Abstract
Hepatocyte-like cells derived from the differentiation of human embryonic stem cells (hES-Hep) have potential to provide a human relevant in vitro test system in which to evaluate the carcinogenic hazard of chemicals. In this study, we have investigated this potential using a panel of 15 chemicals classified as noncarcinogens, genotoxic carcinogens, and nongenotoxic carcinogens and measured whole-genome transcriptome responses with gene expression microarrays. We applied an ANOVA model that identified 592 genes highly discriminative for the panel of chemicals. Supervised classification with these genes achieved a cross-validation accuracy of > 95%. Moreover, the expression of the response genes in hES-Hep was strongly correlated with that in human primary hepatocytes cultured in vitro. In order to infer mechanistic information on the consequences of chemical exposure in hES-Hep, we developed a computational method that measures the responses of biochemical pathways to the panel of treatments and showed that these responses were discriminative for the three toxicity classes and linked to carcinogenesis through p53, mitogen-activated protein kinases, and apoptosis pathway modules. It could further be shown that the discrimination of toxicity classes was improved when analyzing the microarray data at the pathway level. In summary, our results demonstrate, for the first time, the potential of human embryonic stem cell--derived hepatic cells as an in vitro model for hazard assessment of chemical carcinogenesis, although it should be noted that more compounds are needed to test the robustness of the assay.
Keywords: carcinogenicity, systems toxicology, risk assessment, toxicogenomics, computational biology
The inherent capacity of human embryonic stem cells (hESC) to grow indefinitely and to differentiate into all mature cell types of the body makes them extremely attractive for toxicity testing and other applications, such as regenerative medicine, tissue engineering, and drug discovery (Thomson et al., 1998). Although a few factors still limit the general implementation of pluripotent stem cells for clinical applications, their opportunities for use in predictive in vitro assays are immense and fuel further developments of improved cellular models that may increase their relevance for the human situation in vivo and reduce the need of experimental animals in testing of drugs, cosmetics, and other chemical compounds (Jensen et al., 2009).
To improve the assessment of the carcinogenic hazard (and ultimately the risk) due to the exposure to chemicals is a major challenge to public health and customer’s safety. It has been addressed in Europe within the Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) initiative aiming to assess toxicity of an estimated number of 68,000 chemicals (Hartung and Rovida, 2009). Until now, the majority of tests are based on in vivo assays, in particular on the 2-year rodent bioassay for carcinogenicity. Besides the challenge of replacing animal testing (it has been estimated that full compliance with REACH legislation for all endpoints of toxicity will require a grand total of 54 million vertebrate animals and will cost €9.5 billion over the next decade), it has been argued that the effects of chemical exposure differ widely in rodents and humans, and this might lead to a high number of false positive predictions. For example, cholesterol-lowering drugs, such as atorvastatin, fluvastatin, and simvastatin among many other pharmaceutical agents approved as safe drugs for human use by the FDA, were classified as rodent carcinogens (Ward, 2008). Thus, it has been understood that human in vitro assays must be developed for predicting carcinogenic effects of chemicals in human more reliably (Vinken et al., 2008).
High-throughput technologies such as microarrays have opened the way to a systemic understanding of toxicology and carcinogenicity (Waters and Fostel, 2004). Such systems toxicology approaches offer the chance for disclosing valuable mechanistic information on toxic modes of action of substances in the assay system under study, which obviously is promising in view of the urgent need to develop better (in vitro) tests for chemical safety. Mechanistic approaches require the analysis of whole-genome data at the pathway level incorporating knowledge on human interaction networks (Kitano, 2002; Wierling et al., 2007). Pivotal for such approaches is the availability of sufficient information on human pathways, related to cancer initiation and progression, along with computational approaches that combine results from high-throughput experiments with biological networks (Bader et al., 2006; Kamburov et al., 2011).
The liver appears a major target for the effect of carcinogenic compounds in the rodent 2-year bioassay. Although recent reports evidenced the suitability of human carcinoma cell lines for classification of chemicals (Jennen et al., 2010), the applicability of stem cells for such purpose is still unexplored. The feasibility of differentiating hESC to hepatic cells (hES-Hep) using a developmental biology approach (Brolén et al., 2010) has already been shown. In the present work, we demonstrate the application of a hESC-derived cell assay for hazard assessment of carcinogenicity with a panel of 15 chemicals from three different toxicity classes (genotoxic carcinogens [GTX], nongenotoxic carcinogens [NGTX], and noncarcinogens [NC]) using Affymetrix microarray measurements before and after chemical treatment of the cells. We applied an ANOVA model that identified 592 genes highly discriminative for the assayed chemicals. The expression of these response genes as well as the expression of Phase I–III genes derived from hES-Hep is strongly correlated with those in human primary hepatocytes cultured in vitro demonstrating the metabolic competence of the system. Supervised classification analysis with the response genes, also incorporating additional data, achieved a cross-validation performance of > 95%. In order to deduce mechanistic information on the modes of action of the different compounds in hES-Hep, we assigned numerical scores based on expression data to 1695 manually annotated human pathways originating from several pathway resources (Kamburov et al., 2011) that reflect the response of these pathways to the chemicals. Using this approach, we were able to identify discriminative pathway modules, both for individual substances and toxicity classes, in particular in p53, mitogen-activated protein kinase (MAPK), and apoptosis pathways. Moreover, it was observed that the discrimination of the toxicity classes was improved when shifting from the gene to the pathway level analysis of microarray data.
To summarize, our results demonstrate, for the first time, the potential of hESC-derived hepatocyte-like cells as an in vitro model for hazard assessment of carcinogenicity of chemical compounds and open a new application domain for stem cell research in toxicology.
MATERIALS AND METHODS
Cultures of hESC and differentiated hepatocytes.
This study was performed with the commercially available product hES-Hep002 (Cellartis AB, Göteborg, Sweden, http://www.cellartis.com). The hESC line SA002 was derived, cultured, and characterized as previously described (Heins et al., 2004, 2006) with an additional step of enzymatic passage for further expansion before the onset of differentiation. Characterization and start of compound incubation were performed at day 22 after the start of hepatic differentiation.
Other liver cellular models.
RNA from five different human liver cell models was compared with hES-Hep. HepG2 (HB-8065; American Type Culture Collection, Manassas, VA), human fetal liver polyA + RNA (Clontech Laboratories, Inc.), human adult liver tissue, and nonplated and plated adult human primary hepatocytes (hpHep). All human adult liver samples were isolated from two different donors and were obtained as described in Brolén et al. (2010). The isolated human hepatocytes were split into two aliquots. One of these was immediately directed into RNA (nonplated hpHep), and the other one was cultured 48 h in vitro before RNA was isolated (plated hpHep).
Immunocytochemistry.
