Abstract
The ability to adapt to adverse environmental conditions encountered in food and during host infection is a sine qua non for a successful Listeria monocytogenes infection. This ability is likely to depend on complex regulatory pathways controlled by a number of key regulators. We utilized the pORI19 plasmid integration system to analyze the role of six putative regulatory loci in growth under suboptimal environmental conditions and during murine infection. Disruption of loci encoding a topoisomerase III (lmo2756), a putative methyltransferase (lmo0581), and a regulator of the MarR family (lmo1618) revealed roles for the methyltransferase and the MarR regulator in growth under environmental stress conditions. However, plasmid integration into these loci had no impact on virulence potential in the murine model of infection. Disruption of the alternative sigma factor Sigma-H resulted in a mutant that demonstrated reduced growth potential in minimal medium. Murine studies indicated a minor role for this sigma factor in the infectious process. Strikingly, disruption of both perR and fur loci resulted in mutants that are significantly affected in virulence for mice, with the fur mutant demonstrating the greatest reduction in virulence potential. Both perR and fur mutants demonstrated increased resistance to hydrogen peroxide and the fur mutant was sensitive to low-iron conditions. The virulence defect of both fur and perR mutants could be rescued by iron-overload after esculetin treatment of mice, suggesting that the in vivo role of these gene products is to procure iron for bacterial growth.
The gram-positive food-borne pathogen Listeria monocytogenes can cause serious infection in susceptible individuals (49). Mortality rates during common-source epidemics can reach 30% (49), and approximately 28% of deaths in the United States due to bacterial food-borne illness can be attributed to L. monocytogenes infection (33). The bacterium is widely distributed in nature, having been identified in soil, silage, sewage, human and animal feces, slaughterhouse waste, and water (11). An important feature of this pathogen is its ability to withstand the suboptimal conditions encountered during a saprophytic lifestyle, to endure food-processing protocols, and eventually to adapt to the environmental stresses encountered during infection of the host.
In order to establish infection of the host, the pathogen must transit the low pH of the stomach and subsequently survive exposure to bile acids, increased osmolarity, volatile fatty acids, and intense competition with intestinal flora for space and nutrients within the small intestine (7). Furthermore, as an intracellular pathogen L. monocytogenes can invade the intestinal epithelium and cause systemic disease characterized by intracellular growth of the pathogen in target organs (29, 49). During invasion of host cells the organism encounters further suboptimal conditions, including a rapid drop in pH within the host cell phagosome (2). Subsequent lysis of the phagosome releases the pathogen into the host cell cytoplasm where conditions of nutrient and iron starvation prevail (18, 49).
It has previously been reported that the ability of L. monocytogenes to sense changes in its environment and respond to various stresses can have an impact on the virulence of this organism (8, 30, 37, 42). Some information is available concerning effector mechanisms used by the pathogen to survive in vivo stress. The presence of an intact glutamate decarboxylase system is essential for the survival of gastric acid (9). The membrane transporter for the osmolyte carnitine encoded by opuC is required for optimal colonization of the small intestine and subsequent invasive disease (46, 50). In addition, elements of the clp operon are necessary for escape from the host cell phagosome and for growth under the iron-limiting conditions encountered in vivo (42, 43). However, other than PrfA, CtsR, and SigB (5, 35, 36), relatively little is known of regulatory networks that function to coordinate listerial responses to environmental stress in vivo.
The recent publication of the genome sequence of L. monocytogenes EGDe provides an opportunity to increase our understanding of the mechanisms by which the organism coordinates the adaptation to suboptimal environments encountered during infection (17). An interesting feature of the genome is the abundance of putative transcriptional regulators which presumably play a role in adaptation to suboptimal environments encountered at all stages of the pathogenic cycle. Determining the function of each regulator will require detailed postgenomic analysis of regulatory elements involved in the infectious process. These analyses will be facilitated by the development of molecular tools to allow rapid and stable gene disruptions.
We have utilized the pORI19 plasmid integration system first described by Law et al. (27) in Lactococcus lactis, for rapid, specific, and stable gene disruption of potential regulatory systems in L. monocytogenes EGDe. Six genes targeted for disruption by this system included two loci previously identified by an in vivo expression technology approach (a predicted topoisomerase III [lmo2756] and a potential methyltransferase [lmo0581]) (15), a gene encoding the alternative sigma factor Sigma-H (lmo0243), a potentially pH-regulated transcriptional regulator of the MarR family (lmo1618) (39), and genes predicted to encode the ferric uptake regulator homologues Fur (lmo1956) and PerR (lmo1683). All six loci were successfully and stably disrupted, demonstrating the usefulness of the pORI19 system for first-look analysis of regulatory genes in L. monocytogenes. The resultant mutants were analyzed for virulence in a mouse model of infection, and experiments demonstrated that both Fur and PerR are essential for virulence of L. monocytogenes. In-frame deletion mutants subsequently created in perR and fur replicated the phenotypes of the pORI19 mutants and thus validate the plasmid integration methodology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium and Listeria monocytogenes strains were grown in brain heart infusion (BHI) broth (Oxoid) at 37 or 30°C for pVE6007. Antibiotics (Sigma Chemical Co., St. Louis, Mo.) were used as appropriate at the following concentrations: erythromycin (ERY) at 200 μg/ml for E. coli and ERY at 5 μg/ml and chloramphenicol (CHL) at 10 μg/ml for L. monocytogenes. For solid media agar was added to 1.5%. Blood agar plates consisted of blood agar base (Lab M), to which 5% sheep blood was added after an autoclaving step. Tropolone and ferric citrate were solublized in water and added to the media as filter-sterilized stocks. Esculetin was dissolved with 5 N NaOH, and the pH was adjusted to 8.7 with 1 N HCl (10) and filter sterilized.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or primer | Characteristica | Reference |
|---|---|---|
| Strains | ||
| E. coli EC101 | Derivative of E. coli with pWVO1 repA integrated in the chromosome | 27 |
| L. monocytogenes | ||
| EGDe | Serotype 1/2a | W. Goebel |
| EGDpORI19::met | EGDe derivative with an insertion into a putative SAM-dependent methyltransferase gene (lmo0581) | This work |
| EGDpORI19::topB | EGDe derivative with an insertion in the topoisomerase III gene (lmo2756) | This work |
| EGDpORI19::fur | EGDe derivative with an insertion in the fur gene (lmo1956) | This work |
| EGDpORI19::marR | EGDe derivative with an insertion in the marR gene (lmo 1618) | This work |
| EGDpORI19::perR | EGDe derivative with an insertion in the perR gene (lmo1683) | This work |
| EGDpORI19::sigH | EGDe derivative with an insertion in the sigma-H gene (lmo0243) | This work |
| EGDΔfur | Δfur, L. monocytogenes EGDe | This work |
| EGDΔperR | ΔperR, L. monocytogenes EGDe | This work |
| Plasmids | ||
| pORI19 | RepA−, Emr | 27 |
| pVE6007 | Temperature sensitive, RepA+, Cmr | E. Maguin |
Emr, ERY resistance; Cmr, CHL resistance.
