Abstract
2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD) induces hepatic dyslipidemia mediated by the aryl hydrocarbon receptor (AhR). Stearoyl-CoA desaturase 1 (Scd1) performs the rate-limiting step in monounsaturated fatty acid (MUFA) synthesis, desaturating 16:0 and 18:0 into 16:1n7 and 18:1n9, respectively. To further examine the role of Scd1 in TCDD-induced hepatotoxicity, comparative studies were performed in Scd1+/+ and Scd1−/− mice treated with 30 μg/kg TCDD. TCDD induced Scd1 activity, protein, and messenger RNA (mRNA) levels approximately twofold. In Scd1+/+ mice, hepatic effects were marked by increased vacuolization and inflammation and a 3.5-fold increase in serum alanine aminotransferase (ALT) levels. Hepatic triglycerides (TRGs) were induced 3.9-fold and lipid profiling by gas chromatography-mass spectroscopy measured a 1.9-fold increase in fatty acid (FA) levels, consistent with the induction of lipid transport genes. Induction of Scd1 altered FA composition by decreasing saturated fatty acid (SFA) molar ratios 8% and increasing MUFA molar ratios 9%. Furthermore, ChIP-chip analysis revealed AhR enrichment (up to 5.7-fold), and computational analysis identified 16 putative functional dioxin response elements (DREs) within Scd1 genomic loci. Band shift assays confirmed AhR binding with select DREs. In Scd1−/− mice, TCDD induced minimal hepatic vacuolization and inflammation, while serum ALT levels remained unchanged. Although Scd1 deficiency attenuated TCDD-induced TRG accumulation, overall FA levels remained unchanged compared with Scd1+/+ mice. In Scd1−/− mice, TCDD induced SFA ratios 8%, reduced MUFA ratios 13%, and induced polyunsaturated fatty acid ratios 5% relative to treated Scd1+/+ mice. Collectively, these results suggest that AhR regulation of Scd1 not only alters lipid composition but also contributes to the hepatotoxicity of TCDD.
Keywords: 2,3,7,8-tetrachlorodibenzo-ρ-dioxin; stearoyl-CoA desaturase 1; saturated fatty acid; monounsaturated fatty acid; polyunsaturated fatty acid; steatosis; liver
2,3,7,8-Tetrachlorodibenzo-ρ-dioxin (TCDD) and related compounds elicit a broad spectrum of biological responses ranging from effects on development to pathologies affecting specific organ functions as well as the immune and nervous system. These effects are mediated by the aryl hydrocarbon receptor (AhR), a cytosolic ligand-activated basic helix-loop-helix Per/aryl hydrocarbon receptor nuclear translocator (ARNT)/Sim family transcription factor (Croutch et al., 2005; Poland and Knutson, 1982). The proposed mechanism involves ligand binding to the cytosolic AhR, leading to dissociation of chaperone proteins and translocation of the AhR to the nucleus (Pollenz et al., 1994). The activated AhR then heterodimerizes with the ARNT (Hankinson, 1995) to bind dioxin response elements (DREs) located within regulatory regions of target genes (Swanson et al., 1995) recruiting chromatin remodeling complexes and transcriptional coregulators that modulate gene transcription (Beischlag et al., 2002).
Although the range of endogenous functions regulated by the AhR remains uncertain, studies in null mice show it is necessary for proper liver development and mediates the toxicity of TCDD (Bunger et al., 2003; Fernandez-Salguero et al., 1996; Schmidt et al., 1996). TCDD elicits hepatomegaly (Poland and Knutson, 1982) and liver pathologies that include hepatocellular neoplasms (Huff et al., 1991), inflammation, necrosis, steatosis, and the differential expression of lipid metabolism and transport genes in mice (Boverhof et al., 2006; Kopec et al., 2008, 2010a, 2010b). DNA binding by AhR is compulsory for TCDD-induced hepatic steatosis (Bunger et al., 2008).
Stearoyl-CoA desaturase 1 (Scd1) catalyzes the rate-limiting step in monounsaturated fatty acid (MUFA) biosynthesis and is a target for the treatment of metabolic related disorders (Issandou et al., 2009; Jiang et al., 2005). Scd1 desaturates palmitate (16:0) and stearate (18:0) into palmitoleate (16:1n7) and oleate (18:1n9), respectively, which can be metabolized further to other MUFAs and polyunsaturated fatty acids (PUFAs), the primary constituents of membrane lipids, triglycerides (TRGs), phospholipids, and cholesterol esters (Miyazaki et al., 2000; Ntambi and Miyazaki, 2004). Dietary, hormonal, and environmental factors regulate Scd1 messenger RNA (mRNA) expression and protein stability, emphasizing the importance of Scd1 in lipid metabolism (Flowers and Ntambi, 2008; Ntambi and Miyazaki, 2004). Furthermore, Scd1−/− mice are resistant to diet-induced steatosis and exhibit impaired triglyceride synthesis (Miyazaki et al., 2000, 2007; Li et al., 2009). In humans, the Scd1 activity index (serum ratios of 16:1/16:0 or 18:1/18:0) correlates with hypertriglyceridemia and insulin resistance (Stefan et al., 2008; Warensjo et al., 2009) and is predictive of metabolic syndrome phenotypes (Warensjo et al., 2006). Furthermore, single nucleotide polymorphisms have been identified in human Scd1 that are associated with obesity and insulin sensitivity (Warensjo et al., 2007).