Cells were fixed in 4% (wt/vol) paraformaldehyde for 15 min at room temperature, washed twice with PBS, blocked with 5% skim milk in PBS for 30 min, and then incubated with primary antibody at the appropriate dilution in PBS with 1% bovine serum albumin (BSA) and 0.2% Triton X-100 at 4°C overnight. After washing with PBS, the secondary antibody, in PBS with 1% BSA and 0.2% Triton X-100, was applied for 1 h. To visualize the nucleus, 4′,6-diamidino-2-phenylindole at 0.5 μg/ml was included during the secondary antibody incubation. The following primary antibodies were used: rabbit anti-albumin (1:500; Bethyl Lab), mouse anti-α-fetoprotein (AFP) (1:500; Sigma), mouse anti-cytokeratin (CK18) (1:200; DAKO Cytomation), rabbit anti-α1-antitrypsine (α1-AT) (1:200; DAKO Cytomation), rabbit anti-Cyp1A2 (1:100; Biomol), rabbit anti-MRP2 (1:50; Santa Cruz), and rabbit anti-HNF4α (1:300; Santa Cruz). The following secondary antibodies were used: Alexa Fluor 488 donkey anti-rabbit (1:1000; Molecular Probes) and Alexa Fluor 488 donkey anti-mouse (1:1000; Molecular Probes). The cells were finally mounted with DAKO fluorescent mounting medium and visualized and captured using a Nikon Eclipse TE2000-U Fluorescence microscope and Nikon Act-1C for DXM1 200C software.
RNA extraction, reverse transcription, and quantitative PCR.
Cells were isolated in RNAprotect Cell Reagent (Qiagen), and total RNA was extracted using RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. Reverse transcription was performed using 0.6 μg of total RNA in a final volume of 20 μl together with reverse transcriptase (Roche Diagnostics), using a High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems) and an Eppendorf Mastercycler Gradient. Each RNA sample was reverse transcribed in duplicate, and appropriate negative controls were included in each run. TaqMan real-time quantitative PCR (qPCR) assay-on-demand primers from Applied Biosystems (ABI) were used for the following genes: CYP1A2, CYP3A4, CYP2C9, OCT1, OATP2, GSTA1, BCEP, and qPCR were conducted as previously described by Ek et al. (2007). For validation of the microarray data, TPD52, TNFAIP3, and PDCD4 were purchased from ABI. Real-time qPCR was conducted as previously described across the panel of treatments (Brolén et al., 2010).
Cytochrome P450 activity.
Cytochrome P450 (CYP) activity assays were performed by the direct incubation of hES-Hep (day 22) and human primary hepatocytes (plated for 48 h) in monolayer cultures with a cocktail of substrates, each one specific for one CYP enzyme, at following final concentrations: 10μM 7-bupropion, 10μM phenacetin, 10μM diclofenac, 5μM midazolam, and 50μM mephenytoin. After 16 h at 37°C, the supernatant was collected and metabolites formed by cells during the incubation were quantified by HPLC-MS/MS as described in Donato et al. (2010) and Lahoz et al. (2008).
Compound treatment experiments.
Samples were generated by incubating hES-Hep with 15 compounds of three different toxicity classes that were chosen from a previously defined body of model compounds causing genotoxicity and carcinogenicity (Vinken et al., 2008). Genotoxic carcinogens (GTX) were 2-nitrofluorene (2NF), benzo[a]pyrene (BAP), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), aflatoxin B1 (AFL), and cyclophosphamide monohydrate (CYC); nongenotoxic carcinogens (NGTX) were methapyrilene hydrochloride (MPH), piperonylbutoxide (PPX), sodium phenobarbital (SPB), WY-14,643/pirinixic acid (WYE), and tetradecanoyl phorbol acetate (TPA); NC were sodium diclofenac (DIC), D-mannitol (DMA), nifedipine (NFE), clonidine hydrochloride (CLO), and tolbutamide (TOL). Chemicals were obtained from Sigma Chemical Co, Aldrich Chemical Company. The hES-Hep were incubated with compounds at IC10 concentrations for 72 h (Supplementary table 1) and compared with respective control samples (hES-Hep cultured in media supplemented with 0.5% dimethyl sulfoxide [DMSO]). Final concentration of DMSO was 0.5% in all cultures and assays were run in triplicates using three different cell passages.
Determining final exposure concentrations based on the MTT assay.
The cytotoxicity of the 15 compounds investigated was assessed using the MTT assay (reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), following the manufacturer’s instructions (Sigma) (Supplementary figure 1A). Dilution series for each of the 15 chemicals were performed. hESC-derived hepatocyte-like cells cultured in 0.1% gelatin-coated 24-well plates were incubated for 72 h. Incubated cells and controls were washed in PBS, and the MTT assay was started by adding 0.4 ml per well of MTT reagent, dissolved in PBS and diluted to 0.5 mg/ml in phenol red–free media, and incubated for 1 h at 37°C. The supernatants were collected, and the spectrophotometric absorbance was read at 550 nm. From these absorbance data, IC10 at 72 h was calculated and determined with the GraphPad Prism 5 software. The MTT curves displayed data from maximum achievable concentration to zero and were repeated 3–5 times with cells from different passages for each concentration and for all 15 compounds.
Microarray data generation.
Target preparation and microarray hybridization of the Affymetrix Human Genome U133 Plus 2.0 GeneChip arrays were performed by standard methods as described in Jennen et al. (2010). The arrays were scanned by means of an Affymetrix GeneArray scanner. Normalization quality controls, including scaling factors, average intensities, present calls, background intensities, noise, and raw Q values, were within acceptable limits for all chips. Hybridization controls, BioB, BioC, BioD, and CreX, were called present on all chips and yielded the expected increases in intensities.
Gene expression analysis.
Raw microarray data was remapped to the Ensembl version 55 genome build as previously described in Dai et al. (2005). Data was preprocessed with the GC-RMA method. For each gene in each exposure experiment, we computed its detection p value indicating the strength of gene expression and its fold change (log base 2) based on the average values of the three treatments and the three control experimental replicates. Fold changes for all chemical treatments were analyzed with a one-way ANOVA model in order to identify a response gene set that is able to discriminate between the three compound classes. The significance of variance ratios was quantified with the F statistic (p value < 0.05). Additionally, we computed a one-sample Student’s t-test (p value < 0.05) to judge whether the gene’s fold changes were consistent within the same toxicity class.
Pathway response analysis.
As pathway resource, we used the ConsensusPathDB (Kamburov et al., 2011; http://cpdb.molgen.mpg.de), a metadatabase that integrates the content of 22 different interaction databases and that comprises 2144 predefined human pathways, of which 1695 had more than five members and were used for pathway response analysis. We computed a response score Sij for each gene i in each treatment experiment j by
where Rij and Pij are the fold change and the p value, respectively, of gene i when comparing the treatment j microarray replicates against their control replicates. The score for each pathway is the average score derived from all scores of genes assigned to the pathway. In order to make the raw pathway response scores comparable across different treatment experiments, we computed the log2 ratios of the pathway response scores for a particular treatment and the median score of that treatment.
Cross-validation analysis.
We used supervised classification and follow-up cross-validation in order to challenge the response gene set with the problem of classifying chemicals according to the toxicity classes. The analysis was done with a support vector machine with a linear kernel and a penalty factor of 10. To assess the misclassification rates, we used the Leaving-One-Out (LOO) method by removing and subsequent classification of single compound patterns.