DNA manipulations.
Gel extraction was performed by using the Qiagen gel extraction kit (Qiagen). Plasmid DNA isolation was performed by using the Qiagen QIAprep spin miniprep kit (Qiagen). T4 DNA ligase, PCR reagents, and restriction enzymes were purchased from Roche Diagnostics GmbH (Mannheim, Germany) and used according to the manufacturer's instructions. PCRs were performed by using a Hybaid (Middlesex, United Kingdom) PCR express system, and products were cloned into pORI19 and sequenced by using universal M13 primers (Lark Technologies, Inc., Essex, United Kingdom). Oligonucleotide primers (Table 2) for PCR were synthesized by Sigma-Genosys Biotechnologies and Taq DNA polymerase (Biotaq; Bioline) was used for all PCRs.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| MetF | TAGATCTAGAAAGATGCCTGTCATCC |
| MetR | ATGGTACAGTCTAGGGATCCACGCT |
| M-INT | ATTGTCTTGCTGAGCGAACGACC |
| TopF | GTGGTAGGATCCAATGCTAC |
| TopR | ATTTTCGCCCACACTGCAGGC |
| T-INT | CAAAAACATTAGTACTGGCAG |
| FurF | CACAACTTCTAGATGCTAGTTATAA |
| FurR | GAAAAAGCTTAGCGCCTTCTTGT |
| F-INT | GCAACCAAGGGAGGAACTATAAT |
| MarRF | TTTCTAGAAGTACAACGAAAAACAC |
| MarRR | CTGAAGCTTTTGTAGCTCCTTTT |
| MAR-INT | GGATGAAAAATTGGTGAAAGAGG |
| PerRF | ACTCTAGAAGAGGCAGTAGATGTC |
| PerRR | GCGTGGTAAAGCTTAGATGTAGAA |
| P-INT | GGAGTGCATGGCGGTGTCTAAT |
| SigHF | ATTTCTGCAGTAAGTATCAATCGG |
| SigHR | CAGGATCCTCAGCCATTGGTGTA |
| SH-INT | GGAGTAAGGGATTGACGGTATGT |
| HlyF | GACTAATCTAGACAATAAAATCG |
| HlyR | TCGCGTAAGGATCCGAGGTT |
| Hly-INT | CGCCTGCAAGTCCTAAGAC |
| M13F | GTTTTCCCAGTCACGAC |
| M13R | CAGGAAACAGTATGAC |
| M13Rmut | GCGGATAACAATTTCACACAGGA |
| FurSOEA | TCCTTCCTGCAGAAGCTAAAT |
| FurSOEB | AAGTTGTGCTTTAATGCGTCC |
| FurSOEC | GGACGCATTAAAGCACAACTTGGGATTTGCGCGAATTGCAGAb |
| FurSOED | ATTCGGATCCGTTAACTCCT |
| PerRSOEA | AGACTGCAGCTTTGAAATTT |
| PerRSOEB | GCAACTCTAAAAGAGGCAGT |
| PerRSOEC | GCAACTCTAAAAGAGGCAGTTGCTGGTCTAATTAAGGAATb |
| PerRSOED | GATTGTTTTTCTAGACACGCT |
Restriction sites incorporated into primer sequences are underlined.
Overhangs complementary to corresponding SOE primers are underlined.
pORI19 mutagenesis.
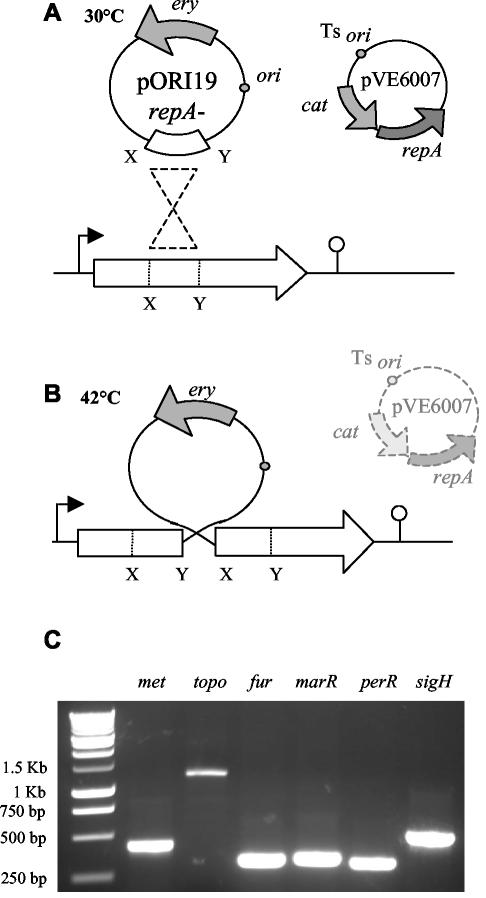
A central portion of the gene of interest was amplified by PCR and cloned into the multiple cloning site of pORI19. Electrotransformation of Escherichia coli EC101 was performed according to standard protocols. After plasmid isolation, electrotransformation of L. monocytogenes EGDe containing pVE6007 (RepA+/temperature sensitive) was performed according to protocols outlined by Park and Stewart (38). pORI19 was maintained in EGDe at 30°C. Loss of pVE6007 was achieved by transferring 10 μl of an overnight culture to BHI broth prewarmed to 42°C and subsequent growth for 16 h at 42°C. Aliquots were then spread-plated onto prewarmed BHI-ERY plates and incubated overnight at 42°C. Loss of pVE6007 (CHL sensitive) was confirmed by replica plating individual colonies onto BHI-ERY and BHI-CHL plates with overnight incubation at 30°C. Integration results in the formation of a stable ERY-resistant mutant and was confirmed by PCR with a forward primer outside the region of integration and a primer for the plasmid (Fig. 1).
FIG. 1.
(A) A fragment of EGDe chromosomal DNA was cloned into the multiple cloning site of pORI19 and maintained in Listeria through the use of the temperature-sensitive plasmid pVE6007, which provides the RepA protein in trans. (B) pVE6007 replication is arrested by increasing the temperature to 42°C. This selects for events in which pORI19 has integrated into the host chromosome by homologous recombination at the point of homology provided by the cloned DNA. (C) The integration event was confirmed by PCR with a primer on the chromosome and a primer on the plasmid. No PCR product was detected in control PCRs with wild-type cells (not shown).
Creation of deletion mutants.
