Abstract
We asked whether Campylobacter jejuni isolated from patients with Guillain-Barré syndrome (GBS) differ from isolates isolated from patients with uncomplicated gastrointestinal infection using DNA microarray analysis. We found that specific GBS genes or regions were not identified, and microarray analysis confirmed significant genomic heterogeneity among the isolates.
Campylobacter jejuni subsp. jejuni (referred to as C. jejuni hereafter) is one of the most common causes of bacterial infectious diarrhea and is estimated to cause approximately 2.5 million cases of infectious diarrhea per year in the United States (5). In most cases of C. jejuni infection, patients develop acute gastroenteritis and resolve the infection without complications. There is a small subset of patients with C. jejuni infection who develop the postinfectious neurologic disorder, Guillian-Barré syndrome (GBS), an acute, immune system-mediated polyneuropathy that leads to ascending paralysis (9, 22). C. jejuni has been implicated as one of the leading infectious agents associated with this syndrome (15).
Although not completely understood, the development of GBS after C. jejuni infection is thought to be related to molecular mimicry by anti-Campylobacter lipooligosaccharide (LOS) antibodies that cross-react with ganglioside epitopes on neural tissue (22). C. jejuni express a number of ganglioside-like moieties in the outer LOS core region, including GM1-like and GD1a-like structures, and some patients with Campylobacter-associated GBS develop antibodies to these and other ganglioside-like structures (14). The resulting antibodies are thought to target relevant epitopes in peripheral nerve tissue. causing demyelination and axonal damage (7, 8).
With the availability of the C. jejuni sequence (18), genomic and expression analyses using a C. jejuni DNA microarray have been made possible (3, 11, 20). In order to better understand the pathogenesis of C. jejuni associated with GBS, we compared a collection of isolates from patients with GBS with isolates from patients with uncomplicated gastrointestinal infection using an open reading frame (ORF)-specific C. jejuni DNA microarray. The aim of this study was to identify unique differences within the genome that were associated with GBS.
Twelve strains of C. jejuni associated with GBS were compared to 12 strains associated with uncomplicated gastrointestinal illness (Table 1). A variety of serotypes are represented in both groups of isolates. Some of the isolates were also previously characterized by multilocus enzyme electrophoresis and represent a number of different electrophoretic types (4, 16).
TABLE 1.
Bacterial isolates used in microarray study
| Isolate designationa | Strain | Country of originb | HS serotype(s) |
|---|---|---|---|
| GBS-associated isolates | |||
| 1 | HB93-6 | P.R. China | 2 |
| 2 | DVL5906 | Denmark | 2 |
| 3 | JHU2 | USA | 4 |
| 4 | DVL5610 | Denmark | 4 |
| 5 | HB93-10 | P.R. China | 5 |
| 6 | HB93-13 (ATCC 700297) | P.R. China | 19 |
| 7 | OH4384 | Japan | 19 |
| 8 | INP7 (ATCC BAA-528) | Mexico | 19 |
| 9 | HB97-34 | P.R. China | 37 |
| 10 | INP16 | Mexico | 37 |
| 11 | INP21 (ATCC BAA-530) | Mexico | 41 |
| 12 | INP59 (ATCC BAA-529) | Mexico | 41 |
| Enteritis-associated isolates | |||
| 14 | DVL5384 | Denmark | 4 |
| 15 | DVL5529 | Denmark | 4 |
| 16 | INP66 | Mexico | 5 |
| 17 | D3002 | USA | 19 |
| 18 | KB1428 | Japan | 19 |
| 19 | INP15 | Mexico | 19 |
| 20 | DVL5443 | Denmark | 37 |
| 22 | DVL5671 | Denmark | 41 |
| 23 | D3007 | USA | 2 |
| 24 | 81-176 | USA | 23, 36 |
| 25 | D3027 | USA | 2 |
| 26 | DVL5558 | Denmark | 41 |
| NCTC 11168 | UK | 2 |