RESULTS
Liver Characteristics of hES-Hep
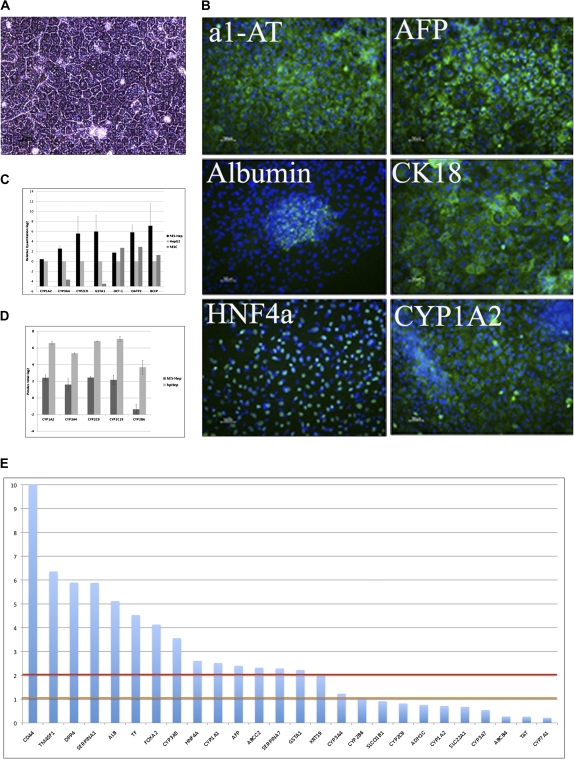
Differentiation of hESC into hepatocyte-like cells was carried out as described in “Materials and Methods”. The hES-Hep were grown at day 22 after the onset of differentiation in homogenous cultures in monolayer displaying a typical hepatic morphology (Fig. 1A) and were positive for important hepatocyte markers and liver-related proteins. For example, α1-AT, AFP, ALB, CK18, and HNF4α were expressed along with CYP1A2 and visualized by immunocytochemistry (ICC) (Fig. 1B). Hepatic phase I enzymes (CYP1A2, CYP3A4, and CYP2C9), phase II enzyme (GSTα1), and phase III transporter proteins (OCT1, OATP2, and BCEP) were detected on mRNA level by real-time qPCR (Fig. 1C). Additionally, results from the CYP activity measurements confirmed the presence of functional enzymatic activity of the most important hepatic CYP enzymes (CYP1A2, CYP3A4, CYP2B6, CYP2C9, and CYP2C19; Fig. 1D) in comparison with hpHep.
FIG. 1.
Hepatocyte-like characteristics of hES-Hep. (A) Phase contrast microscopy demonstrating the typical hES-Hep morphology after 22 days of differentiation. (B) Cells were positive for hepatocyte markers, such as α1-AT, AFP, albumin, CK18, HNF4α, and CYP1A2. (C) qPCR gene expression of hES-Hep show an expression of CYP1A2, CYP3A4, CYP2C9, OCT1, OATP2, GSTA1, and BCEP in comparison with HepG2 and undifferentiated hESC. Y-axis shows log2 values relative to HepG2 and hESC (n = 4). (D) CYP activity of CYP1A2, CYP3A4, CYP2C9, CYP2C19, and CYP2B6 further supporting that these cells are hepatocyte-like. Y-axis scale is Pmoles total log2 (n = 3). hpHep = human primary hepatocytes cultured for 48 h. (E) Significance of gene expression of hepatic markers in untreated hES-Hep. Y-axis displays the negative log10 of the detection p values of the gene expression signals. Horizontal lines indicate the 0.1 (orange) and 0.01 (red) detection levels.
Microarray data from untreated hES-Hep gave insights into the transcriptome of the cells. Remapping of oligoprobes yielded 18,394 Ensembl-annotated genes with sufficient uniquely mapped probe sequences. Of these, 8745 genes (47.54%) were found expressed in hES-Hep (detection p value < 0.01). In addition, 2976 genes were expressed at a lower level (detection p value between 0.01 and 0.1). The actual number of expressed genes in hES-Hep relates to published estimates of gene expression in human hepatocytes found with other technologies. For example, 7475 genes were previously found expressed in human hepatocytes on the basis of public expressed sequence tag data (Huang et al., 2007). Among the expressed genes, we found hepatocyte markers such as CD44, TM4SF1, DPP4, SERPINA1/α1-AT, ALB, TF, FOXA2, CYP3A5, HNF4α, CYP1A1, AFP, ABCC2, SERPINA7, GSTA1, KRT19, CYP3A4, and CYP2B6. Several markers that were tested for hepatocyte-like characteristics of the cells were not detected with microarrays including SLCO1B1, CYP2C9, ADH1C, CYP1A2, SLC22A1, CYP3A7, ABCB4, TAT, and CYP7A1 (Fig. 1E). These genes were either not expressed in the cells, expressed at a level below the detection limit of microarrays, or were not annotated after remapping of the oligoprobes. For example, cytokeratin 18 (CK18) is a hepatic marker that was clearly visible in the ICC screens (cf. Fig. 1B) but was excluded from microarray annotation because of absence of uniquely mappable oligoprobes.
Furthermore, we used Gene Ontology annotation (GO; Ashburner et al., 2000) in order to characterize the functional content of the gene expression in hES-Hep. Interestingly, 4148 (47%) of the expressed genes were associated with “metabolic processes” underlining the high level of metabolic competence of the cells. Closer investigation of the genes associated with drug metabolism yielded gene expression of important phase I, II, and III genes (Supplementary table 2). Although there was fairly consistent expression of phase II and III genes across the panel of treatments, higher variability was observed in cytochrome P450 enzyme expression in particular with the CYP2 family, which is known as a heterogeneous group of enzymes with highly variable substrate specificities (Nelson et al., 1996).
Identification and Characterization of Response Genes
The study design of chemical treatments with the hES-Hep is described in Supplementary figure 1B. We used an ANOVA model (“Materials and Methods”) that identified 592 response genes with significant variation among the three toxicity classes (F test p value < 0.05; Supplementary table 3). These genes achieved a high level of discrimination according to the three toxicity classes (Fig. 2A). The major proportion of response genes was associated with metabolic processes (234 of 592). A GO characterization is visualized in Supplementary figure 2.
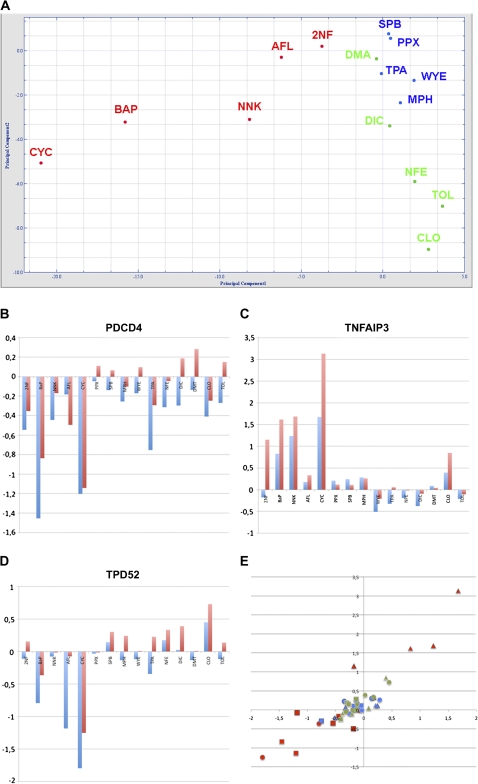
FIG. 2.