In this study, we examined AhR regulation of Scd1- and TCDD-elicited steatosis in Scd1+/+ and Scd1−/− mice. Hepatic fatty acid (FA) profiling with complementary histopathology, enzyme activity assays, and gene expression were assessed in immature Scd1+/+ and Scd1−/− female mice and integrated with AhR ChIP-chip, band shift, and computational data. Our results suggest that the regulation of hepatic lipid transport and metabolism genes by the AhR, including Scd1, is involved in TCDD-induced steatosis in the mouse.
MATERIALS AND METHODS
Animal breeding and genotyping.
B6.129-Scd1tm1Myz/J heterozygous mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and bred at the Michigan State University Laboratory Animal Care facility. Mice were maintained on a 12-h light/dark cycle and housed in autoclaved polycarbonate cages with microisolator lids containing aspen woodchips and nesting material. Animals were allowed free access to Harlan Teklad irradiated F6 rodent diet 7964 (Madison, WI) and autoclaved deionized water throughout the study. On postnatal day (PND), 21 mice were genotyped by ear punch and weaned. All procedures were carried out with the approval of the Michigan State University Institutional Animal Care and Use Committee.
In vivo treatment.
On PND, 28 mice were gavaged with 0.1 ml of sesame oil for a nominal dose of 0 (vehicle control) or 30 μg TCDD/kg body weight. The immature mouse was used to facilitate comparisons with other data sets as well as avoid potential interactions with estrogens produced by developed ovaries. Doses were chosen to elicit moderate hepatic effects while avoiding overt toxicity in long-term studies. Litters were combined with no more than five animals per cage. Animals were sacrificed at 24, 72, and 168 h postdose. Mice were weighed and blood was collected via submandibular vein puncture before sacrifice. Tissue samples were removed, weighed, flash frozen in liquid nitrogen, and stored at −80°C. The right lobe of the liver was fixed in 10% neutral buffer formalin for histological analysis.
Histopathology and clinical chemistry.
Fixed liver tissues were processed as previously described (Kopec et al., 2010b) at the Michigan State University Investigative Histopathology Laboratory, Division of Human Pathology, using a modified version of previously published procedures (Sheehan and Hrapchak, 1980). Serum alanine aminotransferase (ALT) levels were measured by the Michigan State University Diagnostic Center for Population and Animal Health Clinical Laboratory.
Hepatic triglyceride levels.
Frozen liver samples (∼100 mg) were homogenized (Polytron PT2100; Kinematica) in 1 ml of 1.15% KCl. TRGs were extracted from 200 μl of hepatic homogenate with 800 μl of isopropyl alcohol by vortex mixing for 10 min. The samples were centrifuged for 5 min at 800xg at room temperature and the supernatant was collected into separate vials. The concentration of hepatic TRGs was determined by spectrophotometry from 20 μl supernatant with a commercial L-Type Triglyceride M kit (Wako Diagnostics, Richmond, VA) with Multi-Calibrator Lipids as a standard (Wako Diagnostics) according to manufacturer’s protocol. Final results were normalized to the initial weight of the liver sample.
Gas chromatography–mass spectroscopy FA methyl ester hepatic lipid profiling.
Liver samples (∼100 mg, n = 5/group) were homogenized in 40% methanol and acidified with concentrated HCl (∼34 μl). Lipids were extracted with chloroform:methanol (2:1) containing 1mM 2,6-di-tert-butyl-4-methylphenol. The organic phase was removed and protein and aqueous phases were reextracted with chloroform. The organic phases were pooled and solvents evaporated under nitrogen. The samples were resuspended in 3N nonaqueous methanolic HCl and held at 60°C overnight. The next day, samples were cooled to room temperature and 0.9% (wt/vol) NaCl and hexane was added. The organic phase was separated by centrifugation, collected, dried under nitrogen, and resuspended in hexane. Samples were separated and analyzed with an Agilent 6890N gas chromatography (GC) with a DB23 column (30-m length, 0.25 mm id, 0.25-μm film thickness) interfaced to an Agilent 5973 mass spectroscopy (MS). 19:1n9 free fatty acid (FFA) and 19:0 TRG were added as extraction efficiency controls, and 17:1n1 FA methyl ester was spiked in as a loading control (Nu-chek, Elysian, MN). GC/MS data files were converted to Waters MassLynx file format, were analyzed with MassLynx software, and are reported as μmol/g liver tissue or mol%. FA levels are based upon peak areas from total ion chromatograms, and μmol/g is obtained from a linear calculation of a calibration curve normalized to sample weight. Principal component analysis (PCA) of FA abundance was performed in R V2.6.0.
Scd1 activity assay.
Scd1 activity assays were performed as previously described (Attie et al., 2007). Microsomal fractions were isolated by differential centrifugation, and protein was quantified by using the Bradford assay (Bio-Rad). Assays were performed at 37°C for 30 min with 100 μg microsomal protein, 0.03 μCi 14C-stearoyl-CoA (ARC 0756, 50–60 mCi/mmol, American Radiolabeled Chemicals, St Louis, MO) 2mM NADH, and 0.03mM cold stearoyl-CoA in 0.1M phosphate buffer at pH 6.8. The reaction was quenched with 2.5M KOH and saponified at 80°C for 45 min. FAs were acidified with formic acid, extracted with hexane, and dried under nitrogen. FAs were resuspended in 50 μl hexane and separated by 100 g/l AgNO3-impregnated TLC using CHCl3:MeOH:acetic acid:water (90:8:1:0.8 vol/vol) as a developing solvent. TLC plates were dried and exposed to autoradiography film, bands scraped into scintillation fluid, and measured by liquid scintillation counting. Scd1 activity is expressed as nmol/(mg protein × min).
Western blotting.