The splicing-by-overlap-extension (SOE) procedure was used to create nonpolar deletions in L. monocytogenes EGDe. The procedure was carried out as described previously (9, 15). Two pairs of primers were designed (SOEA-SOEB and SOEC-SOED) (Table 2) to amplify two fragments of equal size on either side of the gene to be deleted. The resulting products were mixed in a 1:1 ratio and reamplified with the SOEA and SOED primers. This product was digested and cloned into the temperature-sensitive plasmid pKSV7. The plasmid construct was electroporated into EGDe, and transformants were selected by using BHI agar with 10 μg of CHL/ml. Chromosomal integration of the plasmid at 42°C was selected by serial passage of a transformant in prewarmed BHI-CHL broth and streaking onto prewarmed BHI-CHL agar. Plasmid excision and curing was brought about by continuous passages in BHI broth at 30°C, followed by spread plating onto BHI agar at 30°C. The deletion event was confirmed by PCR with primers upstream of SOEA and downstream of SOED (Table 2). The appropriate deletion event was detected in 1 of 47 colonies for the fur mutation and 1 in 357 colonies for the perR mutation.
Growth curves.
Overnight cultures were centrifuged (12,000 rpm for 5 mins), washed, and resuspended in an equal volume of one-quarter-strength Ringers solution (Merck). A 2% inoculum was added to 10 ml of BHI. Then, 200 μl was added to a 96-well plate, and growth was determined automatically at 600 nm by using a SpectraMax 340 spectrophotometer (Molecular Devices, Sunnyvale, Calif.) for 24 h at 37°C. Acidic and alkaline conditions were created by adjusting BHI to pH 5.5 with 3 M lactic acid and to pH 9.0 with 5 N NaOH. Growth was determined as described above. Overnight cultures (2%) were also inoculated into BHI with 5% ethanol, and growth was determined manually with a Beckman 640 spectrophotometer. Iron deprivation was achieved through the addition of tropolone to BHI (10). After its addition, the broth was left shaking overnight at 37°C to allow the tropolone to bind the iron and then used the following day for inoculation. The addition of esculetin to BHI containing tropolone was used to counteract the iron limitation. Esculetin and tropolone were added to BHI to give a final concentration of 1 mM and 160 μM, respectively. The ability of Listeria to grow in the presence of tropolone with or without esculetin was assessed spectrophotometrically. L. monocytogenes EGDe and fur and perR mutants were also inoculated into BHI and tropolone supplemented with 5 mM ferric citrate. When required, a chemically defined medium (CDM) as described by Premaratne et al. (40) was used. Overnight cultures of the wild-type and all of the mutants were washed with one-quarter-strength Ringers solution, and a 2% inoculum was added to CDM. The following day, 2% was transferred to fresh CDM, and plate count readings were taken at time zero and at 30 h.
Hydrogen peroxide sensitivity assay.
Overnight cultures of wild-type EGDe, fur, and perR mutants were inoculated into fresh BHI and grown to early log phase (i.e., an optical density at 600 nm of 0.15). Cells were harvested and washed with one-quarter-strength Ringers solution. Hydrogen peroxide was added to BHI to give a final concentration of 50 mM, and 1 ml was used to resuspend the pellets. Samples were taken every 30 min for 1.5 h, and survivors were determined by dilution in Ringers solution and plating them onto BHI agar. Overnight cultures of EGDe and fur and perR mutants (2%) were also inoculated into BHI with 22 mM H2O2, and growth was monitored automatically as described above.
Macrophage assay.
All tissue culture reagents were purchased from Gibco Invitrogen Corp. unless otherwise indicated. J774 mouse macrophage cells were used to carry out the in vitro infection assay. Macrophage cells were grown overnight in 24-well tissue culture plates in antibiotic-free Dulbecco modified Eagle medium containing 10% fetal calf serum. For infection, 1 ml of an overnight culture was spun down and washed once with phosphate-buffered saline (PBS) and resuspended in 1 ml of PBS. This was diluted 1/100 with antibiotic-free Dulbecco modified Eagle medium, giving a final concentration of 2 × 107 CFU/ml, and was used to infect the macrophages, giving a multiplicity of infection of 5 bacteria per cultured cell. Plates were centrifuged at 1,500 × g for 10 min to increase contact between bacteria and macrophages and incubated for 1 h in 5% CO2 at 37°C. Subsequently, gentamicin (15 μg/well; Sigma) was added for a further 30 min. Bacterial counts were determined at this stage T0 and at T6 (in hours) by washing the cells twice with 1 ml of PBS and then lysing them by the addition of 250 μl of cold sterile-distilled H2O. Then, 100 μl was removed, serially diluted, and plated onto BHI agar plates that were incubated at 37°C overnight.
Virulence assay.
Overnight cultures were centrifuged, washed once with PBS, resuspended, and subsequently diluted in PBS. In vivo survival was determined by inoculating 8- to 12-week-old BALB/c mice intraperitoneally (i.p.) with 3 × 105 CFU in 200 μl of PBS. The mice were euthanized on the appropriate day postinfection, and bacteria were harvested from the spleen or liver by homogenization of organs in PBS and serial dilution of organ homogenates onto BHI agar and incubation overnight at 37°C. Esculetin was used at a final concentration of 20 μmol and was administered to the mice on the day before inoculation (day −1), the day of infection (day 0), and the day after infection (day +1) as outlined previously (9). When esculetin was incorporated into the assay, the inoculum used to infect the mice was reduced to 3 × 102 CFU.
RESULTS
Strategy for mutant construction.
Mutants were constructed by using the pORI19 integration strategy outlined in Fig. 1 (27). A central region of the target gene was amplified by PCR and cloned into the multiple cloning site of pORI19 (RepA−) by using E. coli EC101 (RepA+) as a cloning host. Plasmid replication was subsequently maintained in L. monocytogenes EGDe through the provision of the RepA protein in trans by the temperature-sensitive plasmid pVE6007. Growth at 42°C results in the curing of pVE6007 and integration of pORI19 by homologous recombination at the site of homology provided by the cloned L. monocytogenes DNA. Replica plating of candidate integrants onto BHI-ERY and BHI-CHL verified that integrants were CHL sensitive and ERY resistant, whereas PCRs with a primer based on the chromosome of EGDe and a primer based upon the plasmid (Table 2) confirmed that integration had taken place at the predicted location (Fig. 1C). In addition, Southern hybridization confirmed that chromosomal integration had taken place at a single location for all integrants (data not shown). The absence of the repA gene in mutant strains will select against excision and extrachromosomal maintenance of pORI19, ensuring stable integrants for subsequent analysis. In silico analysis revealed the presence of a number of putative stem-loop structures within the sequence of pORI19. This is predicted to prevent readthrough and formation of fusion products with adjacent genes.