Response genes. (A) Principal component analysis (PCA) with the 592 response genes. Different compound classes are indicated by colors (red = genotoxic carcinogens (GTX), blue = nongenotoxic carcinogens (NGTX), and green = noncarcinogens (NC)). The two principal components explain 66.2% of the variance (PC1: 56.58%; PC2: 9.595%). PCA was generated with the J-Express 2009 software (MolMine AS, Bergen, Norway). qPCR validation of (B) PDCD4, (C) TNFAIP3, and (D) TPD52 compared with microarray measurements across the panel of substance treatments. Y-axis displays the log2 ratios in treatment versus control experiments of qPCR (red bars) and microarrays (blue bars). (E) Scatter plot of all 45 log2 ratios. Treatments are displayed with colors (red = GTX, blue = NGTX, and green = NC); individual genes are displayed with different shapes (square = PDCD4, triangle = TNFAIP3, and circle = TPD52).
In order to functionally interpret the gene response, we performed overrepresentation analysis using the ConsensusPathDB. Eleven pathways were found overrepresented (Fisher test p value < 0.05; Table 1) monitoring a significant genotoxic response in particular with ATM and p53 pathway modules. This result suggests that the selected gene set contains a large proportion of genes that act in the interplay of DNA damage response, p53 signaling, and apoptosis. Selective validation of microarray results was performed with qPCR as is shown for PDCD4 (Fig. 2B), TNFAIP3 (Fig. 2C), and TPD52 (Fig. 2D). These genes are involved in pathological apoptotic processes in human and were significantly altered in genotoxic treatments in qPCR as well as microarray experiments (Fig. 2E). For example, downregulation of the tumor suppressor PDCD4 on the protein level and its pro-apoptotic effects for TGF-β1-induced apoptosis has been shown previously in human hepatocellular carcinoma (Zhang et al., 2006).
TABLE 1.
Overrepresentation of Pathways with Respect to the 592 Response Genes
| Pathway | Genes in pathway | Overlap with response genes | p Value (Fisher) | Source database | Response genes in pathway |
| Wnt lrp6 signaling | 7 (7) | 3 | 0.00349 | BioCarta | DKK1, FZD1, WNT8A |
| ATM signaling pathway | 20 (18) | 4 | 0.00991 | BioCarta | ATM, GA45A, MDM2, P73 |
| Hop pathway in cardiac development | 4 (4) | 2 | 0.0134 | BioCarta | NKX25, SRF |
| Aurora A signaling | 33 (31) | 5 | 0.0161 | PID | GA45A, MDM2, MPIP2, OAZ1, RASA1 |
| Stabilization of p53 | 5 (5) | 2 | 0.0215 | Reactome | ATM, MDM2 |
| Glycogen breakdown (glycogenolysis) | 14 (14) | 3 | 0.0281 | Reactome | GDE, PHKG1, PHKG2 |
| Cysteine biosynthesis II | 7 (6) | 2 | 0.0313 | HumanCyc | SERA, SERB |
| CREB phosphorylation | 7 (7) | 2 | 0.0424 | Reactome | KS6A3, KS6A5 |
| p53 signaling pathway | 69 (68) | 7 | 0.047 | KEGG | ATM, GA45A, MDM2, P73, PPM1D, SESN1, TNR6 |
| Branched-chain amino acid catabolism | 18 (17) | 3 | 0.0471 | Reactome | AUHM, HIBCH, ODBB |
| p53 signaling pathway | 17 (17) | 3 | 0.0471 | BioCarta | BCL2, GA45A, MDM2 |
The ultimate purpose of the response gene set is to discriminate chemicals capable of triggering carcinogenesis in vivo; thus, it is particularly important to analyze the response of tumor suppressor genes and oncogenes because in vivo carcinogenesis is a complex process that is driven by tight interactions between oncogene activation, tumor suppressor inactivation, and the cell death machinery (Zhivotovsky and Orrenius, 2010). Among the 592 genes, we found eight tumor suppressors (PDCD4, BCL2, SMAD3, FHIT, ATM, TCHP, ITGB5, and RPL10) and five oncogenes (RAB17, RRAS, FAS, MDM2, and GNA15). Mechanisms relating to these genes have been shown previously in the literature, for example, the activation of MDM2 by the downregulation of the tumor suppressor FHIT (Schlott et al., 1999). Downregulation of FHIT and upregulation of MDM2 was observable in four of the five genotoxic treatments (AFL, BAP, NNK, and CYC), whereas such effects were much weaker or not visible with the nongenotoxic and noncarcinogenic treatments. These findings are consistent with recent results from rodent studies, for example, from primary rat hepatocytes (Mathijs et al., 2009), where Mdm2 has also been prominently identified as discriminating between genotoxic and nongenotoxic compounds.
Performance of hES-Hep as a Hazard Assessment Assay
In order to test the potential of hES-Hep for classifying carcinogenic substances, we applied supervised classification with a support vector machine approach and compared performance of the response gene set (N = 592) with the genome-wide approach (N = 18,394). Misclassification rates were computed with a cross-validation procedure (LOO). Additional data was introduced using the same 15 treatments at a lower time point (IC10 at 24 h, see “Materials and Methods”) and a subset of nine of the compounds also with lower concentrations (50% of the IC10 at 24 h and 72 h). The misclassification rate using all data (in total, 15 + 15 + 9 + 9 = 48 experiments) was 6.25% with whole-genome data and dropped to 4.17% with the response genes as readouts (Table 2).
TABLE 2.
Cross-Validation Performance
| n | Whole genome (N = 18,394) |
Response genes (N = 592) |
|||
| Experiments | Misclassification rate | ± | Misclassification rate | ± | |
| 24-h exposure | 24 | 20.83 | 8.47 | 20.83 | 8.47 |
| 72-h exposure | 24 | 20.83 | 8.47 | 4.17 | 4.17 |
| IC 10/2 concentration | 18 | 27.78 | 1.86 | 16.67 | 9.04 |
| IC 10 concentration | 30 | 26.67 | 8.21 | 1.33 | 6.31 |
| All experiments | 48 | 6.25 | 3.53 | 4.17 | 2.92 |
Comparison of Response Gene Expression in hES-Hep and Other Liver Cellular Models
We compared the expression patterns of the 592 response genes with Affymetrix microarray data on other liver-like cellular models. Pearson correlation of the gene expression in hES-Hep was 0.88 as compared with plated primary human hepatocytes, 0.83 as compared with HepG2 cells, 0.80 as compared with human fetal liver, 0.79 as compared with human adult liver, and 0.76 as compared with nonplated primary human hepatocytes (Table 3A). The result points to the fact that the expression of the response genes in hES-Hep highly resembled human hepatic cell systems and was also similar to HepG2 that is a widely used in vitro hazard assessment system. Moreover, correlation of hES-Hep to the other liver systems was even slightly higher than that of HepG2 in all cases. Furthermore, we computed correlation of gene expression of 78 selected Phase I, II, and III genes (cf. Supplementary table 2) and found a significant correlation of hES-Hep with primary plated hepatocytes (0.71) and HepG2 (0.79, plots not shown).
