Abstract
Salmonella enterica subspecies 1 serovar Typhimurium (serovar Typhimurium) induces enterocolitis in humans and cattle. The mechanisms of enteric salmonellosis have been studied most extensively in calf infection models. The previous studies established that effector protein translocation into host cells via the Salmonella pathogenicity island 1 (SPI-1) type III secretion system (TTSS) is of central importance in serovar Typhimurium enterocolitis. We recently found that orally streptomycin-pretreated mice provide an alternative model for serovar Typhimurium colitis. In this model the SPI-1 TTSS also plays a key role in the elicitation of intestinal inflammation. However, whether intestinal inflammation in calves and intestinal inflammation in streptomycin-pretreated mice are induced by the same SPI-1 effector proteins is still unclear. Therefore, we analyzed the role of the SPI-1 effector proteins SopB/SigD, SopE, SopE2, and SipA/SspA in elicitation of intestinal inflammation in the murine model. We found that sipA, sopE, and, to a lesser degree, sopE2 contribute to murine colitis, but we could not assign an inflammation phenotype to sopB. These findings are in line with previous studies performed with orally infected calves. Extending these observations, we demonstrated that in addition to SipA, SopE and SopE2 can induce intestinal inflammation independent of each other and in the absence of SopB. In conclusion, our data corroborate the finding that streptomycin-pretreated mice provide a useful model for studying the molecular mechanisms of serovar Typhimurium colitis and are an important starting point for analysis of the molecular events triggered by SopE, SopE2, and SipA in vivo.
Salmonella enterica is frequently isolated from wildlife and livestock and can cause diseases ranging from mild gastroenteritis to systemic infections. Humans are usually infected by consumption of contaminated animal products. S. enterica subspecies 1 serovar Typhimurium (serovar Typhimurium) infections are among the leading causes of bacterial food poisoning in industrialized countries. Interestingly, an S. enterica serovar can cause different diseases in different animal species (45, 47). For example, serovar Typhimurium causes a self-limiting enterocolitis in cattle and humans but a typhoid-like systemic infection in susceptible mouse strains.
Serovar Typhimurium employs a multitude of virulence factors to colonize and replicate within the host. These factors include a large array of adhesins, superoxide dismutases, flagella, resistance to complement and antimicrobial peptides, and two type III secretion systems (TTSS) (12, 19). These TTSS have attracted much attention because they enable bacteria to inject effector proteins directly into the host cell cytoplasm, where the proteins manipulate host cell signaling (14). Both S. enterica TTSS have been characterized extensively in susceptible mice (40). The TTSS encoded in Salmonella pathogenicity island 1 (SPI-1) plays a small but detectable role in breaching the intestinal epithelium, while the SPI-2 TTSS is essential for replication within phagocytic cells and systemic infection (21). Nevertheless, due to the absence of any apparent intestinal inflammation comparable to enteric salmonellosis in humans, this animal model has yielded little information about the pathological mechanisms of serovar Typhimurium enterocolitis. Therefore, bovine infection models have been established in recent years. In the bovine models, serovar Typhimurium causes pronounced enterocolitis which is thought to mimic the human disease closely. Type III secretion via the SPI-1 TTSS (14, 40, 48) and to some extent type III secretion via the SPI-2 TTSS (3) play an important role in the induction of enterocolitis. Recently, the SPI-1 TTSS effector proteins SipA, SopA, SopB, SopD, SopE, and SopE2 were found to affect fluid secretion and inflammation in the bovine intestine, as determined by oral infection and ligated ileal loop assays (27, 35, 51, 53, 54).
Several SPI-1 effector proteins have been characterized biochemically. SipA binds actin and has a nebulin-like function. It catalyzes actin polymerization and bundling of actin filaments and is required for efficient invasion of tissue culture cells (15, 24, 57, 58). In line with its role in bovine enterocolitis (54), SipA has been identified as a key SPI-1 effector protein for inducing polymorphonuclear granulocyte (PMN) migration across intestinal epithelial cell monolayers (30).
SopE and SopE2 are approximately 70% identical at the amino acid level and act as G-nucleotide exchange factors for host cellular Rho GTPases (1, 4, 22, 37, 41, 52). These proteins have differential preferences for different members of the Rho GTPase family (13), and both of them play a role in host cell invasion. In the case of SopE it has been demonstrated that the activation of host cellular Rho GTPases leads not only to cytoskeletal rearrangements but also to activation of signaling cascades which trigger cytokine production (5, 22).
SopB contains two motives common to inositol polyphosphatases and has profound effects on inositol and phosphatidylinositol levels in host cells (35, 43, 55, 56). Due to its broad spectrum of substrate specificity, the identity of the relevant substrate(s) in vivo and therefore its exact molecular function(s) in pathogenesis are still a matter of discussion. SopB has been found to affect cytoskeletal rearrangements and host cell invasion (26, 33, 43, 55), to mediate phospholipase C activation (55), to facilitate membrane fission (43), to affect chloride homeostasis (11, 35), and to inhibit nuclear mRNA export (11). Experiments with transiently transfected cells or stable cell lines expressing sopB demonstrated that SopB alone is sufficient to induce cytoskeletal rearrangements, to modulate membrane elasticity and chloride homeostasis, and to inhibit nuclear mRNA export (11, 43, 55). Although SopB cooperates with several other effector proteins during host cell infection, these observations indicate that SopB alone is capable of manipulating host cell signaling.
Molecular analysis of effector protein function has revealed how serovar Typhimurium manipulates host cell signaling. However, it is still not clear how the effects translate into an inflammatory response during the course of a real infection. To investigate this, a mouse model for serovar Typhimurium colitis was recently established (2). Upon oral infection with serovar Typhimurium, streptomycin-pretreated mice develop pronounced colitis on top of the well-known systemic infection. The intestinal inflammation develops within 8 to 20 h postinfection (p.i.) and is strongly dependent on the SPI-1 TTSS. This suggests that streptomycin-pretreated mice provide a versatile animal model to study how serovar Typhimurium induces intestinal inflammation. However, the SPI-1 effector proteins required for the initiation of serovar Typhimurium colitis in streptomycin-pretreated mice have not been identified thus far. Identification of these effectors is of considerable interest because streptomycin-mediated diminution of the intestinal microflora most likely affects the intestinal environment, and environmental signals are known to have a great impact on the expression of Salmonella virulence factors like the SPI-1 TTSS (6, 18, 21). Therefore, it was desirable to compare the effects of specific SPI-1 effector proteins in serovar Typhimurium colitis in streptomycin-pretreated mice with the effects observed in bovine models.
In this study, we identified the SPI-1 effector proteins required for initiation of serovar Typhimurium colitis in streptomycin-pretreated mice. We characterized serovar Typhimurium mutants lacking single or multiple effector proteins in terms of virulence in vivo and cell invasiveness in vitro. Our results are in line with previous studies in which the bovine models were used and corroborate the conclusion that streptomycin-pretreated mice are useful surrogate hosts for studying serovar Typhimurium colitis, and provide the experimental basis for a detailed analysis of the pathogenetic functions of several important SPI-1 effector proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Serovar Typhimurium strains were grown for 12 h at 37°C in Luria-Bertani (LB) broth containing 0.3 M NaCl, diluted 1:20 in fresh medium, and subcultured for 4 h with mild aeration. For infection experiments bacteria were washed twice in ice-cold phosphate-buffered saline (PBS) and suspended in cold PBS (108 CFU/50 μl).
Serovar Typhimurium wild-type strain SL1344 (25) and the isogenic derivatives SB161 (ΔinvG) (28), SB225 (sipA::aphT) (29), SB566 (invC::aphT) (10), and SB757 (sopE::pSB1128) (23) were kindly provided by J. E. Galán. The serovar Typhimurium ATCC 14028s derivatives MvP101 (sseD::aphT) (31) and HH110 (ssaV::cat) (M. Hensel, unpublished data) were kindly provided by M. Hensel. Serovar Typhimurium wild-type strain IR715 (ATCC 14028s; nalidixic acid resistant [Nalr]) (42) was provided by A. J. Bäumler.
