Abstract
Staphylococcus epidermidis is a normal constituent of the healthy human microflora, but it is also the most common cause of nosocomial infections associated with the use of indwelling medical devices. Isolates from device-associated infections are known for their pronounced phenotypic and genetic variability, and in this study we searched for factors that might contribute to this flexibility. We show that mutator phenotypes, which exhibit elevated spontaneous mutation rates, are rare among both pathogenic and commensal S. epidermidis strains. However, the study revealed that, in contrast to those of commensal strains, the genomes of clinical S. epidermidis strains carry multiple copies of the insertion sequence IS256, while other typical staphylococcal insertion sequences, such as IS257 and IS1272, are distributed equally among saprophytic and clinical isolates. Moreover, detection of IS256 was found to be associated with biofilm formation and the presence of the icaADBC operon as well as with gentamicin and oxacillin resistance in the clinical strains. The data suggest that IS256 is a characteristic element in the genome of multiresistant nosocomial S. epidermidis isolates that might be involved in the flexibility and adaptation of the genome in clinical isolates.
Staphylococcus epidermidis is a normal constituent of the healthy human skin and mucosal microflora. In recent decades, however, the bacterium has emerged as a nosocomial multiresistant pathogen and is now the most common cause of device-associated infections. Little is known of the factors that have contributed to this development, but the increasing number of immunocompromised patients, the use of indwelling medical devices, and a high selective pressure by antibiotics offer bacteria a novel ecological niche. It is unclear why just staphylococci were able to occupy this niche and by which factors pathogenic S. epidermidis differ from their commensal counterparts. In recent years, it has been shown that the ability to form biofilms on medical devices is a characteristic feature of nosocomial S. epidermidis isolates. Moreover, clinical S. epidermidis isolates exhibit an extraordinarily high phenotypic and genotypic flexibility. Thus, variants of the same parent strain can differ in terms of colony morphology, growth rate, hemolysis, biofilm formation, and antibiotic susceptibility (4, 7). The molecular mechanisms involved in this phenomenon are poorly understood, but it is assumed that the generation of phenotypic and genotypic variants is an evolutionary advantage that helps staphylococci to adapt to changing environmental conditions. The purpose of this study was therefore to search for genetic factors and mechanisms in clinical S. epidermidis that might contribute to this process. Previous studies have shown that staphylococcal biofilm formation is a highly variable factor which is influenced by both regulatory processes and genetic mechanisms such as phase variations, mutations, and chromosomal rearrangements (5, 10, 26, 32-34). The observation that some of these genetic processes are mediated by the action of insertion sequence (IS) elements prompted us to investigate the distribution of common staphylococcal IS elements among S. epidermidis strains of clinical and commensal origin. Moreover, we analyzed the relationship between IS presence, antibiotic resistance, and biofilm formation as well as the spontaneous mutation rate in this important nosocomial pathogen.
Bacterial strains.
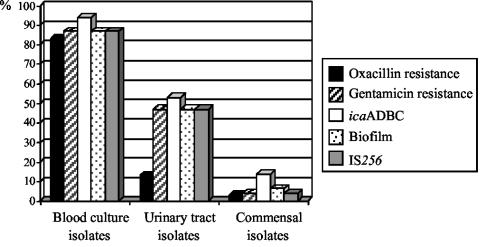
In this study, a total of 230 S. epidermidis strains, 139 of commensal origin and 91 clinical isolates (53 blood culture isolates and 38 isolates from urinary tract infections), were analyzed. Commensal strains were obtained by swabbing of the anterior nares of randomly selected outpatients who attended medical practitioners in the southwestern area of Germany. Patients with a hospitalization record or any other contact with a medical facility during a period of 3 months were excluded from the study. Blood culture isolates were recovered from intravenous catheter-related septicemia, and nosocomial urinary tract isolates were isolated from hospitalized patients suffering from catheter-associated urinary tract infections. Species diagnosis was verified by biochemical characterization using the API-20-Staph (bioMérieux, Marcy l'Etoile, France) system. All strains were tested for oxacillin resistance by growth on Mueller-Hinton agar supplemented with 3% sodium chloride and 6 μg of oxacillin/ml after a prolonged incubation period of 2 days at 30°C. There was a significant difference in terms of oxacillin resistance between clinical and saprophytic isolates (P < 0.001). Forty-four of 53 strains (83%) among the blood culture isolates and 5 of 38 strains (13%) among the urinary tract isolates were found to be resistant to oxacillin. Only 4 of the 139 commensal strains (3%) exhibited resistance to this β-lactam antibiotic (see Fig. 2). mecA-specific PCR confirmed the presence of the resistance-mediating mecA gene in all oxacillin-resistant isolates, while susceptible strains lacked this genetic information (data not shown).