We tested the stability and specificity of pORI19 insertions in L. monocytogenes by creating a pORI19 insertional mutation in the gene encoding listeriolysin (hly). The resulting mutant completely lacked hemolytic activity when tested on blood agar plates, and hemolysis could be restored through provision of hly on a plasmid. After continuous passaging over 48 h in the absence of selective pressure, 100% of pORI19::hly colonies remained hemolysin negative. Over a further 3 days of passaging (approximately 137 total generations), ca. 99.84% of pORI19::hly colonies were hemolysin negative. This represents the appearance of 16 hemolytic colonies per 10,000 CFU. PCR analysis on hemolysin-negative colonies confirmed the presence of the plasmid at the expected location. After a 6-h infection of J774 mouse macrophages, 100% of the survivors remained hemolysin negative. However, during extended infections of J774 cells (over a 24-h period), we could detect a significant number of revertants (∼47%) to wild-type levels of hemolytic activity. The data confirm that the pORI19 system can be used to create plasmid integrations that are stable under in vitro conditions. However, under conditions of extreme selective pressure, such as those encountered within the macrophage phagosome, some reversion to wild-type may occur. However, this may represent a specialized situation, since in subsequent murine infection studies both pORI19::fur and pORI19::perR mutants were stable over the course of a 4-day infection (see below).
We also used the pORI19 hemolysin mutant to investigate the likelihood that pORI19 integration may interfere with the expression of downstream genes through alterations in local DNA topology. Immediately downstream of hemolysin is a gene encoding a zinc metalloproteinase precursor (mpl) whose product is required for activation of phospholipase C (encoded by plcB). Indeed, under appropriate conditions the metalloprotease and plcB are cotranscribed along with actA (45). Efficient expression of plcB can be examined directly by using an assay for phospolipase C (lecithinase) activity on egg yolk agar plates. After plating of the wild-type and hly mutant onto this agar, no difference in the extent of lecithinase activity was observed. This suggests that pORI19 insertion into hemolysin does not affect the expression of the downstream genes responsible for lecithinase activity.
Selection and mutation of potential regulators.
Six loci were chosen for disruption by this method (Fig. 2), and all mutants were created with relative efficiency, indicating the usefulness of this system for functional genomic analysis in L. monocytogenes (Fig. 2). Genes encoding the putative topoisomerase III protein (lmo2756; herein topB) and a predicted methyltransferase (lmo0581; herein met) were identified previously through an in vivo expression technology strategy (15). Another locus (lmo1618; herein marR), identified as a candidate member of the MarR family of transcriptional regulators, was previously shown to be upregulated under low-pH conditions (39) The other three loci were initially identified by searching the prepublication L. monocytogenes EGDe genome sequence for homologues of cognate Staphylococcus aureus or Bacillus subtilis genes. This identified genes predicted to encode the alternative sigma factor Sigma-H (lmo0243; sigH) and the ferric uptake regulator homologues Fur (lmo1956; fur) and PerR (lmo1683; perR). We have ensured that integration takes place in all mutants at a central location, ensuring that at least 26% of the gene is not translated. Although we cannot eliminate the possibility that the interrupted gene is capable of producing a functional product, we have always observed the same phenotype in mutants created by using either pORI19 integration or in-frame deletion strategies (see below). In addition, the met and marR mutants demonstrated discernible phenotypes under in vitro stress conditions (below), suggesting that plasmid insertion has been sufficient to eliminate or reduce gene function. For the predicted topB insertional mutant, a single plasmid integration into the center of the 2,154-bp locus has eliminated a predicted 1,154 bp (54%) of the gene. This is most likely sufficient to reduce or eliminate gene function.
FIG. 2.
Map of genetic loci disrupted during the present study with surrounding regions. All genes are drawn approximately to scale by using the L. monocytogenes EGDe genome sequence data. Genes are numbered according to the National Center for Biotechnology Information annotation scheme. Solid black arrows indicate the genes disrupted in EGDe. Shaded areas within black arrows depict the region of the gene cloned into pORI19. Shaded arrows represent adjacent open reading frames. Stem-loop structures are used to illustrate putative terminator regions.
In four instances (Fur, PerR, the putative topoisomerase III protein [lmo2756], and the putative methyltransferase [lmo0581]), the genes are followed by predicted transcriptional terminators, reducing the likelihood that insertion of a plasmid into these regions will cause polar effects on downstream genes. The gene encoding the putative alternative sigma factor, Sigma-H, is located upstream of a region encoding a putative ribosomal protein (lmo0276) and a predicted preprotein translocase (secE). To rule out the influence of a possible polar effect of the plasmid mutation, we have created a nonpolar SOE deletion mutant in sigH (ΔsigH). This mutant demonstrates an identical phenotype to the pORI19 mutant (data not shown), suggesting that the phenotypes are Sigma-H related. MarR is located upstream of a gene (lmo1617) annotated as encoding a putative multidrug efflux transporter. Further analysis is therefore required to determine whether this locus is also affected by the insertion event.
To determine stability, mutants were grown overnight to stationary phase and then passaged twice daily over four consecutive days in the absence of selective antibiotic pressure. This represents approximately 110 bacterial generations. Subsequently, 100 colonies were replica plated onto BHI and BHI-ERY. For five of the six mutants, all 100 colonies retained the plasmid as evidenced by their resistance to the antibiotic, thus confirming plasmid stability in the absence of antibiotic selection. Interestingly, in the case of the perR mutant we determined that 70% of colonies retained the plasmid, suggesting that some selective pressure is imposed upon this mutant during normal growth over prolonged periods. The data confirm that pORI19 insertion is generally a stable phenomenon but that certain loci may be prone to plasmid curing over long periods. However, in subsequent 4-day virulence experiments where bacterial growth rates are minimal and after exposure to environmental stress where growth is measured over a short time period, both perR and fur mutants were found to retain the plasmid with 100% efficiency.
Physiological analysis of mutants.
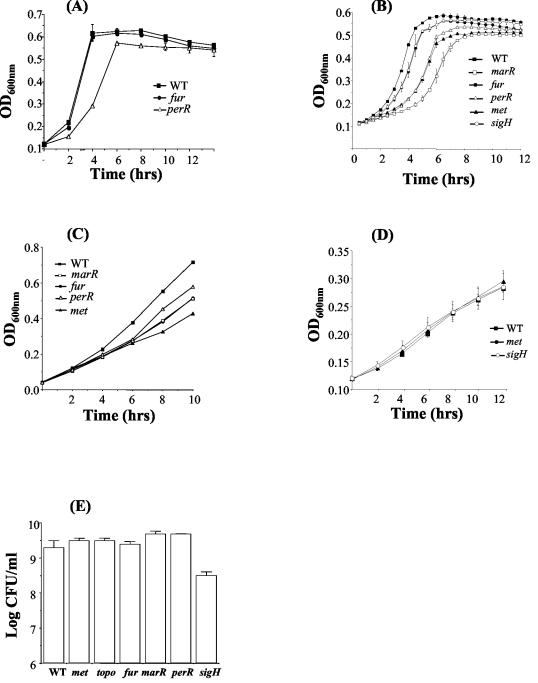
The growth of the six mutants in BHI (pH 7.0) at 37°C was compared to that of the wild type. Under these conditions no difference was detected in either the exponential or stationary phase for five of the six mutants, and all five reached the same final optical density at 600 nm as did the wild type. However, growth of the pORI19::perR mutant was impaired in comparison to the wild type. Indeed, it was noted that pORI19::perR generally resulted in a much smaller colony size on BHI than the wild type. Interspersed among the small colonies at a frequency of 0.9% were large colonies similar in size to those of the wild type. The large colonies do not represent revertants since the same phenomenon was observed with perR deletion mutants and a similar observation has previously been reported in B. subtilis in which the large colonies were termed “pseudorevertants” (6, 20). Figure 3A presents representative data for the wild type and for pORI19::fur and pORI19::perR mutants. The topB, marR, met, and sigH pORI19 integrants demonstrated similar results to those for the wild type and have been omitted for clarity. Overall, the data indicate that plasmid integration does not nonspecifically affect the growth rate of mutants.