Microsomal fractions isolated for activity assays (10 μg each) were separated by 12% SDS-poly acrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane (Millipore Billerica, MA). Scd1 protein was immunoblotted with an Scd1 antibody (SC-14719; Santa Cruz Biotechnology, Santa Cruz, CA). Conventional western blot loading controls, such as Gapdh and Actb are cytosolic proteins, and therefore removed during microsome purification. Epoxide hydrolase (Epxh1, ab76226; Abcam, MA) is a microsomal protein and was used as a reference for loading control. Immunoreactive bands were visualized by chemiluminescence with the Pierce ECL Western blotting substrate (Thermo Scientific Rockford, IL) (Miyazaki et al., 2007). Immunoreactive bands were quantified by densitometry (ImageJ) and normalized to the loading control.
AhR ChIP-chip and DRE motif comparisons.
The genomic locations of AhR enrichment were previously determined by ChIP-chip from hepatic tissue of immature female ovariectomized C57BL/6 mice orally gavaged with 30 μg/kg TCDD (Dere et al., 2011b). The genomic locations of the 5′-GCGTG-3′ DRE core sequences were previously determined in mouse (Dere et al., 2011a). The DRE core along with the flanking upstream and downstream 7 bp were compared with a position weight matrix and matrix similarity scores (MSS) were calculated (Dere et al., 2011a). Regions of AhR enrichment were compared against DRE core sequences across Scd1, Scd2, Scd3, and Scd4 loci. Associated mouse genomic annotation (mm9) was downloaded from the University of California–Santa Cruz Genome Browser (Rhead et al., 2010).
Band shift assays.
Putative DREs with an MSS > 0.8 located from −10 kb upstream from the Scd1 transcriptional start site (TSS) were targeted for band shift assays that were performed as previously described (Maines and Wiley online library, 2002). Equimolar complimentary DNA (cDNA) oligonucleotides (Supplementary table 1) were combined and annealed by heating at 95°C for 5 min and cooling to room temperature. The double-stranded DNA oligonucleotide was labeled with T4 polynucleotide kinase (M0201S; New England Biolabs) and [y-32P]ATP (0135001; MP Biomedicals, Solon, OH). Each reaction contained 3 μg hepatic guinea pig cytosol extract (gift from Dr Michael Denison, Department of Environmental Toxicology, University of California, Davis) × mixed with either 20nM TCDD or an equivalent volume of dimethyl sulfoxide and was incubated for 1 h at room temperature. Guinea pig AhR is readily transformed in vitro and binds DREs with high affinity (Bank et al., 1992). The activated cytosols were combined with HEDG buffer (25mM Hepes, pH 7.7/1mM EDTA/1mM dithiothreitol/10% glycerol) and 1.7 μg polyd(I-C) (Invitrogen) and incubated at room temperature for an additional 15 min. 32P-labeled double-stranded DNA oligonucleotide (10,000 cpm, 0.1–0.3 ng in HEDG buffer) was added to the preincubation mixture and incubated for an additional 15 min. For supershift assays, either 1 μg AhR (ab2769; Abcam) or ARNT (SC-5580; Santa Cruz Biotechnology) antibody was added to the preincubation mixture. Samples were resolved by nondenaturing PAGE and imaged by autoradiography. In competition experiments, unlabeled competitor DNA was added to the preincubation mixture at 100-fold molar excess.
RNA isolation.
RNA was isolated from frozen liver samples with 1.3 ml TRIzol (Invitrogen) according to the manufacturer’s protocol and an additional acid phenol:chloroform extraction as previously described (Boverhof et al., 2006). Total RNA was resuspended in RNA storage solution, quantified by spectrophotometery at A260, and quality assessed by gel electrophoresis.
Quantitative real-time PCR.
Quantitative real-time PCR (QRTPCR) of Scd1, Scd2, Scd3, Scd4, Cyp1a1, Nqo1, Tiparp, Elovl5, Cd36, LdlR, Fabp4, Vldlr, Acaca, and Fasn expression was performed as previously described (Boverhof et al., 2006). The copy number of each sample was standardized to the geometric mean of Gapdh, Hprt, and Actb mRNA levels to control for differences in RNA loading, quality, and cDNA synthesis (Vandesompele et al., 2002). Data are reported as the fold change of standardized treated over standardized vehicle.
Statistical analysis.
Data were analyzed by ANOVA followed by Tukey’s post hoc test in SAS, unless otherwise stated. Differences between treatment groups were considered significant when p < 0.05.
RESULTS
TCDD Effects on Body Weights, Liver Histopathology, and Clinical Chemistry
Consistent with previously reported changes in body weights and liver histopathology changes in mice exposed to TCDD (Boverhof et al., 2006), Scd1+/+ and Scd1−/− intact female mice gavaged with 30 μg/kg TCDD had increased relative liver weight with the greatest increases observed at 168 h (Supplementary table 2). No significant alterations in body weight or body weight gain were detected throughout the study. Histopathological changes were marked by cytoplasmic vacuolization in the periportal and midzonal regions that decreased with time (Supplementary table 3, Supplementary figure 1A–H). In Scd1 wild-type mice, hepatic vacuolization was accompanied by cellular inflammation that increased by 168 h, whereas minimal cellular inflammation was observed at only 168 h in Scd1 null mice. Further analysis of ALT levels, a marker of liver damage identified a 3.5-fold increase in treated wild-type mice at 168 h (Supplementary figure 2I), whereas ALT levels remained unchanged in Scd1 nulls. Increased ALT levels suggest liver damage in treated wild-types only; however, longer term exposure may be necessary to differentiate histopathological differences between the two strains.