FIG. 2.
Antibiotic resistance, biofilm formation, detection of the icaADBC operon, and IS256 presence in clinical and commensal S. epidermidis strains.
Detection of IS256, IS257, and IS1272.
In this study, we wanted to answer the question of whether pathogenic and nonpathogenic S. epidermidis differ with respect to the presence of IS elements in their genomes. We investigated the distribution of three typical IS elements which have been described previously as components of staphylococcal genomes, i.e., IS256, IS257, and IS1272. IS256 was initially described as the flanking region of the composite aminoglycoside resistance-mediating transposon Tn4001 (2). But the element also occurs in multiple, independent copies in the genomes of staphylococci and enterococci (9, 27). In previous studies, it was shown that IS256 can be involved in phase variation of biofilm formation in S. epidermidis (3, 34). IS257 is associated with the trimethoprim resistance-mediating transposon Tn4003 and numerous other resistance genes and plasmids in staphylococci (e.g., cadmium resistance) (6). Isoforms of the element are also detectable on the SCCmec element in S. aureus and S. epidermidis (17). IS1272 is detectable in many staphylococcal species and is prevalent in multiresistant clinical isolates (1, 18). IS elements were detected by IS-specific PCRs and Southern blotting. Amplification of IS256 was performed using primers 5′-TGAAAAGCGAAGAGATTCAAAGC-3′ and 5′-ATGTAGGTCCATAAGAACGGC-3′ (GenBank accession no. of the published sequence, M18086). An IS257-specific gene probe was generated by combining primers 5′-GCTAATTTCGTGGCATGGCG-3′ and 5′-GTTATCACTGTAGCCGTTGG-3′ (accession no. X53952). S. epidermidis RP62A chromosomal DNA was used as a template. IS1272 was amplified with primers 5′-GCTCGTTGAGCTACTTTTC-3′ and 5′-CCTAGAGAAATAGCCAGTAAATG-3′ (accession no. U35635) using S. haemolyticus 206 chromosomal DNA as a template. The gene probes were checked by nucleotide sequencing before they were applied for Southern hybridizations.
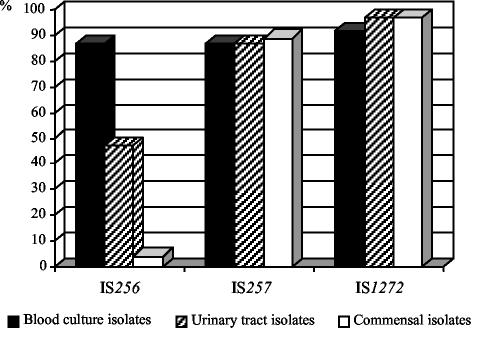
Figure 1 illustrates the distribution of IS256, IS257, and IS1272 among the commensal and clinical strains. Interestingly, IS256 was prevalent in 46 of 53 (87%) blood culture isolates and 18 of 38 (47%) urinary tract isolates but was prevalent in only 6 of 139 (4%) saprophytic strains. Statistical analysis using the chi-square test revealed significant differences in the distribution of IS256 in blood culture isolates (P < 0.001) and urinary tract strains (P < 0.001) compared with that of the commensal strains. In contrast, no significant differences were recorded for the distribution of IS257 and IS1272 among clinical and saprophytic strains, respectively. Here, 47 of 53 (87%) blood culture isolates, 33 of 38 (87%) urinary tract isolates, and 124 of 139 (89%) saprophytic strains carried IS257 copies. Similar results were obtained with respect to IS1272, which was detectable in 49 of 53 (92%) blood culture isolates, 37 of 38 (97%) urinary tract isolates, and 136 of 139 (97%) saprophytic strains. From these data, we conclude that IS256 represents a specific element which is more likely to occur in clinical S. epidermidis strains than in commensal isolates.