The SL1344 derivatives used in this study were constructed by allelic exchange, which yielded in-frame deletions, and/or by phage P22-mediated transduction of antibiotic resistance marker-tagged mutant alleles. We used strains SB757, M200 (sopE2) (41), and MvP101 as donors for phage P22-mediated transduction of the mutant alleles sopE::pSB1128 (Ampr), sopE2::pM218 (Tetr), and sseD::aphT (Kanr), respectively. Strains M556, M557, M704, and M708 were created by P22 phage transduction of the sseD::aphT allele into recipient strains SL1344, SB161, M509 (SL1344 ΔsopB) (33), and M566 (SL1344 ΔsopE ΔsopE2 ΔsopB ΔsipA) (8a), respectively. Strain M719 was obtained by repeated phage transduction of sopE2::pM218, sopE::pSB1128, and sseD::aphT into strain SL1344. In-frame deletions of sopE and sipA were created by allelic exchange by using the suicide plasmids pM608 and pM585 (8a) (see below), respectively. In brief, the appropriate suicide vector was integrated into the serovar Typhimurium chromosome by single recombination, which yielded tetracycline-resistant exconjugants, and this was followed by a second recombination event forced by selection on sucrose agar. Strain M715 was constructed by in-frame deletion of sipA in the chromosome of SL1344 by using the suicide vector pM585 (8a), followed by phage transduction of sseD::aphT. Strain M722 was obtained by sequential phage transduction of sopE2::pM218 and sopE::pSB1128 into strain M715. Strain M716 was created by allelic exchange of sopE in the chromosome of M509 (SL1344 ΔsopB) by using the suicide vector pM608 (8a), followed by phage transduction of the sseD::aphT allele. M707 was obtained by sequential in-frame deletion of sipA and sopE in the chromosome of M509 by using the suicide vectors pM585 and pM608, respectively, followed by phage transduction of sseD::aphT. M717 was constructed by in-frame deletion of sipA in the chromosome of M509 by using the suicide vector pM585, followed by sequential phage transduction of sseD::aphT and sopE2::pM218. For complementation analysis, the suicide plasmids pM706 and pM707 (described below) were integrated into the chromosome of M708 by single recombination, which yielded strains M732 and M734, respectively. These strains express sopE/sopE2 under control of the native chromosomal promoter (Fig. 1). The site of insertion of plasmids pM706 and pM707 was verified by PCR analysis by using the following primer pairs: primer 5′-CAATATCGCCACTTTCAACG plus primer 5′-GTACTAAGCTCTCATGTTTCACG and primer 5′-CGGGATCCTCTTGGCGCGTAGTCCTTC plus primer 5′-GTACTAAGCTCTCATGTTTCACG for pM706; and primer 5′-CTTAAAAGCAGCCATACAGAT plus primer 5′-GTACTAAGCTCTCATGTTTCACG, primer 5′-GCGCAGGCGTTTAGAAGACAGTT plus primer 5′-TGGCATAACCTCTCCTGACA, and primer 5′-AGAAGAACAAAATCCATCAGG plus primer 5′-GCGTTCCAGCATCAGCCACTTG for pM707. All strains were tested for invasiveness in tissue culture cells (COS7) and were compared to appropriate mutant strains.
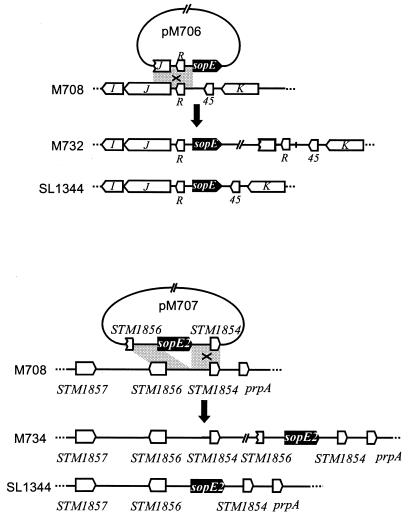
FIG. 1.
Schematic representation of the recombinational events leading to chromosomal vector integration in strains M732 and M734. The shaded areas indicate sequence identity in the plasmid and bacterial chromosome. Designations of the plasmids and Salmonella strains are indicated. The site of homologous recombination that resulted in chromosomal integration of a plasmid is indicated by X and was verified by PCR analysis (see Materials and Methods).
All ATCC 14028s-derived strains used in this study were constructed by phage transduction. We used strains SL1344, SB566, SB225, and HH110 as donor strains for the alleles aadA (conferring streptomycin resistance [Strr] in SL1344), invC::aphT (Tetr), sipA::aphT (Kanr), and ssaV::cat (Cmr), respectively. Strain M741 was created by transduction of the aadA locus into strain IR715. Strain M742 was obtained by phage transduction of invC::aphT, followed by transduction of aadA into IR715. Strain M744 was created by transduction of ssaV::cat, followed by transduction of aadA into IR715. Strain M745 was obtained by sequential phage transduction of invC::aphT, ssaV::cat, and aadA into IR715. Strain M743 was created by sequential phage transduction of sipA::aphT and aadA into the IR715-derived mutant strain M119 (IR715 sopE2::pM218 Tetr) (53). Strain M746 was obtained by sequential phage transduction of sipA::aphT, ssaV::cat, and aadA into M119.
All gene disruptions and deletions were verified by PCR analysis and by Western blot analyses with rabbit polyclonal antisera directed against SopE (34) and SopE2 (41). The stability of the gene disruptions and the retention of plasmids during animal infection were verified by replica plating of the bacterial colonies obtained from intestinal contents and organs (see below).
Recombinant DNA techniques.
Cloning of DNA fragments was performed by using standard protocols (38).
Construction of chromosomal in-frame deletions.
To construct a suicide vector for deletion of sipA, the sequences located directly upstream (primers 5′-GCGGCCGCACCTGGGGTTGAGTCCTAC and 5′-TCTAGAAGGGGGCTGAGTCCTTACAC; 33 cycles of 92°C for 30 s, 53°C for 30 s, and 68°C for 3 min) and downstream (primers 5′-TCTAGAGGCCCGGCTTACGAGTC and 5′-CCCGGGACACCAAGGCACGAG; 33 cycles of 92°C for 30 s, 55°C for 30 s, and 72°C for 3 min) of the sipA coding sequence were amplified by PCR. The PCR products were cloned into pCR-BluntII-Topo (Invitrogen), which yielded pM582 and pM583. The insert of pM582 was cloned into the XbaI sites of pM583, which yielded pM584, and the resulting insert was subcloned into the SmaI/NotI sites of the suicide vector pSB890 (a derivative of pSB377; oriR6K Tetr sacAB) (W.-D. Hardt and J. E. Galán, unpublished data), which yielded the suicide vector pM585, which was used for deletion of sipA.
To construct a suicide vector for deletion of sopE, we amplified the sequences located directly upstream (primers 5′-CGGGATCCTCTTGGCGCGTAGTCCTTC and 5′-GCTCTAGACACGGTAATGATCCTTTTATATGT; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min) and downstream (primers 5′-GCTCTAGACCCTGAACACTGAAAAACCA and 5′-TTTGCGGCCGCGCACTGGATACGCTGAACGA; 33 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 2 min) of the open reading frame by PCR and cloned the PCR products into pBluescriptSKII+ (Stratagene), which yielded pM593 and pM606. The insert of pM606 was cloned by using XbaI/NotI into pM593, which yielded pM607, and the resulting insert was subcloned into the BamHI/NotI sites of pSB890, which yielded the suicide vector pM608.
To construct a suicide vector for deletion of sopE2, we amplified the sequences located directly upstream (primers 5′-CGGGATCCGCGCAGGCGTTTAGAAGACAGTT and 5′-GCTCTAGAAGTCACGGTAGTTCTCCTTTT; 33 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min) and downstream (primers 5′-GCTCTAGAAATGCCTCCTGATGGTAGTAA and 5′-GCGGCCGCGTTCCAGCATCAGCCACTTG; 33 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3.5 min) of the open reading frame by PCR and cloned the PCR products into pBluescriptSKII+ (Stratagene), which yielded pM559 and pM560. The insert of pM560 was cloned by uisng XbaI/NotI into pM559, which yielded pM581, and the resulting insert was subcloned into the BamHI/NotI sites of pSB890, which yielded the suicide vector pM586.
All PCR products and inserts were analyzed by sequencing.
Construction of complementation vectors.
Plasmid pM706 is a suicide vector encoding the sopE promoter region and coding sequence. By single-crossover recombination into the ΔsopE strain M708 this allows chromosomal complementation with a sopE gene under control of the native chromosomal promoter (Fig. 1). The insert of plasmid pSB1120 (Hardt and Galán, unpublished) containing the sopE coding sequence plus 985 bp of upstream sequence was subcloned into the SalI and XbaI sites of suicide vector pGP704 (Ampr) (32), which yielded plasmid pM706. To construct the suicide vector pM707 for chromosomal complementation of sopE2 in the chromosome of strain M708 (Fig. 1), the insert of plasmid pM149 (a pACYC184 derivative) (41) containing the sopE2 coding sequence, 818 bp of upstream sequence, and 444 bp of downstream sequence was subcloned into the XbaI and BamHI sites of suicide vector pSB377 (Tetr) (29), which yielded plasmid pM707. To construct a low-copy-number expression vector for sipA complementation in trans, the sipA gene was cloned from the chromosome of SL1344. In brief, the suicide plasmid pM585 (described above) was integrated into the chromosome of SL1344 by single recombination. Subsequently, integration of pM585 downstream of the sipA coding sequence was verified by PCR, chromosomal DNA of the appropriate clones was isolated by standard techniques (38), and the chromosomal DNA was digested with BamHI, treated with DNA ligase, and electroporated into Escherichia coli CC118λpir, which yielded pM711. Plasmid pM711 harbors the sipA coding sequence and 2,207 bp of upstream sequence. The 2,819-bp HindIII/EcoRV fragment of pM711 harboring the sipA coding sequence and 31 bp of upstream sequence was subcloned into the HindIII and EcoRV sites of the low-copy-number vector pWKS30 (Ampr, lac promoter, pSC101 ori) (49), which yielded pM712.
Gentamicin protection assay.
An analysis of the invasiveness of mutant Salmonella strains for COS7 cells was carried out as described previously (33). COS7 tissue culture cells were grown for 2 days in Dulbecco modified Eagle medium (DMEM) containing 5% fetal bovine serum (FBS) in 24-well dishes to obtain 80% confluence. The culture medium was removed, and 500 μl of Hanks' buffered salt solution was added 3 min before addition of the bacteria. The bacteria were grown as described above. The actual bacterial density was determined by plating appropriate dilutions on LB agar. To start the assay, bacteria were added to COS7 cells at a multiplicity of infection of approximately 15 and incubated for 50 min at 37°C in the presence of 5% CO2. The cells were washed three times with Hanks' buffered salt solution and incubated for 2 h in 500 μl of DMEM containing 5% FBS and 400 μg of gentamicin per ml at 37°C in the presence of 5% CO2. The cells were washed three times with 1× PBS and lysed in 1× PBS-0.1% sodium deoxycholate, and the number of CFU of intracellular bacteria was determined by plating on LB agar. Usually about 25% of the inoculum of the wild-type strain was recovered from COS7 cells at the end of the assay. An analysis of the invasiveness of mutant Salmonella strains for murine embryonic fibroblasts (C57BL/6 genetic background; kindly provided by H. Rüssmann) was carried out as described above for COS7 cells, but the tissue cell culture medium was replaced by DMEM containing 5% FBS, 50 μM 2-mercaptoethanol, and 2 mM glutamine. The numbers given below were determined in at least six independent experiments for each strain. The statistical significance of differences between the invasiveness of different strains was analyzed by using the Mann-Whitney U test.