FIG. 3.
Growth of wild-type L. monocytogenes EGDe (▪), met (▴), fur (•), marR (□), perR (▵), and sigH (○) mutants in BHI at 37°C (A), adjusted to pH 9.0 (B), containing 5% ethanol (C), and adjusted to pH 5.5 (D). (E) Numbers of wild-type and mutant cells after 30 h of growth in CDM. Selected mutants with growth curves similar to the wild type have been eliminated from graphs for clarity. Error bars represent the standard deviations from the mean of triplicate experiments.
An objective of the present study was to establish the effect of specific mutations upon the ability of L. monocytogenes to react to suboptimal stress conditions encountered in vitro and in vivo. Stress conditions analyzed included alterations in pH and ethanol concentrations and growth in minimal medium (Fig. 3). In all cases, mutants that behaved in a similar manner to wild-type cells have been omitted from selected graphs for clarity. Under all stress conditions tested the topB mutant behaved in a manner similar to that of the wild type. Growth of the met mutant was significantly impaired under alkaline conditions and was the most significantly affected mutant in the presence of ethanol, indicating a likely role for the putative methyltransferase under these conditions. The MarR regulator analyzed in the present study was previously demonstrated to be induced in L. monocytogenes by low pH (39). However, although the marR mutant was not affected in growth at low pH, it was dramatically impaired in growth at alkaline pH. Moreover, the marR mutant was impaired in growth in ethanol, indicating a role under general oxidative stress. As stated previously, further study is required to determine whether plasmid integration into the marR or met loci also affects the expression of adjacent genes.
The alternative sigma factor Sigma-H has been shown to be induced in L. monocytogenes by low pH (39) and is expressed upon entry into sporulation in B. subtilis (4, 16). The sigH mutant in L. monocytogenes did not demonstrate a discernible phenotype when grown at an acid pH or in the presence of 5% ethanol but was impaired in growth under alkaline conditions. Although the mutant was not affected in normal growth in complex medium, it was significantly affected in growth in minimal medium and failed to reach wild-type levels after 30 h (Fig. 3E). The data are consistent with a role for Sigma-H in acquisition or utilization of nutrients in minimal medium; a function that is not required in complex media. This phenotype was confirmed by using an in-frame deletion mutant in sigH (data not shown).
In gram-negative organisms Fur plays a role in resistance to low pH (3, 12, 13). In L. monocytogenes the fur mutant was not affected in growth at low pH. However, growth of the fur mutant was significantly affected at alkaline pH and in the presence of ethanol. Susceptibility to ethanol-mediated stress may reflect a role for this transcriptional regulator in general oxidative stress resistance, as has been demonstrated previously for Fur in S. aureus (22, 23).
Virulence studies.
The virulence of the six mutants in comparison to the wild-type was assessed by using the i.p. mouse model (Fig. 4A). The number of bacteria in the spleens of infected mice was determined after 3 days. The topB, met, and marR mutants showed no significant difference from the wild type as determined by the Student t test. The results suggest that these loci are not required for optimal virulence potential and indicate that pORI19 plasmid integration does not nonspecifically affect virulence in mutant strains. The sigH mutant was isolated at fivefold-lower levels (P < 0.05) than the wild type, suggesting a minor, although statistically significant, role for components of this regulon in maintaining optimal virulence potential in L. monocytogenes. In contrast, the fur and perR mutants reached much lower levels than the wild type in the spleens of infected animals (P < 0.05). Numbers recovered from the spleens for these two mutants were 10-fold less than that of the wild-type. Repeat experiments verified that in all cases both pORI19 mutants were isolated at significantly lower levels than the wild-type and indicated that the pathogenesis of the fur mutant was most significantly affected (data not shown).
FIG. 4.
(A) Effect of disruption mutations on the survival of L. monocytogenes EGDe in vivo. Mice were injected i.p. with either the wild type or mutants, and the numbers of bacteria recovered from the spleen were determined 3 days postinoculation. (B) The ability of the pORI19::fur (▵) and pORI19::perR (•) mutants to survive in vivo in comparsion to the wild-type (▪) was assessed over 4 days. Numbers in the spleens of infected animals was determined daily. (C) Confirmation of a role for PerR and Fur in virulence by using in-frame deletion mutants. Δfur and ΔperR mutants were used to infected mice by the i.p. route, and numbers of bacteria in the spleens of infected mice were determined 3 days postinfection. Error bars represent the standard deviations from the mean (n = 4). An asterisk indicates that the means are significantly different from the wild type (P < 0.05).
On the basis of these results a more detailed virulence study was carried out, incorporating the wild type and two of the most significantly affected pORI19 integrants, fur and perR. The bacterial load in the spleens of infected mice was monitored over four consecutive days (Fig. 4B). At day 1 no significant difference was observed between the two mutants and the wild type. This suggests that both perR and fur mutants are not affected in their ability to survive the initial influx of neutrophils and macrophages that infiltrate the peritoneal cavity in response to L. monocytogenes infection (14). On the second day a significant (P < 0.05) difference began to emerge with the wild type reaching higher numbers than the two mutants. A similar trend was observed on days 3 and 4, with the perR mutant failing to reach the same high levels as the wild type in infected spleens. This may reflect, at least in part, the growth defect for the perR mutant seen in complex media. In particular, the fur mutant was significantly impaired in growth potential in the spleens of infected mice and failed to replicate significantly over days 3 and 4 postinfection. The data demonstrate that both Fur and PerR activity are essential for full virulence in L. monocytogenes and that loss of Fur activity in particular severely reduces virulence potential. Listeria mutants isolated from mice were routinely analyzed for the presence of the integrated plasmid (100 colonies analyzed), and no revertants were detected, indicating the inherent stability of the gene disruptions throughout the infection studies.
