to a density of 20–25 Klett units (approximately 5 × 106 cells/mL). As described in ref. 42, the culture was then filtered in 5-mL aliquots onto 0.45-μm, 82-mm-diameter Nylon filters. Each filter was placed on a Petri dish containing YNB5 (minimal medium made with agarose) and 7.5 mg/L methionine. The plates were incubated at 30 °C for 4 h, until the culture reached a density of 80–90 Klett units (approximately 2 × 107 cells/mL). At this time, each filter was transferred to a fresh plate containing YNB5 but no methionine. At specified time points thereafter (0 min, 10 min, 30 min, 1 h, 1.5 h, 2 h, 2.5 h, 3 h, 3.5 h, 4 h, and 6 h), filters from two plates were placed in 50-mL screw-capped tubes and immediately immersed in liquid nitrogen. A third filter was washed and used for Klett readings, Coulter counts, and bud index measurements.
Metabolite Measurements in met13Δ.
During the filter-switching time course for met13Δ described above, samples were simultaneously collected for metabolite extraction. Metabolites were extracted using methanol quenching, and their abundance was measured using LC-MS/MS as described in ref. 43.
Oxygen Consumption Measurements.
A single colony was inoculated into 5 mL of nutrient-limited medium (7.5 mg/L methionine, 40 mg/L leucine, or 13.3 mg/L postassium phosphate) and grown overnight with shaking at 30 °C. The culture was diluted to a density between 1 × 105 and 4 × 105 cells/mL in 300 mL of fresh nutrient-limited medium and grown overnight with shaking at 30 °C. At Klett 20 (≈5 × 106 cells/mL), 250 mL of the culture was transferred to a chemostat vessel equipped with two dissolved-oxygen probes and grown to stationary phase in batch mode (400 rpm, 30 °C) with an airflow rate of 40. The oxygen probes were calibrated in water at 25 °C, so that full probe saturation corresponded to a known dissolved oxygen concentration (0.27 mMol O2/L). Three days after inoculation of the chemostat vessel, cellular oxygen consumption was measured by periodically turning off the chemostat air supply for 20 min and recording the change in dissolved oxygen per unit time during the first 10 min.
Methionine-Depletion Batch-Growth Time Course for met13Δ.
A single colony was inoculated into 100 mL methionine-limited minimal medium and grown with shaking at 30 °C for 32 h. The culture was then diluted to 5 × 105 cells/mL in 500 mL of fresh methionine-limited minimal medium and grown in batch with shaking. Beginning 5 h after dilution and every hour thereafter until the culture reached steady state with respect to density and bud index, samples were removed for bud index, culture density, and cell count measurements, and cell samples for RNA extraction were obtained by filtering and freezing in liquid nitrogen.
Preparation of Cells for Microarray Reference Samples.
In a chemostat, FY4 was grown to steady state in chemostat medium containing limiting phosphate (10 mg/L) at a dilution rate of 0.17 h−1. Cells were harvested by filtering onto a nitrocellulose filter and frozen in liquid nitrogen.
RNA Isolation, Labeling, and Hybridization.
RNA was collected from the frozen filters using phenol-chloroform extraction, purified using an the Qiagen RNeasy kit, labeled with Cy5 CTP (samples) or Cy3 CTP (phosphate-limited chemostat reference) using the Agilent Low-input Linear Amplification kit, and hybridized to 2 × 105k, 4 × 44k, or 8 × 15k Agilent Yeast Oligo V2 microarrays. Arrays were washed and scanned using extended dynamic range according to Agilent protocols. Agilent Feature Extraction Software was used with default settings and linear and lowess dye bias correction.
Expression and Metabolite Data Analysis.
Expression and metabolite data analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sanford Silverman and Viktor Boer for helpful discussions of this work and Olivia Ho-Shing for technical assistance with oxygen consumption measurements. A.A.P. was supported by Ruth Kirschstein Cancer Training Grant T32 CA-009528. Research was supported by National Institute of General Medical Sciences Center for Quantitative Biology Grant P50 GM-071508 and National Institutes of Health Grant GM-046406 (to D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray expression data reported in this paper have been deposited athttp://genomics-pubs.princeton.edu/StarvationSurvival/.
See Author Summary on page 18217.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101494108/-/DCSupplemental.
References
- 1.Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci USA. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unger MW, Hartwell LH. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci USA. 1976;73:1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray DB, Beckmann M, Kitano H. Regulation of yeast oscillatory dynamics. Proc Natl Acad Sci USA. 2007;104:2241–2246. doi: 10.1073/pnas.0606677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 8.Kresnowati MTAP, et al. When transcriptome meets metabolome: Fast cellular responses of yeast to sudden relief of glucose limitation. Mol Syst Biol. 2006;2:49. doi: 10.1038/msb4100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler GL, Trotter EW, Dawes IW, Grant CM. Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J Biol Chem. 2003;278:49920–49928. doi: 10.1074/jbc.M310156200. [DOI] [PubMed] [Google Scholar]
- 10.Yen JL, Su N-Y, Kaiser P. The yeast ubiquitin ligase SCFMet30 regulates heavy metal response. Mol Biol Cell. 2005;16:1872–1882. doi: 10.1091/mbc.E04-12-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 14.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voet D, Voet JG, editors. Biochemistry. 2nd Ed. New York: John Wiley & Sons; 1995. [Google Scholar]
- 17.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T, et al. Dissection of combinatorial control by the Met4 transcriptional complex. Mol Biol Cell. 2010;21:456–469. doi: 10.1091/mbc.E09-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen OS, et al. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J Biol Chem. 2004;279:29513–29518. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- 20.Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: A web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu WS. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006;25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- 23.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 24.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 25.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gresham D, et al. System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae. Genetics. 2011;187:299–317. doi: 10.1534/genetics.110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaeberlein M, et al. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 29.Richie JP, Jr, et al. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8:1302–1307. doi: 10.1096/fasebj.8.15.8001743. [DOI] [PubMed] [Google Scholar]
- 30.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pamplona R, Barja G. Mitochondrial oxidative stress, aging and caloric restriction: The protein and methionine connection. Biochim Biophys Acta. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Sanz A, et al. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. FASEB J. 2006;20:1064–1073. doi: 10.1096/fj.05-5568com. [DOI] [PubMed] [Google Scholar]
- 33.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 34.Sung HJ, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 36.Lin S-J, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 37.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 41.Fox TD, et al. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 42.Brauer MJ, et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci USA. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]