Animal experiments.
Animal experiments were performed as described recently (2) by using specific-pathogen-free female C57BL/6 mice that were 6 to 9 weeks old and were obtained from Harlan (Horst, The Netherlands). For the experiments, animals were housed individually or in groups of up to five animals under standard barrier conditions in individually ventilated cages (Tecniplast, Buguggiate, Italy) equipped with steel grid floors and autoclaved filter paper at the BZL (Zürich, Switzerland). Water and food were withdrawn 4 h before per os (p.o.) treatment with 20 mg of streptomycin (50 μl of a sterile solution). After this, the animals were given water and food ad libitum. Twenty hours after streptomycin treatment, water and food were withdrawn again for 4 h before the mice were infected p.o. with 108 CFU of serovar Typhimurium (50 μl of a suspension in PBS). After this, drinking water was offered ad libitum immediately and food was offered at 2 h p.i. At different times after infection, the mice were sacrificed by cervical dislocation, and tissue samples from the intestinal tract, spleen, and liver were removed for analysis. Animal experiments were approved by the Swiss authorities and were performed according to the legal requirements.
Analysis of serovar Typhimurium loads in the intestine, mesenteric lymph nodes, spleen, and liver.
To analyze colonization, spleens and livers were removed aseptically and homogenized in 4°C PBS containing 0.5% Tergitol and 0.5% bovine serum albumin by using a Potter homogenizer as described recently (2). The numbers of CFU were determined by plating appropriate dilutions on MacConkey agar plates containing streptomycin (50 μg/ml). The minimal detectable levels were 20 CFU/organ in the spleen and 100 CFU/organ in the liver.
Intestinal contents of the cecum were collected on day 2 p.i. and weighed before they were suspended in 500 μl of 4°C PBS. The number of CFU was determined by plating appropriate dilutions on MacConkey agar plates containing streptomycin (50 μg/ml). The minimum detectable level was 10 CFU per sample (between 25 and 150 mg of intestinal contents).
Histological procedures.
Segments of ileum, cecum, and colon were embedded in O.C.T. (Sakura, Torrance, Calif.), snap-frozen in liquid nitrogen, and stored at −80°C. Cryosections (5 μm) were mounted on glass slides, air dried for 2 h at room temperature, and stained with hematoxylin and eosin (HE).
Cecum pathology was evaluated by pathologists in a blinded manner by using 5-μm HE-stained sections and the following histopathological scoring scheme, as previously described (2).
(i) Submucosal edema.
For submucosal edema a score of 0 indicated that there were no pathological changes; a score of 1 indicated that there was mild edema (the submucosa was <0.20 mm wide and accounted for <50% of the diameter of the entire intestinal wall [tunica muscularis to epithelium]); a score of 2 indicated that there was moderate edema (the submucosa was 0.21 to 0.45 mm wide and accounted for 50 to 80% of the diameter of the entire intestinal wall); and a score of 3 indicated that there was profound edema (the submucosa was >0.46 mm wide and accounted for >80% of the diameter of the entire intestinal wall). The widths of the submucosa were determined by quantitative microscopy and were the averages for 30 evenly spaced radial measurements of the distance between the tunica muscularis and the lamina mucosalis mucosae.
(ii) PMN infiltration into the lamina propria.
PMN in the lamina propria were enumerated by examining 10 high-power fields (magnification, ×400; field diameter, 420 μm), and the average number of PMN per high-power field was calculated. The scores were as follows: 0, less than 5 PMN per high-power field; 1, 5 to 20 PMN per high-power field; 2, 21 to 60 PMN per high-power field; 3, 61 to 100 PMN per high-power field; and 4, more than 100 PMN per high-power field. Transmigration of PMN into the intestinal lumen was consistently observed when the number of PMN was more than 60 PMN per high-power field.
(iii) Goblet cells.
The average number of goblet cells per high-power field (magnification, ×400) was determined by examining 10 different regions of the cecal epithelium. The scores were as follows: 0, more than 28 goblet cells per high-power field (in the ceca of the normal specific-pathogen-free mice we observed an average of 6.4 crypts per high-power field, and the average crypt consisted of 35 to 42 epithelial cells, 25 to 35% of which were differentiated into goblet cells); 1, 11 to 28 goblet cells per high-power field; 2, 1 to 10 goblet cells per high-power field; and 3, less than 1 goblet cell per high-power field.
(iv) Epithelial integrity.
For epithelial integrity a score of 0 indicated that no pathological changes were detectable in 10 high-power fields (magnification, ×400); a score of 1 indicated that there was epithelial desquamation; a score of 2 indicated that there was erosion of the epithelial surface (gaps of 1 to 10 epithelial cells per lesion); and a score of 3 indicated that there was epithelial ulceration (gaps of >10 epithelial cells per lesion) (at this stage, there was generally granulation tissue below the epithelium).
The combined pathological score for each tissue sample was determined by adding the averaged scores. The values ranged from 0 to 13 arbitrary units and covered the following levels of inflammation: 0, intestine intact without any signs of inflammation; 1 to 2, minimal signs of inflammation (these scores were frequently obtained for the ceca of specific-pathogen-free mice, and this level of inflammation is generally not considered a sign of disease); 3 to 4, slight inflammation; 5 to 8, moderate inflammation; and 9 to 13, profound inflammation.
Statistical analysis.
Statistical analysis of the individual pathological scores for submucosal edema, PMN infiltration, loss of goblet cells, and epithelial integrity and for the combined pathological score was performed by using the exact Mann-Whitney U test and the SPSS software (version 11.0), as described previously (2). P values of <0.05 were considered statistically significant. Bacterial colonization was analyzed in a similar way. To allow statistical analysis of the bacterial loads, the values used for animals that yielded no CFU were the minimal detectable values (20 CFU for the spleen, 100 CFU for the liver, and between 67 and 400 CFU for the intestinal contents [see above]). After this, the median values were calculated by using Microsoft Excel XP, and a statistical analysis was performed by using the exact Mann-Whitney U test and the SPSS software (version 11.0). P values of <0.05 were considered statistically significant.
RESULTS
SipA plays an important role in murine serovar Typhimurium colitis.
It has recently been shown that the SPI-1 TTSS plays a key role in serovar Typhimurium-induced colitis in streptomycin-pretreated mice (2). Serovar Typhimurium mutants incapable of translocating effector proteins into host cells are strongly attenuated in this animal model and induce only mild signs of intestinal inflammation (2), but previously it was not known which SPI-1 effector proteins are involved. Therefore, we analyzed serovar Typhimurium mutants lacking one or more SPI-1 effector proteins. In order to optimize the sensitivity of the assay, most of the experiments in this study were performed in the serovar Typhimurium mutant M556 (SL1344 sseD::aphT) background, which lacks an essential component of the SPI-2 translocon. This choice was based on previous observations demonstrating that the SPI-2 TTSS can affect enteric disease in the bovine model (3). Later, we confirmed that the results obtained in the M556 genetic background can be reproduced with SPI-2-positive serovar Typhimurium strains (see below).
The effector proteins SopB, SopE, SopE2, and SipA have been shown to contribute to enteric salmonellosis in calves (35, 50, 52-54). To assess the role of these proteins in the murine model, we constructed a series of M556 derivatives carrying in-frame deletions of sopB (M704) or sipA (M715) and a sopEE2 double effector mutant strain (M719) (Table 1) (see Materials and Methods). These strains were characterized by performing murine infection experiments and tissue culture cell invasion assays (see below).
TABLE 1.
Serovar Typhimurium strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Reference |
|---|---|---|
| Serovar Typhimurium | ||
| SL 1344 strains | ||
| SL1344 | Wild type | 25 |
| SB161 | ΔinvG | 28 |
| M556 | sseD::aphT | This study |
| M557 | sseD::aphT ΔinvG | This study |
| M715 | sseD::aphT ΔsipA | This study |
| M704 | sseD::aphT ΔsopB | This study |
| M719 | sseD::aphT sopE::pSB1128 sopE2::pM218 | This study |
| M716 | sseD::aphT ΔsopB ΔsopE sopE2::pM218 | This study |
| M722 | sseD::aphT sopE::pSB1128 sopE2::pM218 ΔsipA | This study |
| M707 | sseD::aphT ΔsopB ΔsopE ΔsipA | This study |
| M717 | sseD::aphT ΔsopB sopE2::pM218 ΔsipA | This study |
| M708 | sseD::aphT ΔsopB ΔsopE ΔsopE2 ΔsipA | This study |
| M732 | sseD::aphT ΔsopB ΔsopE::pM706 ΔsopE2 ΔsipA | This study |
| M734 | sseD::aphT ΔsopB ΔsopE ΔsopE2::pM707 ΔsipA | This study |
| ATCC 14028s strains (IR715 derivatives; Nalr)a | ||
| M741 | aadA (from SL1344) | This study |
| M742 | aadA invC::aphT | This study |
| M744 | aadA ssaV::cat | This study |
| M745 | aadA invC::aphT ssaV::cat | This study |
| M743 | aadA sopE2::pM218 sipA::aphT | This study |
| M746 | aadA ssaV::cat sopE2::pM218 sipA::aphT | This study |
| Plasmids | ||
| pM706 | Ampr, oriR6K, carrying sopE coding and promotor region | This study |
| pM707 | Tetr, oriR6K oriT, carrying sopE2 coding and promotor region | This study |
| pWKS30 | Ampr, lac promotor, pSC101 ori | 49 |
| pM712 | sipA under control of the lac promotor, pWKS30 backbone | This study |
See reference 42.