TCDD Effects on Hepatic Lipid Content in Scd1+/+ Mice
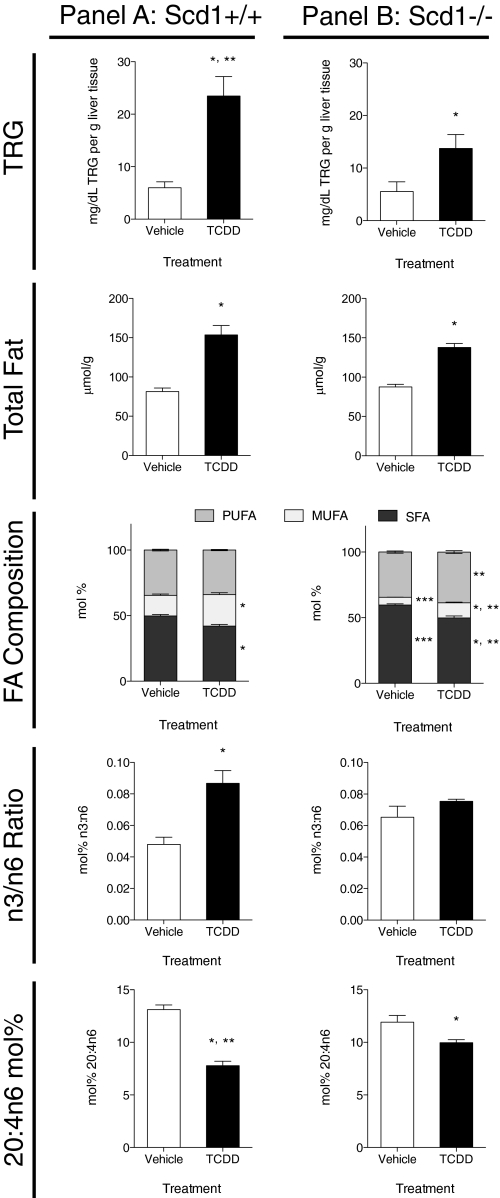
In Scd1+/+ mice, TCDD induced hepatic TRGs and hepatic TFA content 3.9- and 1.9-fold, respectively, compared with vehicle controls (Fig. 1, panel A). Analysis of FA composition by GC-MS identified TCDD-induced alterations in the SFA/MUFA/PUFA molar proportions (Fig. 1, panel A). Overall, TCDD reduced SFA by 8%, increased MUFA by 9%, and had no effect on PUFA proportions (p < 0.05, Fig. 1, see *). Further analysis of individual FA species revealed that palmitate (16:0), stearate (18:0), and lignoceric acids (24:0) represented more than 95% of the total SFA content (Table 1). Interestingly, palmitate (67%) and stearate (16%) are the major SFA constituents of the rodent chow diet (Harlan Teklad rodent diet 7964).
FIG. 1.
Hepatic lipid composition at 168 h in vehicle and 30 μg/kg TCDD-treated Scd1+/+ (panel A) and Scd1−/− (panel B) mice. Total triglycerides (TRGs) were extracted from mouse liver and quantified using a commercial L-Type Triglyceride M kit (Wako Diagnostics). Data are reported as mg/dL TRG per gram of liver tissue. Absolute total hepatic fat was extracted from Scd1+/+ and Scd1−/− mouse liver and analyzed by GC-MS and are reported as μmol/g tissue. Hepatic fat compositions (SFA/MUFA/PUFA ratios) are reported as mol%. SFA (dark gray)/MUFA (light gray)/PUFA (medium gray) proportions are represented. n3/n6 FA ratios are reported as mol %. Arachidonic acid (20:4n6) levels are reported as mol%. Data were analyzed by factorial ANOVA followed by Tukey’s post hoc test. Bars represent mean ± SEM, n = 5 biological replicates. *, p < 0.05 for TCDD compared with vehicle within a genotype; **, p < 0.05 for Scd1−/− TCDD compared with Scd1+/+ TCDD; ***, p < 0.05 for Scd1−/− vehicle compared with Scd1+/+ vehicle.
TABLE 1.