TABLE 3.
Correlation of Expression Patterns of Response Genes in Liver-Like Systems
| A | ||||||
| hES-Hep | Adult liver | Fetal liver | HepG2 | Nonplated PHH | Plated PHH | |
| hES-Hep | 1 | 0.7866 | 0.8049 | 0.8300 | 0.7648 | 0.8790 |
| Adult liver | 0.7866 | 1 | 0.8967 | 0.7493 | 0.9796 | 0.9084 |
| Fetal liver | 0.8049 | 0.8967 | 1 | 0.7910 | 0.8798 | 0.8728 |
| HepG2 | 0.8300 | 0.7493 | 0.7910 | 1 | 0.7524 | 0.8442 |
| Nonplated PHH | 0.7648 | 0.9796 | 0.8798 | 0.7524 | 1 | 0.9028 |
| Plated PHH | 0.8790 | 0.9084 | 0.8728 | 0.8442 | 0.9028 | 1 |
| B | ||||||
| GTX | NGTX | GSE29722 | ||||
| GTX | 1.0000 | 0.9818 | 0.8040 | |||
| NGTX | 0.9818 | 1.0000 | 0.8237 | |||
| GSE29722 | 0.8040 | 0.8237 | 1.0000 | |||
| 2NF | 0.9867 | 0.9914 | 0.8099 | |||
| AFL | 0.9917 | 0.9823 | 0.8013 | |||
| BAP | 0.9935 | 0.9656 | 0.7924 | |||
| CYC | 0.9771 | 0.9292 | 0.7659 | |||
| NNK | 0.9924 | 0.9832 | 0.8036 | |||
| MPH | 0.9719 | 0.9941 | 0.8167 | |||
| PPX | 0.9748 | 0.9948 | 0.8140 | |||
| SPB | 0.9749 | 0.9950 | 0.8186 | |||
| TPE | 0.9783 | 0.9889 | 0.8137 | |||
| WYE | 0.9750 | 0.9921 | 0.8270 | |||
Additionally, we have examined whether the expression signals of the response genes in hES-Hep upon carcinogenic treatments (GTX and NGTX compounds) resemble expression patterns in human liver cancer and incorporated publicly available data (GSE29722) on 10 human liver tumors (Stefanska et al., 2011). The correlation of expression patterns with human liver tumor data was 0.80 for the GTX treatments and 0.82 for the NGTX treatments, with fairly consistent values for the individual treatments (Table 3B).
Pathway Responses to Chemical Treatments in hES-Hep
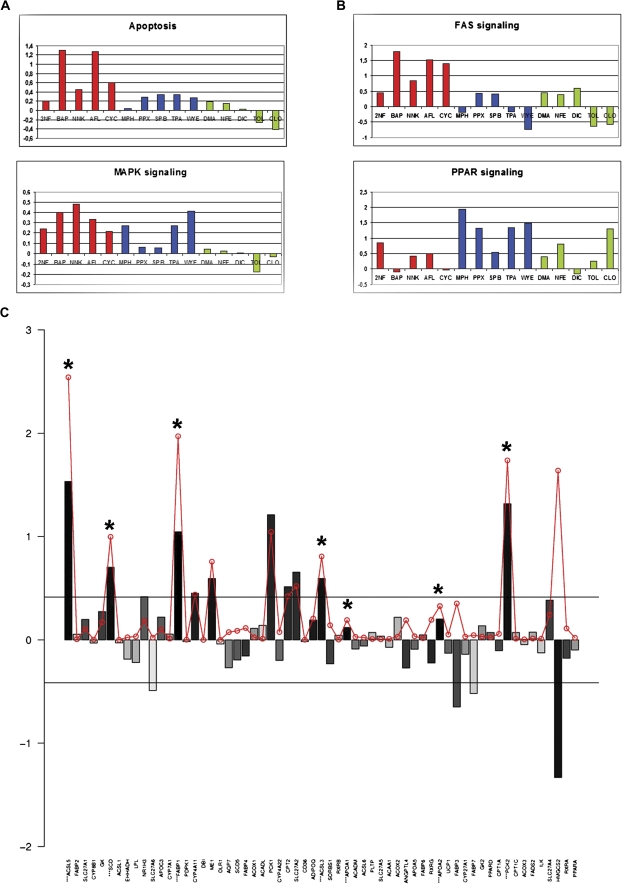
We further aimed to quantify the response of hES-Hep to chemical treatments at the pathway level (“Materials and Methods”) using preannotated pathways with at least five members that were measurable with microarray oligoprobes (1695 of 2144). Global pathway response was highly variable among the 15 treatments. Seven substances induced a strong overall response with respect to many of the 1695 pathways (CYC, CLO, MPH, TOL, BAP, NNK, and TPA), whereas the remaining substances induced a far lower response (data not shown). In order to make the pathway response scores comparable among the different treatments, we divided for each chemical treatment the scores by the median pathway score (see “Materials and Methods”). Interpretation at the pathway level can be driven toward several directions. Firstly, we observed pathway responses distinguishing carcinogenic (either GTX or NGTX) from noncarcinogenic treatments, for example, modules from the apoptosis (Student’s t-test p = 0.0076) and MAPK (p = 0.0003) signaling pathways (Fig. 3A). This finding correlates with published results for genotoxic compounds, for example, for BAP (Chen et al., 2003). Highest response to MAPK signaling among the NGTX treatments was observed for WYE, a PPAR-α ligand that is prototypical for peroxisome proliferators. It has been shown in primary rat hepatocyte cultures that carcinogenic effects on proliferation and apoptosis of peroxisome proliferators require p38 MAP kinase activity (Cosulich et al., 2000), although there is an ongoing debate whether the carcinogenic effects of peroxisome proliferators via PPAR-α extrapolate from the rodent to the human system (Peters, 2008).
FIG. 3.