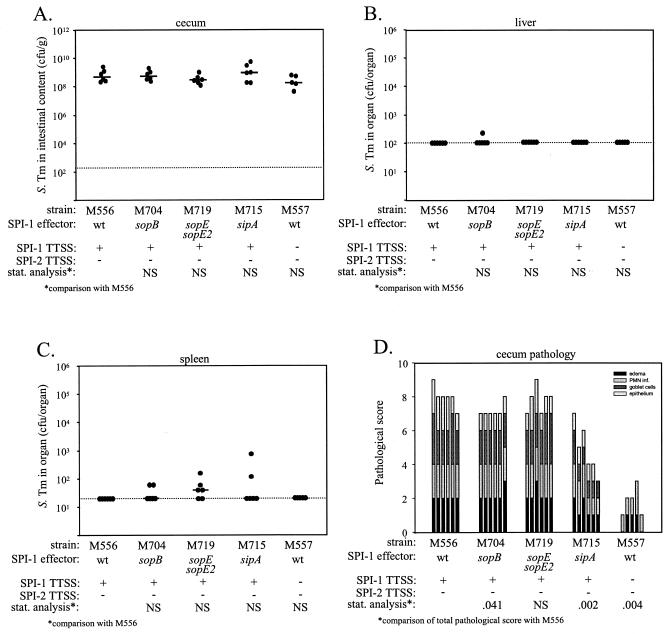
Streptomycin-pretreated C57BL/6 mice (five or six animals per group) were infected with 108 CFU p.o. and sacrificed 2 days p.i. The serovar Typhimurium load in the cecal contents, colonization of the liver and spleen, and pathological changes in the cecum with respect to edema in the submucosa, PMN infiltration, loss of goblet cells, and damage to the intestinal epithelium were analyzed as described in Materials and Methods. M556 (sseD::aphT) was used as a positive control for determination of the contribution of SPI-1 effector proteins to enteric disease, and M557 (ΔinvG sseD::aphT), which is deficient in SPI-1- and SPI-2-dependent type III protein secretion (Ehrbar and Hardt, unpublished data), was used as a negative control.
The densities of the different serovar Typhimurium strains in the cecal contents did not differ significantly (median, 108 to 109 CFU/g) (Fig. 2A). Also, the numbers of bacteria present in the liver and spleen did not differ significantly for the different serovar Typhimurium strains and were below the limit of detection in most cases (Fig. 2B and C). Signs of strong inflammation were evident in the cecal tissues of mice infected with M556 (sseD::aphT), the sopB mutant (M704), and the sopEE2 double effector mutant (M719) (Fig. 2D). These signs included strong edema in the submucosa, massive PMN infiltration, erosion or desquamation of the intestinal epithelium, and loss of goblet cells. In this experiment the total pathological score (P = 0.041), but none of the individual scores for the sopB mutant M704, appeared to be significantly reduced (Fig. 2D and Table 2). Additional experiments confirmed that the pathological scores of M704 did not differ significantly from those of M556 (data not shown). The sipA mutant (M715) caused significantly less inflammation (P = 0.002), indicating that SipA plays an important role in serovar Typhimurium colitis in streptomycin-pretreated mice. M557 (ΔinvG sseD::aphT) was attenuated even more strongly than M715 (P = 0.004) (Fig. 2D). This suggested that effector proteins in addition to SipA might be involved in the induction of serovar Typhimurium colitis.
FIG. 2.
Analysis of sopB and sipA mutants and a sopEE2 double effector mutant of serovar Typhimurium SL1344. Five groups of streptomycin-pretreated mice were infected for 2 days with 108 CFU of serovar Typhimurium strains M556 (six mice), M704 (six mice), M719 (six mice), M715 (six mice), and M557 (five mice). These strains had disrupted SPI-1 effector genes and/or disrupted genes encoding structural components of the SPI-1 and SPI-2 TTSS. (A) Bacterial loads in the cecal contents. (B) Bacterial loads in the liver. (C) Bacterial loads in the spleen. (D) Histopathological analysis. HE-stained sections of cecal tissue were scored with respect to edema in the submucosa, PMN infiltration (PMN inf.), reduction in the number of goblet cells, and desquamation, erosion, or ulceration of the epithelial layer (see Materials and Methods), and the scores were plotted as stacked vertical bars. The total pathological score (sum of the separate scores) was statistically analyzed (stat. analysis) by using the exact Mann-Whitney U test (with comparison to M556). The dotted lines indicate the limits of detection. NS, not statistically significant (P ≥ 0.05); S. Tm, serovar Typhimurium; wt, wild type.
TABLE 2.
Statistical analysis of disease parameters in streptomycin-pretreated mice infected with serovar Typhimurium mutants lacking one or two SPI-1 effector proteinsa
| Comparison |
P (vs M556) for:
|
|||
|---|---|---|---|---|
| M704 | M719 | M715 | M557 | |
| Combined score | 0.041 | NS | 0.002 | 0.004 |
| Edema | NS | NS | NS | 0.004 |
| PMN infiltration | NS | NS | NS | 0.030 |
| Goblet cells | NS | NS | 0.009 | 0.004 |
| Epithelium | NS | NS | 0.041 | 0.004 |
The data in Fig. 2D were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
Identification of additional effector proteins involved in enteric disease.
Tissue culture cell invasion experiments revealed that several SPI-1 effector proteins have at least partially redundant functions and act in concert to manipulate host cells (20, 33, 55, 56). Similarly, several effector proteins have been reported to act in concert in bovine Salmonella infection models (54). This functional redundancy of SPI-1 effector proteins might explain the lack of a detectable phenotype for the sopB and sopEE2 mutants in the murine colitis model (Fig. 2D). Effects of SipA or other effector proteins may have masked an inflammation phenotype associated with sopB, sopE, or sopE2. Thus, the effect of a single effector protein might only be detectable in a mutant lacking all other effector proteins that induce cecal inflammation.
Based on analyses in which bovine infection models were used (54), SopB, SopE, and SopE2 were good candidates to account for the residual cecal inflammation observed with the sipA mutant M715 (Fig. 2D and Table 2).
In fact, preliminary experiments indicated that a serovar Typhimurium SL1344 sipA sopBEE2 quadruple effector deletion mutant (M708) (Table 1) was not capable of causing substantial intestinal inflammation (Fig. 3D and data not shown). This indicated that the effector proteins SipA, SopE, SopE2, and SopB or subsets of these proteins are required for the SPI-1-dependent induction of intestinal inflammation in streptomycin-pretreated mice.
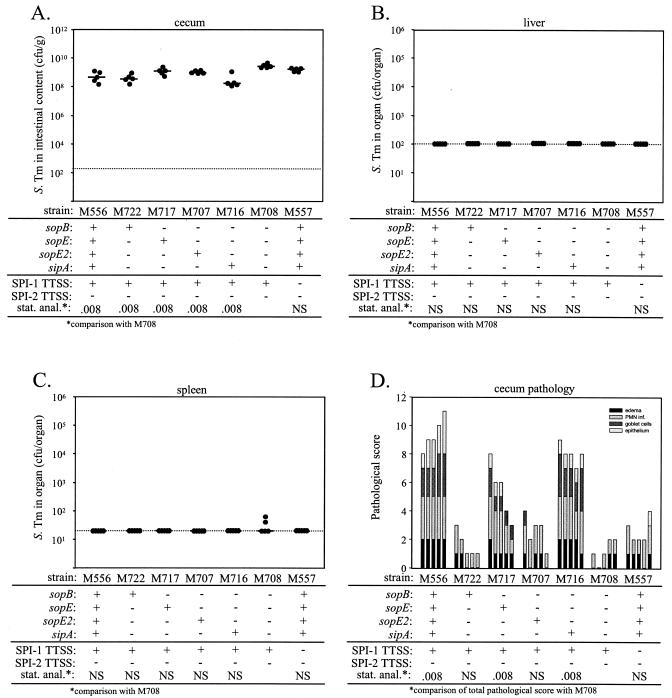
FIG. 3.
Analysis of effector gene triple mutants and a sipA sopBEE2 quadruple effector mutant of serovar Typhimurium SL1344. Seven groups of five streptomycin-pretreated mice were infected for 2 days with 108 CFU of serovar Typhimurium strains M556, M722, M717, M707, M716, M708, and M557. The relevant genotypes are indicated. (A) Bacterial loads in the cecal contents. (B) Bacterial loads in the liver. (C) Bacterial loads in the spleen. (D) Histopathological analysis. HE-stained sections of cecal tissue were scored with respect to edema in the submucosa, PMN infiltration (PMN inf.), reduction in the number of goblet cells, and desquamation, erosion, or ulceration of the epithelial layer (see Materials and Methods), and the scores were plotted as stacked vertical bars. The total pathological score (sum of the separate scores) was statistically analyzed (stat. anal.) by using the exact Mann-Whitney U test (with comparison to M708). The dotted lines indicate the limits of detection. NS, not statistically significant (P ≥ 0.05); S. Tm, serovar Typhimurium.