FIG. 1.
Distribution of IS256, IS257, and IS1272 among 230 S. epidermidis isolates from different origins. The study comprised 53 isolates recovered from blood cultures, 38 isolates obtained from catheter-related urinary tract infections, and 139 commensal strains obtained from nasal swabs of healthy volunteers.
Resistance towards gentamicin and detection of Tn4001 and free IS256 copies.
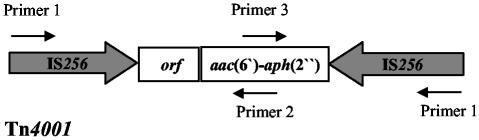
Normally, IS256 is part of the composite transposon Tn4001, which mediates gentamicin resistance by the bifunctional aminoglycoside-modifying enzyme AAC(6′)-APH(2′′). To investigate whether or not the IS256-positive strains carry Tn4001, the isolates were tested for gentamicin resistance by the agar diffusion method according to the Deutsche Industrienorm 58940 guidelines and by an aac(6′)-aph(2′′)-specific PCR using the primers 5′-GTATTAGAATTTTATGGTGG-3′ and 5′-CCATACATTCTTAATATATC-3′ under the following conditions: 1 min at 95°C, 1 min at 44°C, and 1.5 min at 68°C for 30 cycles. The resulting 1,184-bp PCR fragment was visualized by agarose gel electrophoreses and ethidium bromide staining. Resistance towards gentamicin was recorded in 46 of the 53 (87%) blood culture isolates, 18 of the 38 (45%) urinary tract isolates, and 6 of the 139 (4%) commensal strains (Fig. 2). Remarkably, 48 of 70 (68%) of the gentamicin-resistant strains, i.e., 41 of 44 (93%) blood culture isolates, 5 of 5 (100%) urinary tract isolates, and 2 of 4 (50%) oxacillin-resistant commensal strains were concomitantly resistant to oxacillin. In all gentamicin-resistant strains, the aac(6′)-aph(2′′) gene was detectable by PCR, which implicates Tn4001 in mediation of the aminoglycoside-resistant phenotype in the IS256-positive strains. Interestingly, in a recent study on other coagulase-negative staphylococci, Tn4001-like elements were identified in which the IS256 copies at the ends of the transposon were largely truncated and replaced by IS257 (19). To search for such truncated Tn4001 forms, we analyzed the 70 gentamicin-resistant strains by PCR using primers that cover the IS256 and the aac(6′)-aph(2′′) genes (Fig. 3). When the entire Tn4001 transposon is present, PCRs combining primer 1 (5′-TGAAAAGCGA AGAGATTCAAAGC-3′) and primer 2 (5′-CTAAACCGTGCATTTGTCTTA-3′) result in a DNA fragment of approximately 2.5 kb in size, while a PCR using primer 1 and primer 3 (5′-TTTAAGACAAATGCACGGTTTAG-3′) will amplify a 1.7-kb fragment. In 63 of 70 (90%) gentamicin-resistant isolates, both the 2.5- and 1.7-kb fragments were detectable, indicating the presence of the entire Tn4001 transposon in these strains. Interestingly, in seven strains (two from blood cultures, four from urinary tract infections, and one from commensal isolates) no PCR products or smaller fragments were amplified, suggesting that these strains might harbor similar truncated Tn4001 forms, as described recently by Lange et al. (19) (data not shown). However, more-detailed studies are necessary to support this hypothesis.