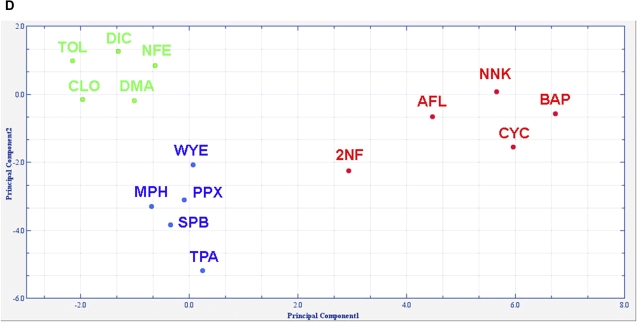
Pathway response analysis. (A) Response scores for apoptosis (top) and MAPK (bottom) pathways distinguishing carcinogenic from noncarcinogenic treatments (red = GTX, blue = NGTX, and green = NC). Y-axis shows relative response with respect to the median response in the respective treatment experiment (log2 scale). (B) Response scores for FAS (top) and PPAR (bottom) distinguishing toxicity classes. (C) Illustration of pathway scoring with PPAR signaling after WYE treatment. 61 genes (x-axis) were measured with microarrays. Bars (y-axis) show the log2 ratio of WYE treatment signals versus controls. Bars are shadowed according to significance of the fold changes when judging the three replicate measurements (dark = significant). Genes with significant fold change (p < 0.05) are displayed with an asterisk on top of the bar. The red circles display the gene score that was computed from the fold change and significance p values as described in “Materials and Methods.” The horizontal lines mark a 1.33 deregulation. (D) PCA derived from 37-pathway response patterns for the 15 substance treatments yields a perfect separation of the toxicity classes. Total variance explained is 69.5% (PC1: 49.9%; PC2: 19.6%).
Secondly, pathway analysis enables to distinguish low-responding and high-responding pathway modules for each toxicity class. High pathway responses among the GTX treatments were observed for p53 and apoptosis pathway modules, whereas NGTX treatments exerted their effects mainly through PI3K/AKT, PPAR, and MAPK signaling (Fig. 3B). NC treatment responses were less specific. In order to identify pathways that appear consistently affected by the different toxicity classes, we computed for each toxicity class and each pathway the coefficient of variance (CV) among the compound pathway response scores. Because we were only interested in responding pathways, we preselected pathways with a positive average relative score meaning that the pathway’s response score is higher compared with the median pathway score on average. With that procedure, we identified 72 pathways “consistently affected” (CV < 0.5) by GTX compounds, 45 pathways “consistently affected” by NGTX compounds, and 35 by NC compounds. Among these, “consistently affected” NGTX pathways were, for example, PPAR signaling (CV = 0.383), glycolysis (CV = 0.258), and mTOR signaling (CV = 0.309) pointing to important human in vivo processes of carcinogenesis. An illustrative example of the gene-wise contributions to the pathway scoring is given with the PPAR signaling pathway (Fig. 3C).
Thirdly, extrapolating gene expression information to the pathway level improved the discrimination of the toxicity classes. We have performed a similar ANOVA approach as for gene expression patterns with the pathway response patterns and were able to achieve a complete separation of the 15 compounds into 3 distinct groups with a subset of 37 pathways (Fig. 3D). Compared with the grouping achieved on the gene level (cf. Fig. 2A), the pathway level shows an increase in performance.
Genotoxic Responses in hES-Hep—Benzo[a]pyrene Case Study
Because we measured a strong overall signal from cancer-related pathways (see above), we were interested whether the hES-Hep system reproduces genotoxic responses known from (rodent) in vivo studies and investigated exemplary the effects of BAP treatment, which is one of the best studied genotoxic carcinogens. BAP is of particular interest because it requires metabolic transformation for exerting its genotoxic effects through BAP diolepoxide by several cytochrome P450 enzymes and, thus, challenges the metabolic competence of the cell system. Upon BAP treatment, we observed upregulation of cytochrome P450 enzymes CYP1A1 (p = 0.006, fold change = 3.57) and CYP1B1 (p = 0.0004, fold change = 6.30) judging the three BAP treatment and control replicates with Student’s t-test. CYP1A1 and CYP1B1 display similar catalytic activities in converting B[a]P-7,8-dihydrodiol to mutagenic metabolites (Chen et al., 2003). Upregulation of these CYPs is through binding of BAP to AHR, a ligand-activated nuclear transcription factor (Hockley et al., 2007; Nebert et al., 2004). AHR was also found upregulated in hES-Hep (p = 0.002, fold change = 1.93). In total, 673 genes were differentially regulated upon BAP treatment (p value < 0.01). These genes monitored carcinogenic response at the interplay of apoptosis (IRAK1/2, BIRC3, FAS, PIK3R2, TNFRSF10D, and IL1B), p53 signaling (FAS, NOXA, RRM2B, PPM1D, DDB2, SESN1/2, PIDD, YWAS, GADD45A, and RIR2B), and DNA damage pathways.
At the pathway level (cf. previous section), the highest response scores to BAP exposure were observed for modules of the apoptosis pathway in particular its induction by FAS ligand and caspase activation (CASP8). Fourteen pathways had a highly elevated pathway score, seven of which were associated with apoptosis (Supplementary table 4). This strong apoptotic response in the human cells reproduced previous findings, for example, in rat liver epithelial cell lines (Huc et al., 2006). Furthermore, we performed Student’s t-test with the pathway response scores and compared all carcinogenic treatments (GTX and NGTX) with the noncarcinogenic treatments (NC). It can be seen in Supplementary table 4 that the apoptotic response observed in BAP-treated cells was fairly stable among other carcinogenic treatments as well, for example, for “FasL/CD95L signaling” (p = 0.085), “sodd/tnfr1 signaling pathway” (p = 0.045), and “death receptor signaling” (p = 0.054).
DISCUSSION
In this study, we have used an in vitro model based on hESC differentiated to hepatocyte-like cells (hES-Hep). It has been previously published by us and others that such cells show similarities as well as differences, depending on the differentiation protocol, compared with HepG2 and different fetal and adult liver samples. hES-Hep were studied at day 22 after onset of differentiation when the cells show an attractive hepatic morphology and express many liver markers at gene, protein, and functional level (Fig. 1). hES-Hep, in general, express higher levels of CYP expression as compared with HepG2 but lower levels in comparison with hpHep, however, still confirming the metabolic competence of the system. An interesting outcome of this study is that the correlation of expression of the response genes in hES-Hep is mostly correlated to that in plated hpHep among the five hepatic cell models used (Table 3). It is worth mentioning, though, that these data monitor the expression of hES-Hep in the untreated state and, thus, give no evidence per se that either of the models is a valid assay system for carcinogenicity hazard prediction because each system can respond differently to chemical treatment.
We challenged the hES-Hep model with a panel of 15 substances from three different toxicity classes. Although these substances exerted different modes of action and different strengths of effects, we were able to identify discriminative classifier sets on the gene (592 genes) and the pathway (37 pathways) levels. In particular, genotoxic substances induced the largest effects in the cells. We exemplified this with benzo[a]pyrene, but similar results were obtained with other GTX compounds. Strong pathway responses were observable in DNA damage response, p53 signaling, and, in particular, apoptosis. It is evident that the prediction of nongenotoxic carcinogenic effects is far more difficult compared with genotoxic and, thus, that the hES-Hep model, as presumably most in vitro models, has a prevalence for the identification of directly acting genotoxins. On the other hand, we observe clear effects related to the hallmarks of cancer (Hanahan and Weinberg, 2011) induced by nongenotoxic substances what emphasizes the usefulness of the cellular model for prediction of carcinogenicity. Additionally, hierarchical clustering of the expression matrix derived from the 592 genes and the 15 treatment experiments (Supplementary figure 3A) revealed five main clusters of genes (Supplementary figure 3B). Among these, cluster 2 (C2, N = 176) exhibited genes basically unaffected by GTX treatments and predominantly upregulated with NC and NGTX substances.