In order to analyze the contribution of SipA, SopE, SopE2, and SopB in more detail, we constructed a comprehensive set of serovar Typhimurium triple effector mutants. Each of these strains retained only one of the four SPI-1 effector proteins suspected to be involved in the induction of colitis. We used the streptomycin-pretreated mouse model to compare the virulence of these triple effector mutants with that of the sipA sopBEE2 quadruple effector mutant M708. M556 (sseD::aphT; SPI-2 defect) and M557 (ΔinvG sseD::aphT; SPI-1 and SPI-2 defects) served as positive and negative controls. Groups of five streptomycin-pretreated C57BL/6 mice were infected p.o. with 108 CFU of the appropriate serovar Typhimurium strains. Two days after infection the animals were sacrificed and analyzed for colonization of the cecum, liver, and spleen, as well as for pathological changes in cecal tissues (Fig. 3). We detected 108 to 1010 CFU of serovar Typhimurium per g in the cecal contents in all groups (Fig. 3A). The numbers of bacteria in livers and spleens were very low in all animals (Fig. 3B and C). In contrast, the different triple effector mutants had quite different capacities to induce intestinal inflammation; compared to the sipA sopBEE2 quadruple effector mutant M708, the two triple effector mutants carrying intact genes for SipA (M716) or SopE (M717) caused significantly greater pathological changes. The triple effector mutant M707 carrying an intact gene for SopE2 caused less severe inflammation than M716 or M717 caused (Fig. 3D), but it had a significantly higher capacity to induce PMN infiltration into the lamina propria than the sipA sopBEE2 quadruple effector mutant M708 (P = 0.032) (Table 3). In contrast, the cecal inflammation caused by the triple effector mutant M722, which carried an intact gene for SopB, did not differ significantly from that caused by M708. These data indicated that SipA, SopE, and SopE2 contribute to elicitation of serovar Typhimurium colitis in streptomycin-pretreated mice. In contrast, we could not detect a phenotype for sopB in the absence of SipA, SopE, and SopE2.
TABLE 3.
Statistical analysis of disease parameters in streptomycin-pretreated mice infected with serovar Typhimurium SL1344 mutants lacking three or four SPI-1 effector proteinsa
| Comparison |
P (vs M708) for:
|
|||||
|---|---|---|---|---|---|---|
| M556 | M722 | M717 | M707 | M716 | M557 | |
| Combined score | 0.008 | NS | 0.008 | NS | 0.008 | NS |
| Edema | 0.008 | NS | NS | NS | 0.016 | NS |
| PMN infiltration | 0.008 | NS | 0.032 | 0.032 | 0.008 | NS |
| Goblet cells | 0.008 | NS | 0.008 | NS | 0.008 | NS |
| Epithelium | 0.008 | NS | NS | NS | 0.008 | NS |
The data in Fig. 3D were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
Complementation analysis verified that SipA, SopE, and SopE2 contribute to murine serovar Typhimurium colitis.
In order to prove that the reduced enteric virulence of the sipA sopBEE2 quadruple effector mutant M708 was attributable to specific deletion of the genes encoding SipA, SopE, and SopE2, we performed a complementation analysis.
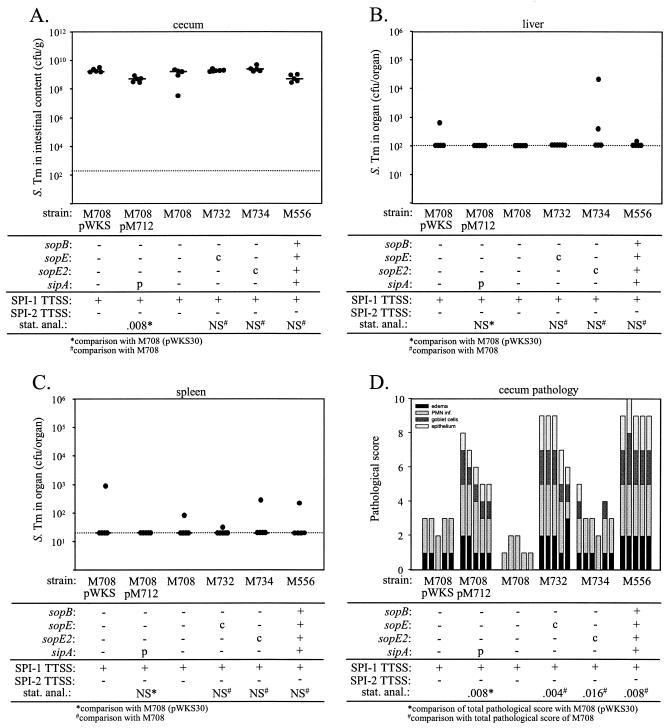
Preliminary experiments were performed to identify the optimal genetic constructs for complementation. Transformation of M708 with a low-copy-number sipA expression plasmid (pM712) could complement this strain's enteric virulence, while various sopE and sopE2 expression plasmids could not (data not shown). For this reason, we constructed the suicide vectors pM706 and pM707, which were integrated into the chromosome of M708 by single-crossover recombination, which yielded M732 and M734, respectively (see Materials and Methods) (Table 1).
To perform the complementation analysis, streptomycin-pretreated C57BL/6 mice were infected p.o. with 108 CFU of M708, M732, M734, M708(pWKS30) (empty control vector), or M708(pM712). Infections with M556 and M557 served as additional controls. The mice were sacrificed at 2 days p.i. and were analyzed for intestinal colonization and for bacterial numbers in the liver and spleen, as well as for pathological changes in the cecal tissue. The cecal contents of the mice from every group generally contained between 108 and 1010 CFU/g (Fig. 4A), and we did not detect significant differences in colonization of the liver and spleen (Fig. 4B and C). The pathological analysis revealed that M708(pM712) induced more severe inflammatory changes than the control strain M708(pWKS30) induced. Similar results were obtained for the chromosomal complementation of sopE and sopE2 (Fig. 4D and Table 4 [compare M708 with M732 and M734]). These data verified that the SPI-1 effector proteins SipA, SopE, and SopE2 contribute to murine serovar Typhimurium colitis and demonstrated that each of these proteins is sufficient to induce inflammation in the absence of the other two proteins and of SopB.
FIG. 4.
Complementation analysis of the sipA, sopE, and sopE2 mutations in serovar Typhimurium SL1344. Groups of five (or six in the case of M734) streptomycin-pretreated mice were infected for 2 days with 108 CFU of serovar Typhimurium strains M708(pWKS30), M708(pM712), M708, M732, M734, and M556 (Table 1). The relevant genotypes are indicated. (A) Bacterial loads in the cecal contents. (B) Bacterial loads in the liver. (C) Bacterial loads in the spleen. (D) Histopathological analysis. HE-stained sections of cecal tissue were scored with respect to edema in the submucosa, PMN infiltration (PMN inf.), reduction in the number of goblet cells, and desquamation, erosion, or ulceration of the epithelial layer (see Materials and Methods), and the scores were plotted as stacked vertical bars. The total pathological score (sum of the separate scores) was statistically analyzed (stat. anal.) by using the exact Mann-Whitney U test [with comparison to M708(pWKS30) or M708]. The dotted lines indicate the limits of detection. c, chromosomal complementation; p, complementation in trans with a plasmid; NS, not statistically significant (P ≥ 0.05); S. Tm, serovar Typhimurium.
TABLE 4.
Statistical analysis of disease parameters in streptomycin-pretreated mice infected with the serovar Typhimurium SL1344 mutant M708: complementation analysisa
| Comparison | P (vs M708/pWKS) for M708/pSipA |
P (vs M708) for:
|
||
|---|---|---|---|---|
| M732 | M734 | M556 | ||
| Combined score | 0.008 | 0.004 | 0.016 | 0.008 |
| Edema | NS | 0.004 | 0.032 | 0.008 |
| PMN infiltration | NS | NS | NS | 0.008 |
| Goblet cells | 0.008 | 0.004 | NS | 0.008 |
| Epithelium | 0.008 | 0.004 | NS | 0.008 |
The data in Fig. 4D were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
Analysis of the effector protein mutants in COS7 tissue culture cell invasion assays.
In our experiments in which the mouse colitis model was used we did not detect a phenotype for sopB mutants. However, the molecular functions of SopB, SipA, SopE, and SopE2 have been characterized quite extensively in vitro, and each of these effector proteins (including SopB) plays a role in Salmonella host cell invasion (1, 22, 23, 26, 33, 41, 55, 58). Interestingly, SopB alone was found to be sufficient to modulate vesicle formation and vacuole sealing, to trigger host cell phosphatidylinositol signaling, to induce profound actin cytoskeletal rearrangements, and to alter nuclear mRNA export (11, 35, 43, 55). To address the apparent differences between our observations obtained with the streptomycin-pretreated mouse model and the established in vitro function of SopB, we performed host cell invasion assays with the serovar Typhimurium mutants described above.
COS7 tissue culture cells were infected (multiplicity of infection, 15 bacteria per cell) with wild-type serovar Typhimurium SL1344, the isogenic mutant with a defective SPI-2 translocon (M556), and the different isogenic strains carrying mutations in one or several of the genes encoding the effector proteins SipA, SopB, SopE, and SopE2. Fifty minutes after infection all extracellular bacteria were killed with gentamicin, and the numbers of internalized bacteria were determined as described in Materials and Methods.
As expected, the invasiveness of strain M556 (sseD::aphT) with a defective SPI-2 translocon did not differ significantly from that of the isogenic wild-type strain (P ≥ 0.05) (Table 5). In contrast, both strains carrying a disrupted SPI-1 TTSS apparatus (SB161 and M557) were about 1,000-fold less invasive than M556 or wild-type serovar Typhimurium strain SL1344. These observations indicate that the SPI-1-mediated invasion of COS7 tissue culture cells by serovar Typhimurium strain SL1344 does not require a functional SPI-2 TTSS.