In order to confirm the role of PerR and Fur in virulence of L. monocytogenes, defined, in-frame deletion mutants were created by using the SOE procedure (see Materials and Methods). The virulence potential of the deletion mutants was analyzed by using the mouse model. Both Δfur and ΔperR mutants demonstrated significantly reduced numbers in the spleens of infected mice relative to wild-type bacteria (Fig. 4C). The data confirm a specific role for PerR and Fur in virulence of L. monocytogenes and indicate the validity of using the pORI19 system for initial postgenomic analysis of genes in this organism.
Macrophage assay.
All six mutants and the wild type were assessed for their ability to grow in cultured J774 macrophages. Cells of this type have previously been utilized to determine specific virulence deficiencies in L. monocytogenes mutants (1). However, in all cases, growth potential was similar for both wild type and mutants (data not shown). In addition, perR and fur pORI19 mutants were further investigated for their ability to grow in mouse macrophages isolated from the peritoneal cavity. No significant difference was observed between these two mutants and the wild-type in primary mouse macrophages (data not shown). The data support the observation that both perR and fur mutants are resistant to the initial influx of macrophages that occurs after i.p. infection of mice. Futhermore, the data suggest that in vitro macrophage studies, which utilize nutrient-rich medium components, may not substitute for murine assays in which sequestration of iron and nutrients comprise a significant hurdle for infecting bacteria.
Sensitivity of perR and fur mutants to hydrogen peroxide and low iron.
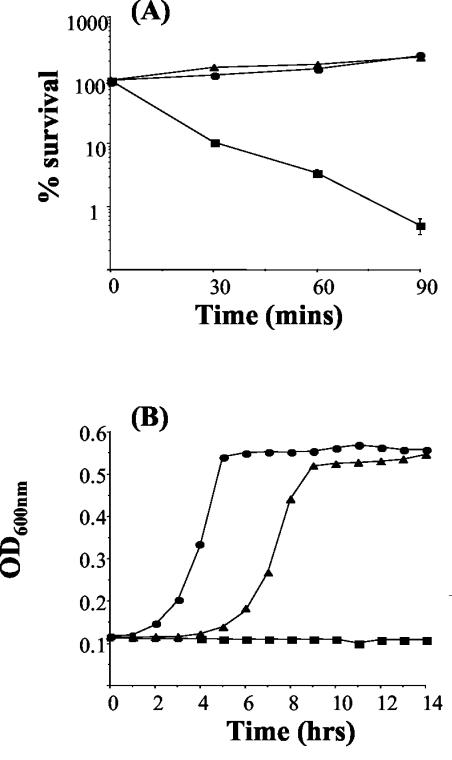
Given the reduced virulence of perR and fur mutants for mice infected by the i.p. route, we analyzed the response of these mutants to further in vivo-associated stress conditions. At lethal levels of hydrogen peroxide (50 mM) numbers of wild-type cells declined significantly over a 90-min period. However, both mutants were not only able to tolerate this level of H2O2 but some growth was observed (Fig. 5A). Furthermore, at levels of hydrogen peroxide (22 mM) that are inhibitory for wild-type cells, both mutants were capable of growth, with the perR mutant exhibiting greater resistance under these conditions than the fur mutant (Fig. 5B). The data indicate that disruption of either perR or fur leads to a significant increase in hydrogen peroxide resistance in L. monocytogenes. This differs from the situation in S. aureus where perR mutants, but not fur mutants, demonstrate increased hydrogen peroxide resistance (22, 23).
FIG. 5.
(A) Effect of 50 mM hydrogen peroxide on L. monocytogenes EGDe (wild-type) (▪), fur (▴), and perR (•) mutants. Cells were in the exponential growth phase. (B) Growth of EGDe (▪), fur (▴), and perR (•) mutants in BHI containing 22 mM hydrogen peroxide. The error bars represent the standard deviations from the means of duplicate experiments.
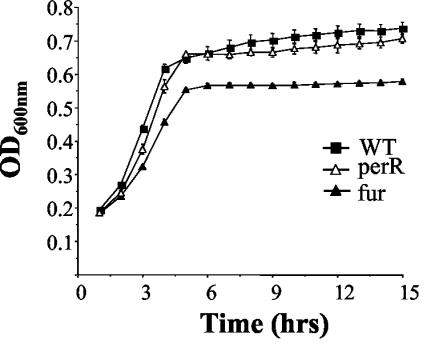
Both Fur and PerR play a role in acquisition and storage of iron in S. aureus (22, 23). We therefore examined the ability of perR and fur mutants to grow in BHI depleted of iron through the addition of tropolone and iron-limited through the addition of low levels (5 mM) of ferric citrate. At this limiting level of iron the perR mutant was capable of normal growth. However, growth of the fur mutant was restricted relative to the wild type, suggesting a significant role for Fur in sequestration of iron in low-iron environments (Fig. 6).
FIG. 6.
Growth of L. monocytogenes EGDe (▪), fur (▴), and perR (▵) mutants in BHI plus 160 μM tropolone plus 5 mM ferric citrate. Error bars represent the standard deviations from the mean of triplicate experiments.
In order to confirm the phenotypes of the pORI19 mutants, deletion mutants were tested for resistance to in vitro stress conditions as described previously. Both deletion mutants exhibited identical phenotypes to the respective pORI19 mutants (data not shown).
Virulence of perR and fur mutants in esculetin-treated mice.
The data from murine infection studies suggest that the growth potential of fur and perR mutants in internal organs is significantly reduced relative to the wild type. In addition, although resistance of the mutants to in vitro hydrogen peroxide activity is actually increased, the fur mutant is sensitive to low-iron conditions. To investigate the possible role of Fur and PerR in overcoming the conditions of iron starvation experienced during infection, we analyzed virulence in mice subjected to iron-overload by using esculetin. Esculetin has a high iron-binding ability and makes iron available for utilization by L. monocytogenes during infection. This results in a substantial increase in sensitivity to infection and a significant reduction in lethal dose (10). Esculetin can supply iron to L. monocytogenes EGDe to facilitate normal growth in complex media depleted of iron (Fig. 7A). Similar results were demonstrated for fur and perR mutants in vitro, suggesting that the mechanism of iron acquisition from esculetin is not controlled by Fur or PerR. This ability of esculetin to restore growth of wild-type L. monocytogenes was similar in medium containing 2 to 10 mM ferric citrate as the sole source of iron (data not shown).
FIG. 7.
(A) Growth of L. monocytogenes EGDe in BHI plus 160 μM tropolone (▪) and BHI plus 160 μM tropolone plus 1 mM esculetin (▴). Both fur and perR mutants demonstrated similar results (data not shown). (B) In vivo survival of L. monocytogenes EGDe and the fur and perR mutants in the presence of esculetin. Esculetin was administered to mice at day −1, day 0, and day +1. The numbers of L. monocytogenes recovered from the spleen were determined 3 days postinoculation. The error bars represent the standard deviations from the mean (n = 4). The asterisk indicates means are significantly different to the wild type (P < 0.05).