DNA microarray analysis was performed blindly, without knowledge of strain designation or clinical outcome. Genomic DNA was prepared using a Wizard Prep kit per the manufacturer's instructions (Promega). All strains were compared to a reference strain composed of DNA from the sequenced strain NCTC 11168 and DNA from the virulence plasmid (pVir) isolated from strain 81-176 (1, 2); the pVir DNA was added to approximate one copy of plasmid per genome. Hybridization was performed using 500 ng of chromosomal DNA from each isolate and the reference array as previously described (11, 19). All strains were hybridized to the microarray in duplicate. After the hybridizations were completed, the microarrays were scanned using an Axon scanner and Genepix 3.0 software (Axon Instruments, Redwood City, Calif.). The data were submitted to the Stanford Microarray Database and retrieved using the following filters: (i) spots with Ch1 pixels > background level plus 1 standard deviation and (ii) values of failed spots are set at 0. The microarray replicates were shown to be reproducible by correlation coefficients and averaged after the data was analyzed and transformed into binary format using a program called Genomotyping Analysis (GACK) (10) (http://cmgm.stanford.edu/falkow/whatwedo/software/software.html). This program calculates an idealized normal distribution curve for each array and assigns a binary value to each data point on the microarray, depending on an estimated probability of a gene being present or absent in a given strain relative to the same probability in reference strain 11168. We designated genes that had an equal probability of being present or absent as present in the analysis. Gene identification used the annotated C. jejuni genome available on the Sanger Centre website (http://www.Sanger.ac.uk/Projects/C_jejuni). The microarray data were subjected to hierarchical clustering between arrays with the CLUSTER program, and the results were presented using the TREEVIEW program (19).
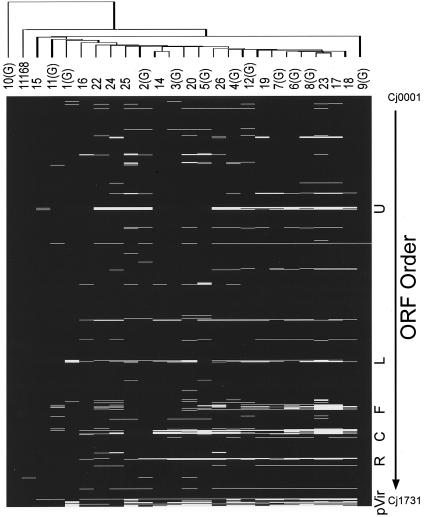
Microarray analysis did not identify discrete groups of isolates or any unique features within the genome of the C. jejuni isolates associated with GBS. Cluster analysis of the groups of isolates with the same serotype was also performed and failed to segregate isolates associated with GBS from isolates associated with gastroenteritis (Fig. 1). The pVir plasmid was not detected in any of the strains studied. However, pVir has been reported to contain several chromosomal homologues, such as fliH and a gene with unknown function that could account for conserved regions within the pVir section of the array (2). Our results suggest that the differences, if any, between strains of C. jejuni that were associated with GBS versus those associated only with enteritis were not at the level of the genome. However, we cannot determine from this study whether the lack of hybridization in various regions represents the absence of a particular gene or nucleotide divergence within an existing gene. Additionally, differences due to the presence of genetic elements in either the GBS- or enteritis-related isolates would not necessarily be detected because of the absence of such elements in the genome of the strain used to construct the microarray.
FIG. 1.
Comparison of C. jejuni isolates to reference strain 11168. Isolates are listed across the top of the schematic by the designations given in Table 1. Strains associated with GBS patients are indicated by (G) after the isolate designation. Conserved genes (black) and divergent or absent genes (white) are indicated. The dendrogram represents a cluster analysis of strains as described in the text. Isolates found within the same node are considered more related than isolates found outside the node. ORF order abbreviations: U, uxaA/sugar modification; L, LOS; F, flagella; C, capsule; R, type I restriction enzyme proteins R, M, and S.