TABLE 5.
Serovar Typhimurium strain SL1344 mutants lacking SPI-1 effector proteins: invasion of COS7 tissue culture cellsa
| Strain | Relevant genotype
|
Invasiveness (%)b | No. of assays |
P vs:
|
||
|---|---|---|---|---|---|---|
| TTSS | Effectors | M556 | M557 | |||
| SL1344 | Wild type | Wild type | 117 | 24 | NS | <0.001 |
| SB161 | invG | Wild type | 0.06 | 23 | <0.001 | NS |
| M556 | sseD | Wild type | 100 | 6 | 0.002 | |
| M557 | sseD invG | Wild type | 0.16 | 6 | 0.002 | |
| M715 | sseD | sipA | 73.2 | 6 | NS | 0.002 |
| M704 | sseD | sopB | 134 | 6 | NS | 0.002 |
| M719 | sseD | sopEE2 | 35.8 | 6 | 0.002 | 0.002 |
| M716 | sseD | sopBEE2 | 0.41 | 6 | 0.002 | NS |
| M722 | sseD | sopEE2 sipA | 32.3 | 6 | 0.004 | 0.002 |
| M707 | sseD | sopBE sipA | 14.2 | 6 | 0.002 | 0.002 |
| M717 | sseD | sopBE2 sipA | 91.2 | 6 | NS | 0.002 |
| M708 | sseD | sopBEE2 sipA | 0.28 | 6 | 0.002 | NS |
The data were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
The invasiveness of M556 was defined as 100% (see Materials and Methods).
Disruption of the gene for SipA (M715) or SopB (M704) had little effect on host cell invasion (P ≥ 0.05) (Table 5), and the sopEE2 double effector mutant was slightly but significantly (P = 0.002) (Table 5) less invasive than the isogenic strain M556. In contrast, the sipA sopBEE2 quadruple effector mutant M708 was attenuated (invasiveness, 0.26%) as strongly as the negative control strain M557 (P ≥ 0.05) (Table 5). Compared to these results, the triple effector mutants carrying an intact gene for SopE (M717), SopB (M722), or SopE2 (M707) were highly invasive (invasiveness, 91 to 14%). This finding was in line with previous less comprehensive analyses (33, 55) and indicated that these three effector proteins were properly synthesized and translocated by the strains used in our study. The triple effector mutant carrying an intact gene for SipA (M716) was only slightly more invasive than the sipA sopBEE2 quadruple effector mutant M708, but the difference was not statistically significant (P ≥ 0.05) (Table 5). Previous studies have also revealed that SipA can slightly increase the efficiency of host cell invasion (57, 58).
The invasiveness of the sipA sopBEE2 quadruple effector mutant M708 was recomplemented by introduction of SopE (pM706) (P = 0.004) or SopE2 (pM707) (P = 0.004) (Table 6). M708 carrying the sipA expression vector pM712 was slightly more invasive than M708 (P = 0.015) (Table 6) or M708 carrying the empty control vector. These data verified that the genes encoding SopE, SopE2, SopB, and SipA were fully functional in the strains used in this study and that the mutations did disrupt gene function. Thus, we excluded the possibility that technical problems (i.e., undetected second-site mutations which might have occurred during strain construction) were the cause of our failure to detect a virulence defect for sopB strains in the streptomycin-pretreated mouse model.
TABLE 6.
Complementation of M708: invasion of COS7 tissue culture cellsa
| Strain | Relevant genotype
|
Invasiveness (%)b | No. of assays |
P vs:
|
||
|---|---|---|---|---|---|---|
| TTSS | Effectorsc | M556 | M708 | |||
| M556 | sseD | Wild type | 100 | 6 | 0.002 | |
| M708 | sseD | sopBEE2 sipA | 0.28 | 6 | 0.002 | |
| M708(pWSK30) | sseD | sopBEE2 sipA | 0.25 | 6 | 0.002 | NS |
| M708(pM712) | sseD | sopBEE2 sipA (pSipA) | 0.48 | 6 | 0.002 | 0.015 |
| M732 | sseD | sopBEE2 sipA (c.E) | 176 | 6 | 0.004 | 0.002 |
| M734 | sseD | sopBEE2 sipA (c.E2) | 9.2 | 5 | 0.004 | 0.004 |
The data were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
The invasiveness of M556 was defined as 100% (see Materials and Methods).
c.E and c.E2, chromosomal complementation of sopE and sopE2, respectively (Fig. 1).
Invasion of primary embryonic murine fibroblasts.
The experiments described above demonstrated that SopB contributes to host cell manipulation in the fibroblast-like cell line COS7, which was derived from green monkey kidney cells (17). However, it is known that the same Salmonella strain can cause different types of disease in different host animals (45, 47), and we have found that streptomycin-pretreated mice develop strong intestinal inflammation but (compared to the bovine model and human disease) only mild secretory responses upon infection with serovar Typhimurium (2). Interestingly, SopB is a key effector protein in the induction of fluid secretion in bovine ileal loop infections (16, 27, 35, 54). This raised the question of whether murine cells are less sensitive to the effects of SopB than cells from primates or calves are. To address this question, we prepared primary embryonic fibroblasts from C57BL/6 mice. Using these cells, we analyzed invasion of wild-type serovar Typhimurium SL1344, a sopB mutant (M704), the triple effector mutants harboring intact genes for just one of the effector proteins (SopB, SopE, SopE2, and SipA), and the sipA sopBEE2 quadruple effector mutant M708. Overall, the results obtained in this assay were very similar to the results obtained with the COS7 cell line. (i) Serovar Typhimurium strains SL1344 (wild type) and M556 (sseD::aphT) were much more invasive than mutants with a disrupted SPI-1 TTSS apparatus (SB161 and M557) (Table 7). (ii) The sipA sopBEE2 quadruple effector mutant M708 was only slightly more invasive than SB161 or M557. (iii) The triple effector mutants harboring intact genes encoding SopB (M722), SopE (M717), or SopE2 (M707) were significantly more invasive (12 to 55%) than the sipA sopBEE2 quadruple effector mutant M708 (2.5%) (Table 7). (iv) The triple effector mutant harboring an intact gene for SipA (M716) was slightly more invasive than the sipA sopBEE2 quadruple effector mutant M708. In conclusion, these data demonstrated that SopB, SopE, and SopE2 are about equally efficient at triggering invasion of murine and primate cells. Therefore, the lack of a phenotype for sopB strains in the murine colitis model could not be attributed to a general insensitivity of murine cells to the effects of SopB.
TABLE 7.
Serovar Typhimurium strain SL1344 mutants lacking SPI-1 effector proteins: invasion of primary embryonic murine fibroblastsa
| Strain | Relevant genotype
|
Invasiveness (%)b | No. of assays |
P vs:
|
||
|---|---|---|---|---|---|---|
| TTSS | Effectors | M556 | M557 | |||
| SL1344 | Wild type | Wild type | 82.1 | 7 | NS | 0.006 |
| SB161 | invG | Wild type | 0.71 | 7 | 0.001 | NS |
| M556 | sseD | Wild type | 100 | 7 | 0.006 | |
| M557 | sseD invG | Wild type | 0.74 | 4 | 0.006 | |
| M704 | sseD | sopB | 60.0 | 7 | NS | 0.006 |
| M719 | sseD | sopEE2 | 71.4 | 7 | NS | 0.006 |
| M716 | sseD | sopBEE2 | 5.5 | 7 | 0.001 | 0.006 |
| M722 | sseD | sopEE2 sipA | 42.9 | 7 | 0.017 | 0.006 |
| M707 | sseD | sopBE sipA | 12.5 | 4 | 0.006 | 0.029 |
| M717 | sseD | sopBE2 sipA | 55.7 | 4 | NS | 0.029 |
| M708 | sseD | sopBEE2 sipA | 2.5 | 6 | 0.001 | 0.042 |
The data were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
The invasiveness of M556 was defined as 100% (see Materials and Methods).
Role of sopB from serovar Typhimurium strain ATCC 14028s in murine colitis.
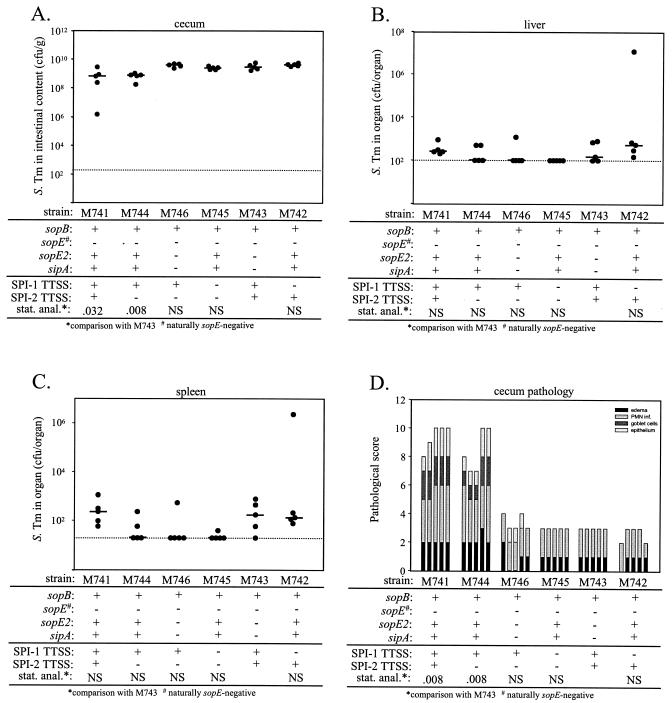
The effector protein SopB has been studied quite extensively. Genes coding for this SPI-1 effector protein are present in all known Salmonella lineages (33), and functional studies have identified a role in host cell invasion in all Salmonella strains analyzed, including S. enterica serovar Dublin strain 2229 (16) and serovar Typhimurium strains ATCC 14028s and SL1344 (33, 43, 54, 55). So far, the role of SopB in enterocolitis has been studied in the bovine model by using serovar Dublin strain 2229 (16, 27, 35) and serovar Typhimurium strain ATCC 14028s (39, 44, 54). SopB from serovar Typhimurium strain SL1344 has not been analyzed yet in the bovine model. We hypothesized that the absence of a sopB phenotype in the murine colitis model might be related to some difference between Typhimurium strains SL1344 and ATCC 14028s which could not be detected in tissue culture experiments. To address this hypothesis, we constructed several mutants in the genetic background of M741, a wild-type serovar Typhimurium ATCC 14028s strain which carries the streptomycin resistance marker (aadA) from SL1344 (Table 1). ATCC 14028s does not carry the SopEφ phage and is therefore naturally SopE deficient (53). We constructed the sipA sopE2 double effector mutant M746, which harbors a functional gene for SopB, and compared it to wild-type ATCC 14028s and ATCC 14028s mutants with a disrupted SPI-1 and/or SPI-2 TTSS (Table 1) using the streptomycin-pretreated mouse model (Fig. 5 and Table 8) and tissue culture cell invasion assays (see below).