Total Hepatic Lipid Composition (μmol/g liver) in Scd1+/+ and Scd1−/− Mice 168 h Post 30 μg/kg TCDD Dose
| Lipids | Scd1+/+ Vehicle | Scd1+/+ TCDD | Scd1−/− Vehicle | Scd1−/− TCDD |
| Total FAs | 81.4 ± 10.4 | 153.6 ± 26.7* | 87.6 ± 7.3 | 137.8 ± 11.1* |
| SFA | 40.6 ± 6.5 | 64.6 ± 11.1* | 52.3 ± 5.3 | 68.8 ± 7.1* |
| Palmitic acid (16:0) | 19.9 ± 2.6 | 29.2 ± 3.7* | 19.8 ± 2.6 | 27.8 ± 2.8* |
| Stearic acid (18:0) | 17.0 ± 3.1 | 20.2 ± 2.2*,** | 19.6 ± 2.1 | 27.1 ± 3.2* |
| Arachidic acid (20:0) | 0.064 ± 0.006 | 0.066 ± 0.010 | 0.117 ± 0.076 | 0.093 ± 0.011 |
| Behenic acid (22:0) | 0.269 ± 0.044*** | 0.175 ± 0.017 | 0.425 ± 0.140 | 0.292 ± 0.056 |
| Lignoceric acid (24:0) | 3.33 ± 1.01*** | 14.9 ± 7.3* | 12.4 ± 1.9 | 13.4 ± 1.9 |
| MUFA | 12.7 ± 1.5*** | 36.8 ± 7.8*,** | 5.1 ± 0.5 | 15.8 ± 0.01* |
| Palmitoleic acid (16:1n7) | 1.24 ± 0.26*** | 2.39 ± 0.64*,** | 0.20 ± 0.03 | 0.46 ± 0.08 |
| Oleic acid (18:1n9) | 11.2 ± 1.4*** | 32.9 ± 6.9*,** | 4.68 ± 0.44 | 14.7 ± 1.8* |
| Eicosenoic acid (20:1n9) | 0.10 ± 0.015*** | 0.72 ± 0.22*,** | 0.040 ± 0.008 | 0.27 ± 0.056* |
| Erucic acid (22:1n9) | 0.028 ± 0.005 | 0.056 ± 0.009* | 0.031 ± 0.005 | 0.064 ± 0.009* |
| Nervonic acid (24:1n9) | 0.135 ± 0.043 | 0.802 ± 0.353*,** | 0.189 ± 0.047 | 0.277 ± 0.040 |
| PUFA | 28.1 ± 3.4 | 52.2 ± 10.1* | 30.1 ± 2.4 | 53.2 ± 5.3* |
| Total n3 FAs | 1.30 ± 0.37 | 4.23 ± 1.56* | 1.85 ± 0.49 | 3.70 ± 0.36* |
| Total n6 FAs | 26.7 ± 3.09 | 47.6 ± 8.5* | 28.2 ± 2.0 | 49.2 ± 4.9* |
| Linoleic acid (18:2n6) | 14.8 ± 1.83 | 31.7 ± 5.67* | 16.7 ± 1.65 | 31.7 ± 3.53* |
| Eicosadienoic acid (20:2n6) | 0.31 ± 0.042 | 1.26 ± 0.26* | 0.30 ± 0.040 | 1.20 ± 0.25* |
| Dihomo-γ-linolenic acid (20:3n6) | 0.95 ± 0.10 | 2.60 ± 0.69* | 0.786 ± 0.111 | 2.52 ± 0.37* |
| Arachidonic acid (20:4n6) | 10.6 ± 1.38 | 12.0 ± 2.66 | 10.4 ± 0.88 | 13.7 ± 1.06* |
| Docosapentaenoic acid (22:2n6) | 0.015 ± 0.007 | 0.035 ± 0.012* | 0.018 ± 0.004 | 0.042 ± 0.008* |
| Eicosatetraenoic acid (20:4n3) | 0.033 ± 0.005 | 0.162 ± 0.057* | 0.032 ± 0.006 | 0.153 ± 0.046* |
| α-Linolenic acid (18:3n3) | 0.458 ± 0.101 | 1.36 ± 0.30* | 0.045 ± 0.088 | 1.09 ± 0.25* |
| Timnodonic acid (20:5n3) | 0.614 ± 0.240 | 0.593 ± 0.231 | 0.629 ± 0.234 | 0.694 ± 0.146 |
| Docosapentaenoic acid (22:5n3) | 0.193 ± 0.078 | 2.12 ± 1.04* | 0.743 ± 0.232 | 1.76 ± 0.24* |
Note. Data were analyzed by factorial ANOVA followed by Tukey’s post hoc test, n = 5 biological replicates. *, p < 0.05 for TCDD versus Vehicle within the same genotype; **, p < 0.05 for +/+ TCDD versus −/− TCDD; ***, p < 0.05 for +/+ Vehicle versus −/− Vehicle.
Increases in absolute hepatic levels of MUFAs were primarily due to a threefold increase of oleic acid (18:1n9), representing >85% of all MUFAs (Table 1). Increases in accumulation of palmitoleic acid (16:1n7, increased 1.9-fold), eicosenoic acid (20:1n9, increased 7.2-fold), erucic acid (22:1n9, increased twofold), and nervonic acid (24:1n9, increased sixfold) accounted for the remaining 15% of MUFAs.
Although TCDD did not alter PUFA levels expressed as mol%, absolute hepatic levels were increased 1.8-fold by TCDD (Table 1). In general, TCDD treatment increased all PUFAs (absolute hepatic levels) examined except for timnodonic acid (20:5n3). Linoleic acid (18:2n6) and arachidonic acid (20:4n6, AA) were the dominant PUFAs, representing ∼60% and ∼25% of PUFAs, respectively. The n-3 and n-6 PUFAs exhibit anti- and proinflammatory effects, respectively (Marszalek and Lodish, 2005). The n3/n6 ratio was induced by TCDD and accompanied by the depletion of AA (Fig. 1, panel A).
TCDD Effects on Hepatic Lipid Content in Scd1−/− Mice
Scd1 performs the rate-limiting step in MUFA synthesis, and its deficiency has been reported to impair TRG synthesis and protect from diet-induced hepatic steatosis. In Scd1−/− mice, TCDD increased TRGs 2.5-fold and TFA content 1.6-fold compared with vehicle controls. However, TRG levels were 42% lower in Scd1−/− mice when compared with treated wild-type mice, yet there was no difference in TFA levels between genotypes (Fig. 1, panel B; p < 0.05, Fig. 1, see **).
Examination of the SFA/MUFA/PUFA ratios in Scd1−/− mouse liver revealed that TCDD reduced SFA proportions and increased relative MUFA and PUFA levels compared with vehicle controls. Scd1 deficiency also induced SFA by 8%, reduced MUFA by 13%, and induced PUFA by 5% compared with treated wild-type mice, consistent with loss of Scd1 activity (Fig. 1, panel B; p < 0.05, Fig. 1, see **).