It has been emphasized that toxicity pathways leading to carcinogenesis in humans are not yet fully characterized (Cohen, 2010) and inherently different from those in rodents (Ward, 2008). Carcinogenic effects induced in rodents by specific compounds might thus not easily been extrapolated to the human situation leading to a large amount of false positive predictions. This is particularly true for the class of nongenotoxic carcinogens that show a diversity of modes of action, tissue and species specificity, and absence of genotoxicity what makes predicting their carcinogenic potential extremely challenging (Hernandez et al., 2009). For example, WYE and SPB are both compounds with clear carcinogenic effects in rodents but with no consensus regarding the human situation. In our pathway analysis, these compounds were jointly highly responding to the BioCarta pathway “srebp control of lipid biosynthesis”, among others, which potentially links their expression responses to human cancer. It has been shown that several steps in lipid synthesis promote tumor development, for example, through involvement of mTOR signaling (Hsu and Sabatini, 2008; Laplante and Sabatini, 2009).
Statistical classification is the standard way of judging the quality of a hazard assessment assay, however, the panel of substances must be enlarged for such an approach. Although there is a considerable risk of overfitting the data because of the small number of compounds, our results serve as a proof of principle demonstrating that toxicity classes can be discriminated with the hES-Hep system with high confidence (Table 2). An alternative to the statistical approach would consist in the development of discriminative mechanisms based on preselected genes. We have shown evidence that the identified response gene set exhibits multiple carcinogenic effects and even increases cross-validation performance compared with the genome-wide approach.
Furthermore, following-up the discriminating pathways will stimulate future mechanistic approaches. Pathway scoring (Fig. 3C) increases the performance of compound discrimination as can be seen by comparing Figures 2A and 3D. This increase of performance is effected by the pregrouping of expression patterns that copes better with the inherent variation of gene expression measurements and confirms previous similar findings, for example, in the context of disease classification (Lee et al., 2008). We have shown that different pathway modules could be associated with individual chemicals and toxicity classes. These pathway modules were highly discriminative, and they were linked to carcinogenesis comprising, for example, apoptosis (p = 0.0065), MAPK (p = 0.0013), and PPAR (p = 0.0157) signaling. The role of PPAR signaling for rodent carcinogenesis is evident due to the fact that PPAR-α activation leads to increased proliferation, decreased apoptosis, and activation of reactive oxygen species leading to hepatocellular carcinoma as a long-term response, whereas in human, this effect has not been proven so far (Michalik et al., 2004). On the other hand, non–DNA reactive mechanisms, such as production of active oxygen species and lipid peroxidation that are influenced by PPAR signaling, can induce genotoxicity in humans by secondary effects (Ellinger-Ziegelbauer et al., 2009).
We also checked the consistency of expression of the response genes within each toxicity class with a one-sample Student’s t-test. 421 (of 592) genes were identified as consistently differentially expressed (Supplementary figure 4). These genes indicate potential mechanisms of carcinogenesis, for example, FBXW7, a known tumor suppressor whose activation could be interpreted as a cell protection mechanism, TAP1 that was related to E2F and the apoptosis pathway in HepG2 liver cells (Li et al., 2010), TRIAP1 that was related to p53 signaling (Felix et al., 2009; Park and Nakamura, 2005), or CXCL10 that was related to prolonged tumor growth delay in CT26 and 4T1 tumor models (Wang et al., 2010). Furthermore, 301 (of 592) genes were selectively altered within a specific toxicity class; of these, 153 (51%) were specific for GTX, 60 (20%) for NGTX, and 88 (29%) for NC substances. These genes build a rich basis for extrapolation of further mechanistic information and ultimately, a mechanistic response model, for chemical carcinogenesis in the hES-Hep model.
To summarize, we have demonstrated that hESC technology has high potential for developing in vitro hazard assessment assays for carcinogenicity of chemicals as was shown with three representative toxicity classes. We have quantified and identified discriminative carcinogenic pathway responses based on modules of the apoptosis, MAPK, and p53 signaling pathways that build the basis for a mechanistic understanding of chemical carcinogenesis. Although the hES-Hep model needs further refinement and, additional challenges with more compounds, in order to meet a broader acceptance, this study paves the way toward use of stem cell–derived liver cells for toxicity testing. In the future, this may lead to less use of animal experiments and increased possibilities to avoid bringing harmful chemicals to the market.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
European Commission under its 6th Framework Programme with the grant carcinoGENOMICS (LSHB-CT-2006-037712); the Beatriu de Pinos postdoctoral fellowship program (2008 BP-A 00184); the German Ministry for Education and Research under its MedSys program (PREDICT 0315428A); and the Max Planck Society; Ministerio Ciencia e Inovación/Instituto de Salud Carlos III for a “Miguel Servet” contract (CP08/00125 to A.L.).