FIG. 3.
Scheme of Tn4001 and position of primers used for the detection of the transposon. (Please note that the image is not drawn to scale.)
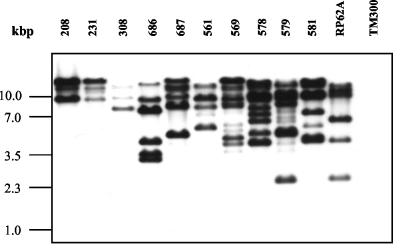
Southern hybridization of EcoRI-restricted chromosomal DNA with an IS256-specific gene probe revealed in 76% (53 of 70) of the aac(6′)-aph(2′′)-positive strains additional IS256-specific fragments which were unrelated to Tn4001, suggesting the existence of multiple free IS256 copies of the element in these genomes (Fig. 4). The number of IS256-hybridizing fragments ranged from 1 (Tn4001-associated) up to 11. Recently, it has been shown that IS256 transposes by a mechanism which involves circularization of the element and tandem dimer formation at the Tn4001 ends (21, 24). Although the molecular mechanism of IS256 transposition is not yet fully understood, it is tempting to speculate that it involves replication of the element. The two flanking IS256 copies at the ends of Tn4001 might therefore represent the origin of the multiple copies in S. epidermidis genomes.
FIG. 4.
Detection of multiple IS256 copies by IS256-specific Southern hybridization of EcoRI-digested chromosomal DNA of different gentamicin-resistant S. epidermidis strains (208 to 581). S. epidermidis RP62A and S. carnosus TM300 were used as positive and negative controls, respectively.
icaADBC detection, biofilm formation, and IS256.
Previous studies have shown that IS256 can influence the expression of biofilm formation in S. epidermidis. Staphylococcal biofilm formation is mainly mediated by the expression of the icaADBC operon, which encodes enzymes for the production of the polysaccharide intercellular adhesin PIA (16, 22). PIA expression undergoes phase variation, and in a substantial part of the variants this process is mediated by the alternating insertion and excision of IS256 in an insertion hot spot of the ica operon (34). Moreover, the element seems to be involved in large rearrangements of the S. epidermidis genome which also affect biofilm formation and aminoglycoside resistance expression (32). To investigate a possible association between the icaADBC operon, biofilm formation, and IS256, all strains were tested for the presence of the icaADBC operon and biofilm formation as described previously (3, 33). The ica operon was detected in 94% (50 of 53) of the blood culture isolates and 53% (20 of 38) of the urinary tract isolates but in only 14% (20 of 139) of the saprophytic strains (Fig. 2). Statistical analysis of the data revealed that the ica operon is significantly more prevalent in strains from clinical origin than in commensal isolates (P < 0.001). After growth in tryptic soy broth supplemented with 3% sodium chloride, 47 of the 53 (87%) blood culture isolates, 18 of the 38 (47%) urinary tract strains, and 9 of the 139 (6.5%) commensal strains formed a detectable biofilm on polystyrene tissue culture plates (Fig. 2). A significant correlation was found between icaADBC presence and biofilm expression in the clinical strains (P < 0.01). Thus, 47 of the 50 (94%) ica-positive blood culture strains, 16 of the 20 (80%) ica-positive urinary tract isolates, and 7 of the 20 (35%) ica-positive commensal strains formed a visible biofilm on polystyrene tissue culture plates. Among the blood culture isolates, 3 isolates remained biofilm negative even though they carried the entire icaADBC operon, which was also the case with 4 urinary tract isolates and 13 ica-positive commensal strains. Among the commensal and urinary tract isolates, two strains were identified which exhibited a biofilm without the presence of the icaADBC operon, indicating that other factors might be involved in S. epidermidis biofilm formation as well. The data confirmed previous results from different studies indicating that biofilm formation and icaADBC presence are highly discriminating factors between clinical and commensal S. epidermidis (11, 12, 33). Interestingly, we observed that the majority of clinical strains carrying the icaADBC operon concomitantly harbored IS256 in their genomes (P ≤ 0.05). Thus, among the 50 ica-positive blood culture isolates, 88% (44 of 50) concomitantly carried IS256. A similar situation was observed with the urinary tract isolates. Here, we found among the 20 ica-positive strains 17 (85%) which were IS256 positive. But only 3 of the 20 ica-positive commensal strains (15%) harbored IS256 in their genomes (Fig. 2).