In the present study, the presence of systemic esculetin during infection rescued any virulence defect associated with the perR mutant (Fig. 7B). Furthermore, whereas the virulence of the fur mutant was not completely restored in the presence of esculetin, the difference between the wild type and mutant was not as evident as for normal mice. The data suggest that the growth deficiency of the fur and possibly the perR mutants is most likely related to an inability to sequester iron during infection. When exogenous iron is supplied in the form of esculetin the virulence defect of the fur mutant is ameliorated and that of the perR mutant is eliminated.
DISCUSSION
In this study we demonstrate that the pORI19 system first implemented in L. lactis by Law et al. (27) can be used to disrupt genes quickly and efficiently in L. monocytogenes, allowing rapid analysis of the roles of these genes in pathogenesis. The approach creates specific and stable gene disruptions that do not interfere with normal growth, do not nonspecifically affect virulence for mice, and can provide the basis for a “first look” functional analysis of the recently published L. monocytogenes genome (17). We have utilized a pORI19 mutant in the gene encoding listeriolysin (hly) to demonstrate that the system creates stable gene disruptions that do not appear to affect expression of downstream genes. As in the current study, genes of interest identified through pORI19 mutagenesis can subsequently be deleted by using a more time-consuming methodology to provide confirmation of mutant phenotypes.
Given the large number of genes in the L. monocytogenes genome that provide a putative regulatory function (17), we have targeted a number of such loci to determine their specific roles in adaptation to environmental stress and virulence potential. Loci were chosen based upon previous characterization as in vivo inducible (a putative topoisomerase III and a hypothetical S-adenosylmethionine (SAM)-dependent methyltransferase) (15), acid inducible (sigH and predicted marR family transcriptional regulator) (39), or due to the involvement of homologous loci in the pathogenesis of S. aureus (perR and fur) (22, 23).
The closest homologues of the predicted L. monocytogenes SAM-dependent methyltransferase (lmo0581) are hypothetical proteins in Bacillus anthracis and L. lactis subspecies lactis and ipa-19d in B. subtilis (26). Methyltransferases play a role in DNA methylation, protein-protein signaling, and biosynthesis of cellular components (25, 41, 48). Given the predicted role for DAM-dependent methyltransferases in regulation of bacterial virulence (19) and the fact that expression of this locus is induced during infection (15), we subjected the methyltransferase pORI19 insertional mutant to in vitro and in vivo analysis. Disruption of the region encoding the predicted methyltransferase resulted in extreme sensitivity to ethanol, a potent inducer of oxidative stress. However, the strain was not affected in virulence potential, either in mouse macrophage cells or in the murine model of infection, suggesting that this locus is not important for pathogenesis or that another gene in the genome can functionally substitute for lmo0581.
The topB gene is predicted to encode topoisomerase III, a type 1 topoisomerase with potent decatenating activity. The gene was previously identified as in vivo-inducible in L. monocytogenes as a result of an in vivo expression technology strategy (15). In other pathogens relaxation of DNA supercoiling has been shown to influence expression of virulence factors (32). Plasmid insertion into the topB gene did not affect cell viability, survival of specific environmental stress conditions or pathogenesis. It has been suggested that in E. coli topoisomerase III plays a role similar to that of topoisomerase IV, the principal decatenating enzyme in the cell (21) and as such mutations in this gene will allow the cell to remain viable. The current study suggests that although these putative methyltransferase and topoisomerase III genes are expressed by L. monocytogenes during infection, plasmid integration at these loci does not affect virulence potential.
Previous studies have demonstrated a correlation between acid tolerance and virulence potential in L. monocytogenes and other pathogens (34, 37, 44). We therefore analyzed the in vivo role of two transcriptional regulators, Sigma-H and a MarR family regulator previously shown to be acid inducible by using two-dimensional gel analysis (39). Disruption of marR significantly affected growth in ethanol and alkaline stress, but not at low pH or during infection. This locus is located upstream of a gene (lmo1617) annotated as encoding a putative multidrug efflux transporter, and the genes are not separated by a terminator. Further analysis is therefore required to determine whether the stress-sensitive phenotype established here for the MarR mutation is due to elimination of MarR activity alone or to the added effect of reducing or eliminating expression from lmo1617.
In B. subtilis Sigma-H is expressed upon entry into sporulation (4, 16). The sigma factor did not appear to play a role in L. monocytogenes growth in ethanol or low pH stress but was necessary for growth under alkaline conditions and was necessary for efficient growth in minimal medium. This growth defect in minimal medium has been confirmed by using a deletion mutant in sigH (data not shown). The pORI19::sigH mutant demonstrated a marginal (fivefold) reduction in virulence potential, as measured by growth in the spleens of infected mice. In Mycobacterium tuberculosis, Sigma-H has a subtle effect on the infectious process, influencing lethality for mice and immunity to infection but not growth in murine organs or within macrophages (24). From our study it is evident that Sigma-H does not have a dominant role in L. monocytogenes pathogenesis and that the marginally restricted growth of the sigH mutant in the spleens of infected mice may reflect an inability to acquire and utilize nutrients in vivo.
Strikingly, disruption of either perR or fur loci by using pORI19 integration significantly reduced the virulence of L. monocytogenes in the murine model of infection. This phenomenon was confirmed by creating in-frame deletion mutants in perR and fur. The data demonstrate that PerR and Fur are essential regulators facilitating in vivo growth of the pathogen and are absolutely required for full virulence potential. The results reflect recent studies in S. aureus demonstrating that both ferric uptake repressor analogues are required for virulence in the murine skin abscess model of infection (22, 23). Unlike the S. aureus fur mutant, mutation of this locus in L. monocytogenes did not affect normal growth in complex media. Interestingly, the L. monocytogenes PerR mutants demonstrated a growth defect in complex media. In addition, both perR and fur pORI19 mutants of L. monocytogenes were affected in growth in ethanol and under alkaline stress conditions. Given the role of Fur in the acid tolerance of gram-negative pathogens (3, 12, 13), it is interesting that the L. monocytogenes fur mutant failed to show a growth defect at low pH.
After murine infection, early translocation of both fur and perR mutants to the spleens of infected mice was similar to that seen for the wild type. This reflects resistance to the influx of neutrophils and macrophages that occurs after i.p. infection (14). Indeed, both mutants were unaffected in growth in J774 tissue cultured mouse macrophages or in primary mouse macrophages. In S. aureus the perR, but not fur, mutant demonstrates increased resistance to hydrogen peroxide, most likely through increased expression of catalase (22, 23). In L. monocytogenes, both fur and perR mutants demonstrated significantly increased hydrogen peroxide resistance. Given these findings, together with evidence that catalase is not essential for L. monocytogenes virulence (28), it is unlikely that the virulence defect of both fur and perR mutants is a result of a poor oxidative stress response.