Supplementary Material
Acknowledgments
We thank Axel Rasche for computational support and Mathieu Vinken for advise on chemical substances. We disclose that Gabriella Brolén, Gustav Eriksson and Petter Björquist are employees of Cellartis AB and Hans Gmuender is an employee of Genedata AG.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Cary MP, Sander C. Pathguide: A pathway resource list. Nucleic Acids Res. 2006;34(Database issue):D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brolén G, Sivertsson L, Björquist P, Eriksson G, Ek M, Semb H, Johansson I, Andersson TB, Ingelman-Sundberg M, Heins N. Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. J. Biotechnol. 2010;145:284–294. doi: 10.1016/j.jbiotec.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Chen S, Nguyen N, Tamura K, Karin M, Tukey RH. The role of AH receptor and p38 in benzo[a]pyrene-7,8-dihydrodiol and benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-induced apoptosis. J. Biol. Chem. 2003;278:19526–19533. doi: 10.1074/jbc.M300780200. [DOI] [PubMed] [Google Scholar]
- Cohen SM. Evaluation of possible carcinogenic risk to humans based on liver tumors in rodent assays: The two-year bioassay is no longer necessary. Toxicol. Pathol. 2010;38:487–501. doi: 10.1177/0192623310363813. [DOI] [PubMed] [Google Scholar]
- Cosulich S, James N, Roberts R. Role of MAP kinase signalling pathways in the mode of action of peroxisome proliferators. Carcinogenesis. 2000;21:579–584. doi: 10.1093/carcin/21.4.579. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato MT, Montero S, Castell JV, Gómez-Lechón MJ, Lahoz A. Validated assay for studying activity profiles of human liver UGTs alter drug exposure: Inhibition and induction studies. Anal. Bioanal. Chem. 2010;396:2251–2263. doi: 10.1007/s00216-009-3441-1. [DOI] [PubMed] [Google Scholar]
- Ek M, Söderdahl T, Küppers-Munther B, Edsbagge J, Andersson TB, Björquist P, Cotgreave I, Jernström B, Ingelman-Sundberg M, Johansson I. Expression of drug metabolizing enzymes in hepatocyte-like cells derived from human embryonic stem cells. Biochem. Pharmacol. 2007;74:496–508. doi: 10.1016/j.bcp.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Fostel JM, Aruga C, Bauer D, Boitier E, Deng S, Dickinson D, Le Fevre AC, Fornace AJ, Jr, Grenet O, et al. Characterization and interlaboratory comparison of a gene expression signature for differentiating genotoxic mechanisms. Toxicol. Sci. 2009;110:341–352. doi: 10.1093/toxsci/kfp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix RS, Colleoni GW, Caballero OL, Yamamoto M, Almeida MS, Andrade VC, Chauffaille Mde L, Silva WA, Jr, Begnami MD, Soares FA, et al. SAGE analysis highlights the importance of p53csv, ddx5, mapkapk2 and ranbp2 to multiple myeloma tumorigenesis. Cancer Lett. 2009;278:41–48. doi: 10.1016/j.canlet.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hartung T, Rovida C. Chemical regulators have overreached. Nature. 2009;460:1080–1081. doi: 10.1038/4601080a. [DOI] [PubMed] [Google Scholar]
- Heins N, Englund MC, Sjöblom C, Dahl U, Tonning A, Bergh C, Lindahl A, Hanson C, Semb H. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells. 2004;22:367–376. doi: 10.1634/stemcells.22-3-367. [DOI] [PubMed] [Google Scholar]
- Heins N, Lindahl A, Karlsson U, Rehnström M, Caisander G, Emanuelsson K, Hanson C, Semb H, Björquist P, Sartipy P, et al. Clonal derivation and characterization of human embryonic stem cell lines. J. Biotechnol. 2006;122:511–520. doi: 10.1016/j.jbiotec.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Hernandez LG, van Steeg H, Luijten M, van Benthem J. Mechanisms of non-genotoxic carcinogens and importance of a weight of evidence approach. Mutat. Res. 2009;682:94–109. doi: 10.1016/j.mrrev.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Hockley SL, Arlt VM, Brewer D, Te Poele R, Workman P, Giddings I, Phillips DH. AHR- and DNA-damage-mediated gene expression responses induced by benzo(a)pyrene in human cell lines. Chem. Res. Toxicol. 2007;20:1797–1810. doi: 10.1021/tx700252n. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Huang J, Hao P, Zhang YL, Deng FX, Deng Q, Hong Y, Wang XW, Wang Y, Li TT, Zhang XG, et al. Discovering multiple transcripts of human hepatocytes using massively parallel signature sequencing (MPSS) BMC Genomics. 2007;8:207. doi: 10.1186/1471-2164-8-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huc L, Rissel M, Solhaug A, Tekpli X, Gorria M, Torriglia A, Holme JA, Dimanche-Boitrel MT, Lagadic-Gossmann D. Multiple apoptotic pathways induced by p53-dependent acidification in benzo[a]pyrene-exposed hepatic F258 cells. J. Cell. Physiol. 2006;208:527–537. doi: 10.1002/jcp.20686. [DOI] [PubMed] [Google Scholar]
- Jennen DG, Magkoufopoulou C, Ketelslegers HB, van Herwijnen MH, Kleinjans JC, van Delft JH. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- Jensen J, Hyllner J, Björquist P. Human embryonic stem cell technologies and drug discovery. J. Cell. Physiol. 2009;219:513–519. doi: 10.1002/jcp.21732. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39(Database issue):D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- Lahoz A, Donato MT, Castell JV, Gomez-Lechon MJ. Strategies to in vitro assessment of major human CYP enzyme activities by using liquid chromatography tandem mass spectrometry. Curr. Drug Metab. 2008;9:12–19. doi: 10.2174/138920008783331112. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput. Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ni GX, Zhang P, Zhang ZX, Li W, Wu Q. Characterization of E2F3a function in HepG2 liver cancer cells. J. Cell. Biochem. 2010;111:1244–1251. doi: 10.1002/jcb.22851. [DOI] [PubMed] [Google Scholar]
- Mathijs K, Brauers KJ, Jennen DG, Boorsma A, van Herwijnen MH, Gottschalk RW, Kleinjans JC, van Delft JH. Discrimination for genotoxic and nongenotoxic carcinogens by gene expression profiling in primary mouse hepatocytes improves with exposure time. Toxicol. Sci. 2009;112:374–384. doi: 10.1093/toxsci/kfp229. [DOI] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: Complex stories. Nat. Rev. Cancer. 2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, et al. P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Park W, Nakamura Y. p53CSV, a novel p53-inducible gene involved in the p53-dependent cell-survival pathway. Cancer Res. 2005;65:1197–1206. doi: 10.1158/0008-5472.CAN-04-3339. [DOI] [PubMed] [Google Scholar]
- Peters JM. Mechanistic evaluation of PPARα-mediated hepatocarcinogenesis: Are we there yet? Toxicol. Sci. 2008;101:1–3. [Google Scholar]
- Schlott T, Ahrens K, Ruschenburg I, Reimer S, Hartmann H, Droese M. Different gene expression of MDM2, GAGE-1, -2 and FHIT in hepatocellular carcinoma and focal nodular hyperplasia. Br. J. Cancer. 1999;80:73–78. doi: 10.1038/sj.bjc.6690324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han ZG, Szyf M. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891–5913. doi: 10.1158/0008-5472.CAN-10-3823. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vinken M, Doktorova T, Ellinger-Ziegelbauer H, Ahr HJ, Lock E, Carmichael P, Roggen E, van Delft J, Kleinjans J, Castell J, et al. The carcinoGENOMICS project: Critical selection of model compounds for the development of omics-based in vitro carcinogenicity screening assays. Mutat. Res. 2008;659:202–210. doi: 10.1016/j.mrrev.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Wang P, Yang X, Xu W, Li K, Chu Y, Xiong S. Integrating individual functional moieties of CXCL10 and CXCL11 into a novel chimeric chemokine leads to synergistic antitumor effects: A strategy for chemokine-based multi-target-directed cancer therapy. Cancer Immunol. Immunother. 2010;59:1715–1726. doi: 10.1007/s00262-010-0901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM. Value of rodent carcinogenesis bioassays. Toxicol. Appl. Pharmacol. 2008;226:212. doi: 10.1016/j.taap.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Waters MD, Fostel JM. Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 2004;5:936–948. doi: 10.1038/nrg1493. [DOI] [PubMed] [Google Scholar]
- Wierling C, Herwig R, Lehrach H. Resources, standards and tools for systems biology. Brief Funct. Genomic Proteomic. 2007;6:240–251. doi: 10.1093/bfgp/elm027. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-β1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Cell-cycle and cell death in disease: Past, present and future. J. Intern. Med. 2010;268:395–409. doi: 10.1111/j.1365-2796.2010.02282.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.