FIG. 5.
Analysis of the contribution of sopB from serovar Typhimurium strain ATCC 14028s to murine colitis. Six groups of five streptomycin-pretreated mice were infected for 2 days with 108 CFU of serovar Typhimurium strains M741, M744, M746, M745, M743, and M742. The relevant genotypes are indicated. (A) Bacterial loads in the cecal contents. (B) Bacterial loads in the liver. (C) Bacterial loads in the spleen. (D) Histopathological analysis. HE-stained sections of cecal tissue were scored with respect to edema in the submucosa, PMN infiltration (PMN inf.), reduction in the number of goblet cells, and desquamation, erosion, or ulceration of the epithelial layer (see Materials and Methods), and the scores were plotted as stacked vertical bars. The total pathological score (sum of the separate scores) was statistically analyzed (stat. anal.) by using the exact Mann-Whitney U test (with comparison to M743). The dotted lines indicate the limits of detection. NS, not statistically significant (P ≥ 0.05); S. Tm, serovar Typhimurium.
TABLE 8.
Statistical analysis of disease parameters in streptomycin-pretreated mice infected with serovar Typhimurium ATCC 14028s mutants lacking the effector proteins SopE2 and SipAa
| Comparison |
P (vs M743) for:
|
||||
|---|---|---|---|---|---|
| M741 | M744 | M746 | M745 | M742 | |
| Combined score | 0.008 | 0.008 | NS | NS | NS |
| Edema | 0.008 | 0.008 | NS | NS | NS |
| PMN infiltration | 0.008 | 0.008 | NS | NS | NS |
| Goblet cells | 0.008 | 0.008 | NS | NS | NS |
| Epithelium | 0.008 | 0.008 | NS | NS | NS |
The data in Fig. 5D were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05). ATCC 14028s is naturally SopE deficient.
Streptomycin-pretreated C57BL/6 mice (five animals per group) were infected with the ATCC 14028s strains, and the bacterial densities in the cecal contents, livers, and spleens, as well as intestinal inflammation, were analyzed at 2 days p.i. With the serovar Typhimurium ATCC 14028s derivatives we observed bacterial densities similar to those in the cecal contents, livers, and spleens of the SL1344 derivatives (see above) (2). At 2 days p.i. M741 (wild-type ATCC 14028s, Strr) induced strong intestinal inflammation, including PMN infiltration, epithelial damage, edema, and the loss of goblet cells (Fig. 5D). Quantitative analysis of the inflammatory changes in the cecal tissues revealed that the sipA sopE double effector mutants M746 and M743 were unable to induce pronounced inflammation (Fig. 5D). It is important to note that the sipA sopE double effector mutants M746 and M743 still harbored a functional sopB gene (see below). Nevertheless, the low residual inflammatory responses observed with M746 and M743 did not differ significantly (P > 0.05) from the responses observed with M741 derivatives carrying a disrupted SPI-1 TTSS (M742 and M745). This is in line with the results obtained with the serovar Typhimurium SL1344 strains and supports the notion that sopB does not play a role in the murine colitis model.
We also compared the virulence of ATCC 14028s mutants carrying an intact or disrupted SPI-2 TTSS. The same levels of intestinal inflammation were observed with M741 and M744 (Fig. 5D) (P > 0.05). Similarly, M746, M743, M745, and M742 were attenuated to similar degrees and produced the same level of residual inflammation (Fig. 5D and Table 8). In conclusion, the data confirmed that translocation of the effector protein SipA and/or SopE2 via the SPI-1 TTSS is a key virulence mechanism in the induction of murine serovar Typhimurium colitis and that the results of our analysis were not affected by the presence of a functional SPI-2 TTSS.
To further characterize the serovar Typhimurium ATCC 14028s mutants, we performed COS7 tissue culture cell invasion assays (Table 9). The results were very similar to the invasion results obtained with serovar Typhimurium strain SL1344. Most importantly, both sipA sopE2 double effector mutants (M746 and M743; intact sopB) were highly invasive (43 or 50%) (Table 9), which suggested that the sopB gene was functional in these two strains. Together with our studies of the SL1344 strain, these data confirmed that SipA, SopE, and SopE2 are important Salmonella virulence factors in vitro and in vivo. In contrast, SopB plays an important role in virulence assays in vitro, but it does not seem to be required for induction of intestinal inflammation in the streptomycin-pretreated mouse model.
TABLE 9.
Serovar Typhimurium strain ATCC14028s mutants lacking SPI-1 effector proteins: invasion of COS7 tissue culture cellsa
| Strain | Relevant genotype
|
Invasiveness (%) | No. of assays |
P vs:
|
||
|---|---|---|---|---|---|---|
| TTSS | Effectors | M741 | M742 | |||
| M741b | Wild type | Wild type | 100 | 6 | 0.002 | |
| M742 | invC | Wild type | 0.14 | 6 | 0.002 | |
| M744 | ssaV | Wild type | 94.2 | 6 | NS | 0.002 |
| M745 | ssaV invC | Wild type | 0.16 | 6 | 0.002 | NS |
| M746c | ssaV | sopE2 sipA | 43.2 | 6 | 0.002 | 0.002 |
| M743c | Wild type | sopE2 sipA | 49.5 | 6 | 0.002 | 0.002 |
The data were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
M741 is a derivative of ATCC 14028s carrying the streptomycin resistance allele from SL1344. The invasiveness of M741 was defined as 100%.
The invasiveness of M746 does not differ significantly from that of M743.
DISCUSSION
Serovar Typhimurium colitis in streptomycin-pretreated mice has been shown to depend on a functional SPI-1 TTSS (2). However, the SPI-1 effector proteins involved have not been identified. Here, we show that the effector protein genes sipA, sopE, and sopE2 play a role in the induction of inflammatory responses in the intestines of streptomycin-pretreated mice. Disruption of the sipA gene resulted in significant attenuation. Further mutagenesis and complementation experiments revealed that the residual capacity of the sipA mutant to induce colitis was attributable to sopE and sopE2.
It should be noted that many experiments in the present study were performed with serovar Typhimurium strains with a disrupted SPI-2 translocon. In this way we ensured that subtle SPI-1-mediated mechanisms were not masked by the onset of the SPI-2-dependent systemic disease. However, our results were subsequently confirmed with SPI-2-proficient strains. Also, it should be kept in mind that the murine colitis model requires antibiotic treatment. In spite of these limitations, our observations are in line with findings obtained with the bovine model and support the notion that streptomycin-pretreated mice offer a useful animal model to study the pathogenetic mechanism of serovar Typhimurium colitis.
SopE contributes to intestinal inflammation in calves and streptomycin-pretreated mice. Interestingly, SopE is not present in all Salmonella species (23, 34, 36). In the sopE-positive serovar Typhimurium strains SopE is encoded by a P2-like prophage (34). Interestingly, serovar Typhimurium strains carrying this prophage have been associated with a major epidemic in the 1970s and 1980s in England and the former German Democratic Republic (34). This suggested that acquisition of the additional sopE gene by lysogenic conversion was an important step in the emergence of these epidemic serovar Typhimurium clones. Experiments with the bovine model have demonstrated that sopE and sopE2 are involved in the elicitation of intestinal inflammation and that lysogenic conversion by SopEφ can increase the virulence of the naïve serovar Typhimurium strain ATCC 14028s (48, 53, 54). The present analysis provides an experimental framework to study how SopE enhances Salmonella virulence.
In bovine ligated ileal loop assays, serovar Dublin sopB mutants are significantly attenuated for induction of PMN influx and fluid secretion (16, 27, 35, 50). Serovar Typhimurium sopB mutants exhibit similar, although much milder, virulence defects in bovine ligated ileal loops (39, 54). However, in orally infected calves, disruption of serovar Typhimurium sopB did not affect the severity of diarrhea and intestinal inflammation (44). In line with the latter observation, we could not detect a virulence defect for the serovar Typhimurium SL1344 sopB mutant. Similarly, the sipA sopEE2 triple effector mutant was attenuated to the same extent as the sopA sopBEE2 quadruple effector mutant. We confirmed these results using the same serovar Typhimurium strain (ATCC 14028s) which had been used for the previous studies with the bovine model (see above). Therefore, the phenotypes of sopB mutants are strikingly similar in streptomycin-pretreated mice and orally infected calves (44).