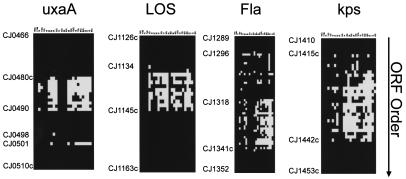
The divergent regions within the C. jejuni genome involve well-defined regions (Fig. 2). Compared with strain 11168, GBS- and enteritis-associated strains had 91.4 and 90.9% ORFs in common, respectively. Compared with enteritis-associated strains, GBS-associated isolates had 89% ORFs in common. Approximately 11% of the ORFs contained in strain 11168 were absent in the GBS- and enteritis-associated strains. Many of the genes that were divergent or absent in the GBS- and enteritis-associated strains were in the same region, regardless of how the strains were compared. These regions of the genome include genes involved in production of the capsule, flagella, and LOS. In addition, there were areas of heterogeneity in the uxaA locus involved with sugar transport and metabolism of hexuronates leading to the formation of d-2-keto-3-deoxy-d-gluconate and is part of the exu regulon (12). There were other less-defined areas of heterogeneity in some strains compared to the genome sequence that were mostly ORFs without an identified function thus far. Variation in type I modification and restriction genes were also observed among the isolates. While the reasons for this variation are unknown, such variation has been observed by our group (Erin Gaynor, personal communication) and we can only speculate that since C. jejuni is naturally transformable, it may have evolved multiple mechanisms to avoid incorporating foreign DNA. Taken together, these results suggest that many C. jejuni strains undergo extensive surface protein modification. These results are similar to findings obtained from microarray analyses of a series of unrelated strains, as well as in bacteria that were epidemiologically linked (3, 11).
FIG. 2.
Detailed view of four major variable regions identified by DNA microarray analysis of C. jejuni isolates associated with GBS and enteritis. Isolates are listed across the top of the schematic by the designations given in Table 1. Strains associated with GBS patients are indicated by (G) after the isolate designation. Conserved genes within the genome relative to reference strain 11168 (black) and genes that are divergent or absent from the reference strain (white) are indicated. Fla, flagella; kps, capsule.
A comparison of C. jejuni strains associated with GBS has not previously been studied by DNA microarray analysis. We are also not aware of studies using subtractive hybridization between GBS- and enteritis-related isolates that could reveal additional differences not detected with the microarray approach in the present study. Certain serotypes of C. jejuni are known to be overrepresented in cases of campylobacter-induced GBS, such as HS:19 and HS:41, suggesting that strains expressing these serotypes might contain specific virulence factors relevant to GBS (15). Previous molecular typing studies have not, however, identified unique features of GBS-associated strains (6), although the sialyltransferase gene, cstII, has been identified as being overrepresented in GBS-related strains of C. jejuni (16, 17, 21). On the other hand, there is evidence from a recent study that C. jejuni isolates associated with GBS preferentially express GD1a-like structures, regardless of the bacterial serotype (17). Possibly, posttranslational modification of the surface structures might account for the discordant results, suggesting that GBS-associated strains might encode regulatory or modification loci that were not detected by microarray analysis. Such modifications might be an important mechanism in the development of molecular mimicry. Alternatively, differential expression of Campylobacter virulence factors might be operative and would be disclosed by expression analysis. Host susceptibility is likely an important factor in the development of GBS (13), and further characterization of the interaction between the host and C. jejuni is needed to better understand the pathogenesis of Campylobacter-induced GBS.
Acknowledgments
We thank Huong Ung for technical assistance. We also thank Erin Gaynor for helpful discussions.
This work was supported in part by the Howard Hughes Postdoctoral Research Fellowship for Physicians and the Minority Medical Faculty Development Award from the Robert Wood Johnson Foundation (E.E.L.), Stanford University Digestive Disease Center (grant DK56339) (S.F. and L.S.T.), and the National Institutes of Health (grant NS-31528) (I.N.).