Determination of the spontaneous mutation rate of commensal and clinical S. epidermidis.
The contribution of an elevated mutation rate to the virulence and adaptation of pathogens has been intensively and controversially discussed during recent years (8, 14, 15). Mutator phenotypes are supposed to have adaptive advantages and play a role in the pathogenesis of bacterial infections as well as in the development of antibiotic resistance (20, 23). Recently, hypermutable strains were identified in macrolide-resistant S. aureus isolates from cystic fibrosis patients (25). Moreover, defects in mismatch repair systems which result in an elevated mutation rate have been shown to contribute to the development of vancomycin resistance in S. aureus (29). Therefore, we wanted to investigate whether or not mutator phenotypes were detectable among our S. epidermidis isolates. Moreover, we wanted to elucidate possible differences in the mutation rates between the antibiotic-susceptible commensal strains and the multiresistant clinical strains. For this purpose, we determined the spontaneous mutation rate in the rpoB gene, which confers resistance to rifampin (23). The mutation rate per cell and generation was calculated as described previously (13, 34). Thus, a single bacterial colony was diluted in 45 ml of phosphate-buffered saline. One hundred microliters of this suspension was plated on agar plates to determine the inoculum size, and another 100-μl aliquot was used to inoculate 50 ml of Luria-Bertani broth, which was grown at 37°C to an optical density at 600 nm of 1.3 to 1.5. From this bacterial culture, 100-μl aliquots and appropriate dilutions were spread on Mueller-Hinton agar plates containing 10 μg of rifampin/ml. CFU were counted after a 24-h incubation at 37°C. The mutation frequency (P) to rifampin resistance per cell and generation was calculated using the formula
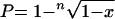 |
where n is the number of generations and x is the number of rifampin-resistant colonies/total number of plated colonies. The number of generations (n) was determined using the equation
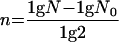 |
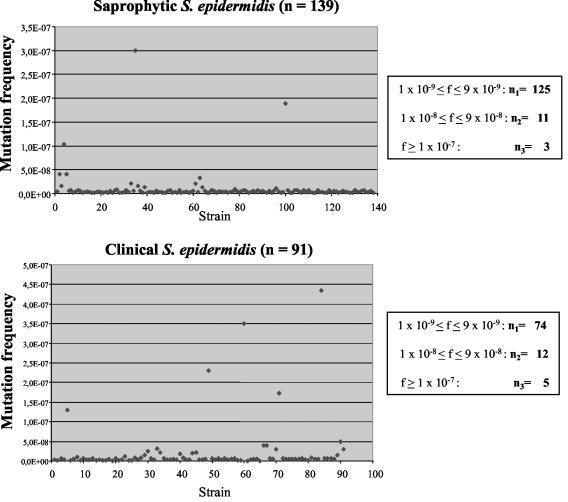
where N is the total number of bacterial cells in the culture and N0 is the number of bacterial cells in the inoculum. Figure 5 indicates that the average mutation rate in the majority of the isolates is relatively low (approximately 10−9). Of the 230 strains tested, 23 (10%) exhibited slightly elevated mutation rates of 10−8, and only 8 strains (3.5%) had a spontaneous frequency greater than 10−7. No difference was detectable between commensal and clinical strains with respect to the mutation rate. Moreover, we found no correlation between the slightly higher mutation rate and the presence of IS256 in these strains. The data demonstrate that in both pathogenic, multiresistant S. epidermidis strains and nonpathogenic, antibiotic-susceptible S. epidermidis strains mutator phenotypes are rare. These findings suggest that in S. epidermidis strains from device-associated infections, point mutations might play a minor role in the pathogenesis of these infections.