Why do sopB mutants not show virulence defects in oral infection experiments? Species differences can probably be excluded. All tissue culture experiments performed with primate and murine cells and bovine ligated ileal loop assays (39, 54) suggest that cells having diverse mammalian origins are principally responsive to SopB. Might expression of sopB play a role? For tissue culture and ligated ileal loop experiments, Salmonella spp. are pregrown in culture broth under SPI-1-inducing conditions. In contrast, upon oral infection, the bacteria grow under the specific conditions present in the host's intestine. Therefore, it is conceivable that sopB expression in the intestine is simply insufficient to produce a detectable response in the current oral infection models. However, it should be noted that expression of sopB is controlled by the same regulator (InvF/SicA) as sopE, which does exhibit a virulence phenotype both in tissue culture and in oral infection models (8, 9, 46, 53, 54). Additional work is required to determine the molecular mechanism that explains why sopB mutants do not show virulence defects in bovine and murine oral infection models.
Similar to findings obtained with the bovine model (54), we found that SipA elicits pronounced murine colitis in the absence of SopE, SopE2, and SopB. Furthermore, SopE induced colitis independent of SipA, SopE2, and SopB, and also SopE2 could induce mild inflammation in the absence of SipA, SopE, and SopB. Therefore, SopE, SipA, and to a limited extent SopE2 can induce intestinal inflammation independent of each other and SopB. Interestingly, these effector proteins have quite different cellular targets; SopE and SopE2 are potent activators of host cell Rho GTPases (4, 7, 13, 22, 37, 41), while SipA acts as an actin binding and bundling protein (15, 24, 57, 58). In spite of these fundamental functional differences, SopE-induced inflammation and SipA-induced inflammation are histologically very similar; both include epithelial damage, loss of goblet cells, submucosal edema, PMN infiltration into the lamina propria, and transmigration of PMN into the intestinal lumen. Future work should identify the molecular mechanisms responsible for this observation.
Overall, the results presented here are in line with results obtained with the bovine model. SipA has a key function in elicitation of intestinal inflammation in calves and streptomycin-pretreated mice. SopE (in SL1344) and to a lesser extent SopE2 play a role in both models. The histopathologies induced by SipA, SopE, and/or SopE2, including massive PMN infiltration and epithelial damage, are very similar in the murine and bovine models. Interestingly, the overall contribution of the serovar Typhimurium SPI-1 TTSS to intestinal inflammation seems to vary significantly in different bovine ileal loop and oral infections. In bovine ileal loop assays, serovar Typhimurium mutants with a disrupted SPI-1 TTSS are clearly attenuated but still induce appreciable inflammation (54). However, in orally infected calves these mutants do not induce appreciable inflammation (44). A similar degree of attenuation is observed in the streptomycin-pretreated mouse model. Serovar Typhimurium mutants lacking the effector proteins SipA, SopE, and SopE2 (this study) or an essential component of the SPI-1 TTSS apparatus (2) did not induce obvious intestinal inflammation. Due to this low background inflammation streptomycin-pretreated mice are a sensitive surrogate host model to analyze the inflammatory cascades triggered by the serovar Typhimurium effector proteins SipA, SopE, and SopE2 in the host intestine. Further analyses should profit from the completed mouse and Salmonella genome sequences and from the multitude of immunological reagents and knockout mouse strains available today.
Acknowledgments
We are grateful to Günther Paesold for discussions, to Natalie Schlegel for critical reading of the manuscript, to Andrew Macpherson for scientific discussions, for help with the histopathological evaluation, and for comments on the manuscript, and to Burkhardt Seifert for advice on the statistical analysis. We thank R. Zinkernagel and H. Hengartner for generous support of our animal work at the BZL Zurich.
This work was funded in part by a grant from the Swiss National Foundation (to W.-D.H.).
Editor: B. B. Finlay
REFERENCES
- 1.Bakshi, C. S., V. P. Singh, M. W. Wood, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bispham, J., B. N. Tripathi, P. R. Watson, and T. S. Wallis. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun. 69:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchwald, G., A. Friebel, J. E. Galan, W. D. Hardt, A. Wittinghofer, and K. Scheffzek. 2002. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. M., S. Bagrodia, R. A. Cerione, and J. E. Galan. 1999. Requirement of p21-activated kinase (PAK) for Salmonella typhimurium-induced nuclear responses. J. Exp. Med. 189:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. Inoculum composition and Salmonella pathogenicity island 1 regulate M-cell invasion and epithelial destruction by Salmonella typhimurium. Infect. Immun. 66:724-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criss, A. K., D. M. Ahlgren, T. S. Jou, B. A. McCormick, and J. E. Casanova. 2001. The GTPase Rac1 selectively regulates Salmonella invasion at the apical plasma membrane of polarized epithelial cells. J. Cell Sci. 114:1331-1341. [DOI] [PubMed] [Google Scholar]
- 8.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Ehrbar, K., A. Friebel, S. I. Miller, and W.-D. Hardt. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950-6967. [DOI] [PMC free article] [PubMed]
- 9.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, Y., S. R. Wente, and P. W. Majerus. 2001. Overexpression of the inositol phosphatase SopB in human 293 cells stimulates cellular chloride influx and inhibits nuclear mRNA export. Proc. Natl. Acad. Sci. USA 98:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friebel, A., H. Ilchmann, M. Aelpfelbacher, K. Ehrbar, W. Machleidt, and W. D. Hardt. 2001. SopE and SopE2 from Salmonella typhimurium activate different sets of Rho GTPases of the host cell. J. Biol. Chem. 276:34035-34040. [DOI] [PubMed] [Google Scholar]
- 14.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 15.Galkin, V. E., A. Orlova, M. S. VanLoock, D. Zhou, J. E. Galan, and E. H. Egelman. 2002. The bacterial protein SipA polymerizes G-actin and mimics muscle nebulin. Nat. Struct. Biol. 9:518-521. [DOI] [PubMed] [Google Scholar]
- 16.Galyov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Watson, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25:903-912. [DOI] [PubMed] [Google Scholar]
- 17.Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 18.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman, E. A., and H. Ochman. 1997. How Salmonella became a pathogen. Trends Microbiol. 5:343-349. [DOI] [PubMed] [Google Scholar]
- 20.Gruenheid, S., and B. B. Finlay. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422:775-781. [DOI] [PubMed] [Google Scholar]
- 21.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 22.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 23.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higashide, W., S. Dai, V. P. Hombs, and D. Zhou. 2002. Involvement of SipA in modulating actin dynamics during Salmonella invasion into cultured epithelial cells. Cell. Microbiol. 4:357-365. [DOI] [PubMed] [Google Scholar]
- 25.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 26.Hong, K. H., and V. L. Miller. 1998. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J. Bacteriol. 180:1793-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, M. A., M. W. Wood, P. B. Mullan, P. R. Watson, T. S. Wallis, and E. E. Galyov. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun. 66:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 29.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. USA 97:12283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina, E., P. Paglia, T. Nikolaus, A. Muller, M. Hensel, and C. A. Guzman. 1999. Pathogenicity island 2 mutants of Salmonella typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect. Immun. 67:1093-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirold, S., K. Ehrbar, A. Weissmüller, R. Prager, H. Tschäpe, H. Rüssmann, and W. D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 183:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris, F. A., M. P. Wilson, T. S. Wallis, E. E. Galyov, and P. W. Majerus. 1998. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA 95:14057-14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knuppel, W. Rabsch, W. D. Hardt, and H. Tschape. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 37.Rudolph, M. G., C. Weise, S. Mirold, B. Hillenbrand, B. Bader, A. Wittinghofer, and W. D. Hardt. 1999. Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for Rho GTPases. J. Biol. Chem. 274:30501-30509. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santos, R. L., R. M. Tsolis, S. Zhang, T. A. Ficht, A. J. Baumler, and L. G. Adams. 2001. Salmonella-induced cell death is not required for enteritis in calves. Infect. Immun. 69:4610-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 41.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 42.Stojiljkovic, I., A. J. Baumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terebiznik, M. R., O. V. Vieira, S. L. Marcus, A. Slade, C. M. Yip, W. S. Trimble, T. Meyer, B. B. Finlay, and S. Grinstein. 2002. Elimination of host cell PtdIns(4, 5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 4:766-773. [DOI] [PubMed] [Google Scholar]
- 44.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsolis, R. M., R. A. Kingsley, S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261-274. [PubMed] [Google Scholar]
- 46.Tucker, S. C., and J. E. Galan. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzzau, S., D. J. Brown, T. Wallis, S. Rubino, G. Leori, S. Bernard, J. Casadesus, D. J. Platt, and J. E. Olsen. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 49.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 50.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883-891. [DOI] [PubMed] [Google Scholar]
- 51.Wood, M. W., M. A. Jones, P. R. Watson, A. M. Siber, B. A. McCormick, S. Hedges, R. Rosquist, T. S. Wallis, and E. E. Galyov. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritidis. Cell. Microbiol. 2:293-303. [DOI] [PubMed] [Google Scholar]
- 52.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, S., R. L. Santos, R. M. Tsolis, S. Mirold, W.-D. Hardt, L. G. Adams, and A. J. Bäumler. 2002. Phage mediated horizontal transfer of the sopE1 gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for calves. FEMS Microbiol. Lett. 217:243-247. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, S., R. L. Santos, R. M. Tsolis, S. Stender, W. D. Hardt, A. J. Baumler, and L. G. Adams. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-260. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. USA 96:10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092-2095. [DOI] [PubMed] [Google Scholar]