In S. aureus, PerR and Fur also regulate iron uptake and storage (22, 23). We determined that the L. monocytogenes fur mutant is significantly affected in growth under iron-limiting conditions. Since iron limitation plays a major role in limiting outgrowth of L. monocytogenes during infection (47), we examined growth of L. monocytogenes mutants in the spleens of mice supplied with iron-bound esculetin. Under these conditions the virulence defect of the perR mutant was completely abolished, whereas the virulence potential of the fur mutant was increased relative to the wild type. Collectively, the data suggest that the reduction in virulence potential of the fur and perR mutants is due to deregulation of iron uptake or storage during infection rather than disruption of the oxidative stress response. A recent study in S. aureus has identified the surface-expressed Fur-regulated isd (iron-regulated surface determinant) gene products as a means of acquiring heme-associated iron during infection (31). We have identified homologues of the S. aureus Isd system as likely components of the Fur regulon in L. monocytogenes through analysis of the genome for predicted Fur binding regions (data not shown). Further study is under way to determine the nature of these and other genes regulated by Fur and PerR in L. monocytogenes.
In conclusion, we have demonstrated that the pORI19 insertional mutagenesis system can be used to create stable disruptions of genes in L. monocytogenes. Subsequent analysis of mutants in a murine model of infection permits rapid investigation of the role of specific genes in the pathogenesis of L. monocytogenes. As in the present study, confirmation of a role in virulence can be obtained through subsequent creation of in-frame deletions.
Acknowledgments
This study was funded by the Irish government under the National Development Plan 2000-2006.
Editor: V. J. DiRita
REFERENCES
- 1.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beauregard, K. E., K. D. Lee, R. J. Collier, and J. A. Swanson. 1997. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J. Exp. Med. 186:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijlsma, J. J. E., B. Waidner, A. H. M. van Vliet, N. J. Hughes, S. Häg, S. Bereswill, D. J. Kelly, C. M. J. E. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm, K., J. Kreft, M. T. Ripio, and J. A. Vazquez-Boland. 1996. Regulation of virulence gene expression in pathogenic Listeria. Microbiologia 12:219-236. [PubMed] [Google Scholar]
- 6.Casillas-Martinez, L., A. Driks, B. Setlow, and P. Setlow. 2000. Lack of a significant role for the PerR regulator in Bacillus subtilis spore resistance. FEMS Microbiol. Lett. 188:203-208. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury, R., G. K. Sahu, and J. Das. 1996. Stress response in pathogenic bacteria. J. Biosci. 21:149-160. [Google Scholar]
- 8.Cotter, P. D., N. Emerson, C. G. M. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal trasnduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 10.Coulanges, V., P. André, and D. J. M. Vidon. 1996. Esculetin antagonizes iron-chelating agents and increases the virulence of Listeria monocytogenes. Res. Microbiol. 147:677-685. [DOI] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, J. W. 1991. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J. Bacteriol. 173:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gahan, C. G., and J. K. Collins. 1995. Non-dystrophic 129 REJ mice are susceptible to i.p. infection with Listeria monocytogenes despite an ability to recruit inflammatory neutrophils to the peritoneal cavity. Microb. Pathog. 18:355-364. [DOI] [PubMed] [Google Scholar]
- 15.Gahan, C. G. M., and C. Hill. 2000. The use of listeriolysin to identify in vivo induced genes in the gram-positive intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 36:498-507. [DOI] [PubMed] [Google Scholar]
- 16.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 18.Goetz, M., A. Bubert, G. Wang, I. Chico-Calero, J. A. Vazquez-Boland, M. Beck, J. Slaghuis, A. A. Szalay, and W. Goebel. 2001. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc. Natl. Acad. Sci. USA 98:12221-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 20.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacterial. 185:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiasa, H., R. J. Digate, and K. J. Marians. 1994. Decatenating activity of Escherichia coli DNA gyrase and topoisomerase I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269:2093-2099. [PubMed] [Google Scholar]
- 22.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 11:8330-8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolosova, N., D. Sherman, D. Karlson, and N. Dudareva. 2001. Cellular and subcellular localization of S-adenosyl-l-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiol. 126:956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lrr, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Law, J., G. Buist, A. Haandrikann, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leblond-Francillard, M., J. L. Gaillard, and P. Berche. 1989. Loss of catalase activity in Tn1545-induced mutants does not reduce growth of Listeria monocytogenes in vivo. Infect. Immun. 57:2569-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Low, J. C., and W. Donachie. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153:9-29. [DOI] [PubMed] [Google Scholar]
- 30.Marron, L., N. Emerson, C. G. M. Gahan, and C. Hill. 1997. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl. Environ. Microbiol. 63:4945-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gomicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 32.McNairn, E., N. Ni Bhriain, and C. J. Dorman. 1995. Overexpression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: effect on virulence gene expression. Mol. Microbiol. 15:507-517. [DOI] [PubMed] [Google Scholar]
- 33.Mead, P., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Chapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 35.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair, S., I. Derré, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 37.O'Driscoll, B., C. G. M. Gahan, and C. Hill. 1996. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 62:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 39.Phan-Thanh, L., and F. Mahouin. 1999. A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis 20:2214-2224. [DOI] [PubMed] [Google Scholar]
- 40.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rando, R. R. 1996. Chemical biology of protein isoprenylation/methylation. Biochim. Biophys. Acta 1300:5-16. [DOI] [PubMed] [Google Scholar]
- 42.Rouquette, C., M. T. Ripio, E. Pellegrini, J. M. Bolla, R. I. Tasion, J. A. Vasquez-Boland, and P. Berche. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977-987. [DOI] [PubMed] [Google Scholar]
- 43.Rouquette, C., C. de Chastellier, S. Nair, and P. Berche. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235-1245. [DOI] [PubMed] [Google Scholar]
- 44.Saklani-Jusforgues, H., E. Fontan, and P. L. Goossens. 2000. Effect of acid-adaptation on Listeria monocytogenes survival and translocation in a murine intragastric infection model. FEMS Microbiol. Lett. 193:155-159. [DOI] [PubMed] [Google Scholar]
- 45.Shetron-Rama, L. M., H. Marquis, H. G. Bouwer, and N. E. Frietag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70:1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleator, R. D., J. Wouters, C. G. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sword, C. P. 1966. Mechanisms of pathogenesis in Listeria monocytogenes infection. I. Influence of iron. J. Bacteriol. 92:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urnov, F. D. 2002. Methylation and the genome: the power of a small amendment. J. Nutr. 132:2450S-2456S [DOI] [PubMed] [Google Scholar]
- 49.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wemekamp-Kamphuis, H. H., J. A. Wouters, R. D. Sleator, C. G. M. Gahan, C. Hill, and T. Abee. 2002. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]