Further analysis of individual FA species identified increases of 18:0, whereas 16:1n7, 18:1n9, and 20:1n9 were reduced in treated null mouse livers compared with treated wild-type mice (Table 1; p < 0.05, Fig. 1, see **). Surprisingly, MUFA increases in Scd1 null mice were primarily due to a 3.2-fold induction of 18:1n9. Similar to Scd1+/+ mice, overall PUFA levels were induced by TCDD in Scd1−/− mice. In contrast, the n3/n6 ratios were not altered and the molar ratio of AA ([nmol/g AA]/[nmol/g total FA detected]) was higher compared with treated wild-type mice (Fig. 1, panel B).
Principal Component Analysis
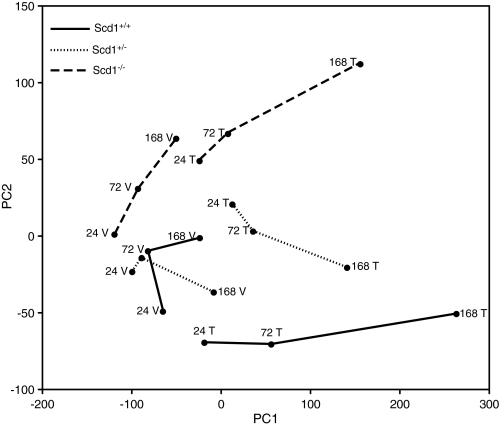
PCA was performed to characterize the trends exhibited by GC/MS-FA Methyl Ester (FAMES) data. PCA of hepatic TFAs indicated a clear time-, treatment-, and Scd1 dose–dependent effect on hepatic FA composition (Fig. 2). PC1 and PC2 accounted for 95% of the cumulative proportion of the variance with vehicles clustering along PC1, treated groups separating along PC1, and genotype separating along PC2. All treated animals exhibited a similar temporal trajectory.
FIG. 2.
PCA of GC-MS lipid profiles from TCDD (T) or Vehicle (V) treated Scd1+/+, Scd1+/−, and Scd1−/− mice 24, 72, and 168 h postdose. PCA was performed in R as described in the “Materials and Methods.” PC1 and PC2 accounted for 95% of the cumulative proportion of the variance with vehicles clustering along PC1, treated groups separating along PC1, and genotype separating along PC2. Dashed lines (Scd1−/−), dotted lines (Scd1+/-), solid lines (Scd1+/+), n = 5 biological replicates.
TCDD Effects on the Scd1 Desaturation Index
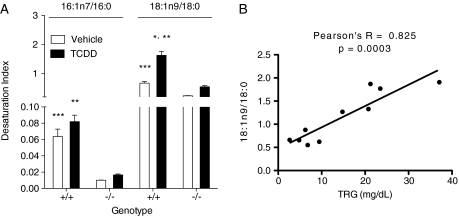
Scd1 is the primary hepatic Δ9 desaturase, metabolizing 16:0 and 18:0 into 16:1n7 and 18:1n9, respectively. TCDD induced the hepatic 18:1n9/18:0 ratio in Scd1+/+ mice compared with vehicles (Fig. 3A). In humans, positive correlations between the Scd1 desaturation index and TRG levels have been reported (Attie et al., 2002). Similarly, TRG levels increased with the 18:1n9/18:0 ratio in TCDD-treated Scd1+/+ mice (Pearson’s r = 0.825, p = 0.0003) (Fig. 3B).
FIG. 3.
Scd1 activity (desaturation) index (A) in female TCDD (T) or vehicle (V) treated Scd1+/+ and Scd1−/− mice. The desaturation index is the ratio of palmitoleic (16:1n7) or oleic acid (18:1n9) to the precursors palmitic (16:0) or stearic acids (18:0), respectively. Bars represent mean ± SEM, n = 5 biological replicates. *, p < 0.05 for TCDD compared with vehicle within a genotype; **, p < 0.05 for Scd1−/− TCDD compared with Scd1+/+ TCDD; ***, p < 0.05 for Scd1−/− vehicle compared with Scd1+/+ vehicle. Data were analyzed by factorial ANOVA followed by Tukey’s post hoc test. (B) Correlation analysis was performed in Graphpad Prism 5.0a between the hepatic triglyceride (TRG) levels and the 18:1n9/18:0 desaturation index in Scd1+/+ mice gavaged with 30 μg/kg with TCDD or vehicle for 168 h.
TCDD Effects on Hepatic Lipid Metabolism Gene Expression
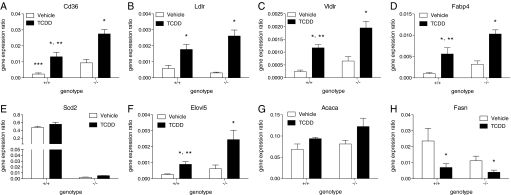
To further examine TCDD’s effects on hepatic lipid composition in Scd1+/+and Scd1−/− mice, QRT-PCR was used to quantify mRNA levels for genes involved in lipid metabolism. Consistent with TCDD-induced increases in hepatic lipid levels, the FA transport genes Cd36, Ldlr, Vldlr, and Fabp4, were induced threefold to fivefold in both genotypes (Fig. 4A–D). Furthermore, Cd36, Vldlr, and Fabp4 mRNA were significantly increased in treated Scd1-/- mice compared with Scd1+/+ mice and may explain increases in 18:1n9 levels, the primary MUFA (81.3% of all MUFAs) in Harlan Teklad rodent diet 7964, in null mice.
FIG. 4.