FIG. 5.
Spontaneous mutation frequencies per cell and generation of the commensal and clinical S. epidermidis strains toward rifampin resistance.
Conclusions.
Staphylococcus epidermidis is now a common pathogen which has been successfully established in the hospital environment. The majority of nosocomial S. epidermidis infections are catheter-related bloodstream infections, but the organism has also now emerged as a cause of urinary tract infections, preferentially in elderly, hospitalized patients carrying indwelling urinary tract catheters (28, 30, 31). Because staphylococci are also natural inhabitants of the skin, it is often difficult to decide whether an isolate represents the causative agent of an infection or an unspecific contamination of the specimen. Since this is specifically true for urinary tract infections, we also included in this study a group of nosocomial S. epidermidis isolates obtained from proven catheter-associated urinary tract infections from hospitalized patients. The data indicate that, in addition to the known differences in terms of biofilm formation and icaADBC presence, other factors exist which seem to be characteristic for pathogenic S. epidermidis isolates, from both line-associated septicemia and catheter-related urinary tract infections. Thus, a high proportion of the clinical isolates is resistant to oxacillin and gentamicin, and multiresistance in S. epidermidis is often accompanied by the presence of the biofilm-mediating icaADBC operon, which is expressed in the majority of the strains. The existence of ica-positive and ica-negative S. epidermidis strains raises the question of the origin of the biofilm-mediating icaADBC operon. It is conceivable that ica-positive S. epidermidis represent a single clone which acquired this genetic information and from which all biofilm-forming strains have evolved. Another possibility is the spread of the genes by horizontal gene transfer into different genetic backgrounds, and finally, it is possible that the ica-negative S. epidermidis represent deletion mutants in which the ica genes got lost. So far, there is no evidence to support any of these hypotheses, and more experimental work is needed to answer this important question in the future.
In contrast to other pathogens, where elevated mutation rates were proven to play a role in pathogenesis (20, 23), our study revealed no clue for an involvement of mutator phenotypes in device-associated S. epidermidis infections. However, the strains differed significantly with respect to the occurrence of IS elements in their genomes. Specifically, the presence of IS256 seems to be a feature of pathogenic S. epidermidis strains. In our strain collection, IS256 was exclusively associated with Tn4001, but the element also occurred independently in multiple free copies in these strains. In general, mobile genetic elements, such as insertion sequences, transposons, phages, and genomic islands, are common components of microbial genomes. Together with point mutations, homologous recombination, and horizontal gene transfer, mobile DNA elements are driving forces for the generation of novel genetic and phenotypic variants. It is tempting to speculate that the presence of multiple IS256 copies might play a role in the flexibility of the genome of multiresistant, biofilm-forming S. epidermidis isolates. This model could represent an advantage in the rapid adaptation of the bacterium to changing environmental conditions, and the underlying genetic mechanisms and effects therefore merit further detailed analyses.
Acknowledgments
This work was supported by grant SFB479 of the University of Würzburg.
We thank Stephanie Waeckerle for excellent technical assistance and Jörg Hacker for helpful discussions and for his support.