Editor: V. J. DiRita
REFERENCES
- 1.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engberg, J., I. Nachamkin, V. Fussing, G. M. McKhann, J. W. Griffin, J. C. Piffaretti, E. M. Nielsen, and P. Gerner-Smidt. 2001. Absence of clonality of Campylobacter jejuni in serotypes other than HS:19 associated with Guillain-Barre syndrome and gastroenteritis. J. Infect. Dis. 184:215-220. [DOI] [PubMed] [Google Scholar]
- 5.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 6.Fujimoto, S., B. M. Allos, N. Misawa, C. M. Patton, and M. J. Blaser. 1997. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barre syndrome. J. Infect. Dis. 176:1105-1108. [DOI] [PubMed] [Google Scholar]
- 7.Hafer-Macko, C., S.-T. Hsieh, C. Y. Li, T. W. Ho, K. A. Sheikh, D. R. Cornblath, G. M. McKhann, A. K. Asbury, and J. W. Griffin. 1996. Acute motor axonal neuropathy: an antibody-mediated attack on axolemma. Ann. Neurol. 40:635-644. [DOI] [PubMed] [Google Scholar]
- 8.Hafer-Macko, C., K. A. Sheikh, C. Y. Li, T. W. Ho, D. R. Cornblath, G. M. McKhann, A. K. Asbury, and J. W. Griffin. 1996. Immune attack on the Schwann cell surface in acute inflammatory demyelinating polyneuropathy. Ann. Neurol. 39:625-635. [DOI] [PubMed] [Google Scholar]
- 9.Ho, T. W., G. M. McKhann, and J. W. Griffin. 1998. Human autoimmune neuropathies. Annu. Rev. Neurosci. 21:187-226. [DOI] [PubMed] [Google Scholar]
- 10.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 29 October 2002, posting date. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:research0065.1-0065.17. [Online.] http://genomebiology.com/2002/3/11/research/0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 12.Lin, E. C. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella. ASM Press, Washington, D.C.
- 13.Magira, E. E., M. Papaioakim, I. Nachamkin, A. K. Asbury, C. Y. Li, T. W. Ho, J. W. Griffin, G. M. McKhann, and D. S. Monos. 2003. Differential distribution of HLA-DQβ/DRβ epitopes in the two forms of Guillain-Barre syndrome, acute motor axonal neuropathy (AMAN) and acute inflammatory demyelinating polyneuropathy (AIDP): identification of DQβ epitopes associated with susceptibility to and protection from AIDP. J. Immunol. 170:3074-3080. [DOI] [PubMed] [Google Scholar]
- 14.Moran, A. P., and M. M. Prendergast. 2001. Molecular mimicry in Campylobacter jejuni and Helicobacter pylori lipopolysaccharides: contribution of gastrointestinal infections to autoimmunity. J. Autoimmun. 16:241-256. [DOI] [PubMed] [Google Scholar]
- 15.Nachamkin, I., B. M. Allos, and T. W. Ho. 2000. Campylobacter jejuni infection and the association with Guillain-Barre syndrome, p. 155-175. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 16.Nachamkin, I., J. Engberg, M. Gutacker, R. J. Meinersmann, C. Y. Li, P. Arzarte Barbosa, E. Teeple, V. Fussing, T. W. Ho, A. K. Asbury, J. W. Griffin, G. M. McKhann, and J. C. Piffaretti. 2001. Molecular population genetic analysis of Campylobacter jejuni HS:19 associated with Guillain-Barre syndrome and gastroenteritis. J. Infect. Dis. 184:221-226. [DOI] [PubMed] [Google Scholar]
- 17.Nachamkin, I., J. Liu, M. Li, H. Ung, A. P. Moran, M. M. Prendergast, and K. Sheikh. 2002. Campylobacter jejuni from patients with Guillain-Barre syndrome preferentially express a GD1a-like epitope. Infect. Immun. 70:5299-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 19.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. S. Tompkins, and S. Falkow. 2000. A whole genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum, A., N. Van Den Braak, P. Godschalk, C. W. Ang, B. Jacobs, M. Gilbert, W. W. Wakarchuk, H. Verbrugh, and H. Endtz. 2001. A Campylobacter jejuni gene associated with immune-mediated neuropathy. Nat. Med. 7:752-753. [DOI] [PubMed] [Google Scholar]
- 22.Yuki, N. 2001. Infectious origins of, and molecular mimicry in, Guillain-Barre and Fisher syndromes. Lancet Infect. Dis. 1:29-37. [DOI] [PubMed] [Google Scholar]