QRTPCR of hepatic lipid transport, modification, and biosynthesis genes in Scd1+/+ (+/+) and Scd1−/− (−/−) mice gavaged with 30 μg/kg TCDD or sesame oil vehicle for 24 h. The gene expression ratio is the total quantity normalized to the geometric mean of Hprt, Actb, and Gapdh mRNA levels. Genes are indicated by official gene symbols. Error bars represent the SEM, n = 5. *, p < 0.05 for TCDD compared with vehicle within a genotype; **, p < 0.05 for Scd1+/+ TCDD compared with Scd1−/− TCDD; ***, p < 0.05 for Scd1+/+ vehicle compared with Scd1−/− vehicle. Data were analyzed by factorial ANOVA followed by Tukey’s post hoc test.
Four isoforms, Scd1-4, exist in the mouse and may underlie increases in 18:1n9. However, Scd3 is reported to be expressed by sebocytes in skin, the preputual gland, and the Harderian gland (Miyazaki et al., 2006), whereas Scd4 expression is limited to the heart (Miyazaki et al., 2003). Neither Scd3 or Scd4 mRNA were detected in the liver of either TCDD-treated genotype. Scd2 is primarily expressed in adipose tissue but also in the neonatal mouse liver. Scd2 mRNA was expressed in wild-type mice but not induced by TCDD (Fig. 4E). Surprisingly, Scd2 mRNA levels were scarce (100-fold lower) in null mice compared with wild-types. The lack of Scd2 mRNA expression in nulls suggests Scd1 deletion also affected Scd2 expression.
TCDD-mediated increases in the PUFAs eicosatetraenoic acid (20:4n3) and docosapentaenoic acid (22:5n3) were consistent with the twofold induction of Elovl5 mRNA (Fig. 4F) an elongase that catalyzes the elongatation of very long-chain PUFAs (Guillou et al., 2010). The lipogenic gene Acaca, which provides malonyl-CoA for FA biosynthesis, was not altered by TCDD (Fig. 4G). Fasn, a long-chain FA synthetase mRNA was reduced 2.5- to 3-fold by TCDD in both genotypes (Fig. 4H), suggesting that increases in hepatic lipid content are not due to de novo lipogenesis. Finally, the well-characterized TCDD-inducible genes Cyp1a1, Nqo1, and Tiparp exhibited comparable induction in wild-type and null mice (Supplementary figure 2).
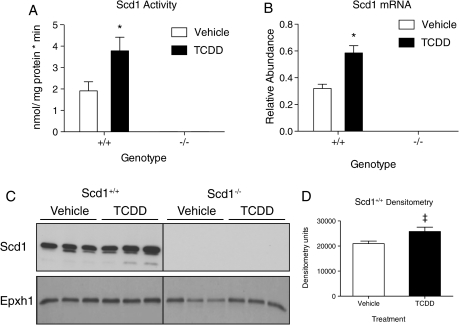
TCDD Effects on Scd1 Activity, mRNA, and Protein
The effects of TCDD on Scd1, activity, mRNA, and protein levels were examined. Scd1 enzyme activity was measured by following the conversion of 14C 18:0 to 14C 18:1n9. Consistent with increases in the 18:1n9/18:0 desaturation index, TCDD induced Scd1 activity twofold in wild-type mice compared with vehicles at 24 h (Fig. 5A). QRTPCR also showed that TCDD induced Scd1 mRNA levels 1.5-fold in Scd1+/+ mice (Fig. 5B). Furthermore, western blots showed an increase in Scd1 immunoreactivity in treated Scd1+/+ microsomal fractions (Figs. 5C and 5D). Induction in Scd1 activity, mRNA, and protein were not detected in control or TCDD-treated Scd1−/− mice.
FIG. 5.
Scd1 activity, mRNA, and protein levels in Scd1+/+ and Scd1−/− mice treated with 30 μg/kg TCDD (T) or sesame seed oil (V) 24 h postdose. (A) Scd1 activity. Hepatic microsomes (100 μg, n = 5) isolated from mice were incubated with 0.03 μCi 14C-stearoyl-CoA (14C 18:0), NADH, and stearoyl-CoA. 14C 18:0 was separated from 14C 18:1 by silver ion chromatography and radioactivity measured by scintillation counting. Scd1 activity is expressed as nmol 14C 18:0 converted to 14C 18:1 per mg Scd1 protein per min. (B) QRT-PCR of Scd1 mRNA (n = 5). Expression is represented as a ratio of the total quantity of Scd1 normalized to the geometric mean of Hprt, Actb, and Gapdh. (C) Scd1 Western blot. Hepatic Scd1 protein (n = 3) was detected in 10 μg of microsomes. Epxh1 was used as a microsomal protein reference for loading control. (D) Densitometry. Densitometry was determined with ImageJ from Scd1 bands and normalized to Ephx1 bands. ‡ p = 0.08. For A, B, and D bars represent mean ± SEM, *, p < 0.05 for T compared with V within a genotype; **, p < 0.05 for Scd1−/− T compared with Scd1+/+ T within a time point; ***, p < 0.05 for Scd1−/− V compared with Scd1+/+ V within a time point. Data were analyzed by factorial ANOVA followed by Tukey’s post hoc test.