Editor: J. N. Weiser
REFERENCES
- 1.Archer, G. L., J. A. Thanassi, D. M. Niemeyer, and M. J. Pucci. 1996. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 40:924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 3.Cho, S. H., K. Naber, J. Hacker, and W. Ziebuhr. 2002. Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int. J. Antimicrob. Agents 19:570-575. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrance, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J. Infect. Dis. 161:1153-1169. [DOI] [PubMed] [Google Scholar]
- 5.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crupper, S. S., V. Worrell, G. C. Stewart, and J. J. Iandolo. 1999. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181:4071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deighton, M., S. Pearson, J. Capstick, D. Spelman, and R. Borland. 1992. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J. Clin. Microbiol. 30:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denamur, E., S. Bonacorsi, A. Giraud, P. Duriez, F. Hilali, C. Amorin, E. Bingen, A. Andremont, B. Picard, F. Taddei, and I. Matic. 2002. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184:605-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyke, K. G., S. Aubert, and N. el Solh. 1992. Multiple copies of IS256 in staphylococci. Plasmid 28:235-246. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick, F., H. Humphreys, E. Smyth, C. A. Kennedy, and J. P. O'Gara. 2002. Environmental regulation of biofilm formation in intensive care unit isolates of Staphylococcus epidermidis. J. Hosp. Infect. 52:212-218. [DOI] [PubMed] [Google Scholar]
- 11.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 13.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud, A., I. Matic, M. Radman, M. Fons, and F. Taddei. 2002. Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob. Agents Chemother. 46:863-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud, A., M. Radman, I. Matic, and F. Taddei. 2001. The rise and fall of mutator bacteria. Curr. Opin. Microbiol. 4:582-585. [DOI] [PubMed] [Google Scholar]
- 16.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, N., S. Urasawa, N. Uehara, and N. Watanabe. 1999. Distribution of insertion sequence-like element IS1272 and its position relative to methicillin resistance genes in clinically important staphylococci. Antimicrob. Agents Chemother. 43:2780-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange, C. C., C. Werckenthin, and S. Schwarz. 2003. Molecular analysis of the plasmid-borne aacA/aphD resistance gene region of coagulase-negative staphylococci from chickens. J. Antimicrob. Chemother. 51:1397-1401. [DOI] [PubMed] [Google Scholar]
- 20.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 21.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 184:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 24.Prudhomme, M., C. Turlan, J. P. Claverys, and M. Chandler. 2002. Diversity of Tn4001 transposition products: the flanking IS256 elements can form tandem dimers and IS circles. J. Bacteriol. 184:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclercq. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 26.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, L. B., and S. H. Marshall. 1994. Insertion of IS256-like element flanking the chromosomal β-lactamase gene of Enterococcus faecalis CX19. Antimicrob. Agents Chemother. 38:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 29.Schaaff, F., A. Reipert, and G. Bierbaum. 2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagenlehner, F. M., S. Krcmery, C. Held, I. Klare, W. Witte, A. Bauernfeind, I. Schneider, and K. G. Naber. 2002. Epidemiological analysis of the spread of pathogens from a urological ward using genotypic, phenotypic and clinical parameters. Int. J. Antimicrob. Agents 19:583-591. [DOI] [PubMed] [Google Scholar]
- 31.Wagenlehner, F. M., A. Niemetz, A. Dalhoff, and K. G. Naber. 2002. Spectrum and antibiotic resistance of uropathogens from hospitalized patients with urinary tract infections: 1994-2000. Int. J. Antimicrob. Agents 19:557-564. [DOI] [PubMed] [Google Scholar]
- 32.Ziebuhr, W., K. Dietrich, M. Trautmann, and M. Wilhelm. 2000. Chromosomal rearrangements affecting biofilm production and antibiotic resistance in a Staphylococcus epidermidis strain causing shunt-associated ventriculitis. Int. J. Med. Microbiol. 290:115-120. [DOI] [PubMed] [Google Scholar]
- 33.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziebuhr, W., V. Krimmer, S. Rachid, I. Loessner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]