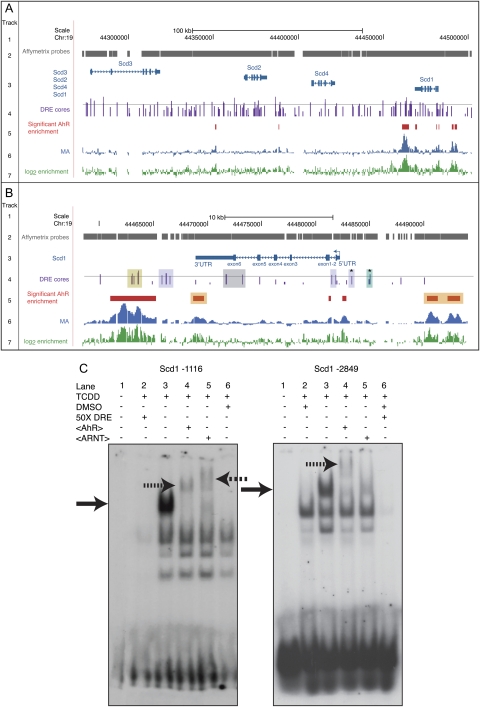
Putative DRE Distribution and AhR Enrichment at Scd1 Loci
To further examine AhR regulation of Scd1, Figure 6A summarizes the ChIP-chip analysis of AhR enrichment at Scd1-4 genomic loci induced by TCDD (Dere et al., 2011b). The moving average (MA) value visualizes the enriched genomic regions within the Scd1-4 loci, whereas the log2 enrichment illustrates the fold change for each Affymetrix probe. Six AhR-enriched regions (up to 5.7-fold, FDR <0.01, red bar) ranging from 70 to 4200 bp were associated with Scd1 (Fig. 6A). AhR enrichment was located within 10-kb upstream region through the 3′-UTR. In contrast, the adjacent ∼150-kb genomic region spanning Scd2, Scd3, and Scd4 exhibited only two AhR-enriched regions (2- to 2.7-fold).
FIG. 6.
DRE distribution and TCDD-inducible AhR enrichment within Scd loci. (A) Genomic region spanning Scd1, Scd2, Scd3, and Scd4. (B) Scd1 genomic region only. Genomic DRE distributions and regions of AhR enrichment induced by TCDD were previously determined (Dere et al., 2011a; Dere et al., 2011b). Track 1: scale and chromosome position. Track 2: probe tiling across the Affymetrix 2.0R mouse array. Track 3: gene organization including TSS (closed arrow), exons (closed boxes), introns, and direction of transcription (solid arrowhead line). Track 4: location of DRE cores (5′-GCGTG-3′). Height of vertical bars indicate MSS for the 19 bp DRE sequence (Dere et al., 2011a). The horizontal line indicates the MSS = 0.8. MSSs greater than 0.8 are considered putative functional DREs. The asterisk (*) denotes 19 bp DRE sequences that bound TCDD-activated AhR in band shift assays. Track 5: regions of significant (FDR < 0.01) AhR enrichment in genome-wide ChIP-chip assays. Tracks 6 and 7: histograms depicting the signal intensities for the MA(blue, track 6) and log2 fold enrichment (green, track 7) values for regions exhibiting AhR enrichment in genome-wide ChIP-chip assays. The above tracks were modified from the University of California–Santa Cruz genome browser. (B) Scd1 genomic region only. Track 4: yellow shading, putative DRE-AhR enrichment overlap; purple shading, putative DREs lacking Affymetrix tiling probes; gray shading, putative DREs that do not overlap with AhR enrichment; and green shading, putative DRE in an AhR region that failed to meet the FDR cutoff of 0.01. Track 5: orange shading, AhR-enriched regions that do not overlap with putative DREs. (C) Representative band shift assays with putative, functional DREs (track 4, asterisks [*]). The DRE position is numerically indicated relative to the Scd1 TSS. Solid arrows indicate a TCDD-inducible band shift. Hatched or dashed arrows indicate an AhR or ARNT supershift, respectively.
AhR is proposed to regulate gene expression through DNA binding at DREs containing the core sequence 5′-GCGTG-3′. DREs with MSS ranging from 0.69 to 0.93 were identified within the genomic region spanning Scd genomic loci (Dere et al., 2011a). Of the 39 DREs possessing a putative functional (high scoring) MSS (>0.8; track 4, horizontal line), 16 were located 10 kb upstream of the Scd1 TSS through the Scd1 3′-UTR (Fig. 6B). Five of these 16 high-scoring Scd1 DREs overlapped with regions of AhR enrichment (track 4, yellow shading). Notably, seven DREs that did not overlap with regions of significant AhR enrichment coincided with regions lacking tiling probes (track 4, purple shading). One DRE core fell within a twofold AhR-enriched region that failed to meet the FDR cutoff of 0.01 (track 4, green shading). Moreover, three AhR-enriched regions lacked any DRE cores (track 5, orange shading), consistent with promoter- and genome-wide ChIP-chip studies reporting 50% overlap between DRE cores and AhR enrichment as well as other studies suggesting AhR interaction with DNA independent of ARNT (Beischlag et al., 2008; Dere et al., 2011a,b; Klinge et al., 2000; Marlowe et al., 2008).
Band Shift Assays
Two DRE cores (Fig. 6B, track 4, indicated by*) within 10 kb upstream of the Scd1 TSS and with an MSS > 0.8 exhibited band shifts with TCDD-activated guinea pig cytosol (Fig. 6C). The addition of AhR or ARNT antibodies to the incubation resulted in a supershift, confirming AhR binding to these putative DREs. Nucleotides flanking the DRE core sequence modulate binding affinity and coactivator recruitment and may underlie differences in the banding pattern between the two DREs (Bank et al., 1995; Gillesby et al., 1997; Lusska et al., 1993; Shen and Whitlock, 1992). These results, in addition to mRNA, protein, and activity levels, support AhR recruitment to and regulation of Scd1 by TCDD.
DISCUSSION
Previous studies examining the role of Scd1 in hepatic diseases have focused on dietary or genetic modulation of lipid metabolism (Attie et al., 2002; Miyazaki et al., 2001; Popeijus et al., 2008; Sampath et al., 2007; Warensjo et al., 2007) but not the effects of environmental contaminants. In this study, AhR-mediated induction of Scd1 by TCDD and subsequent effects on hepatic FA composition were examined in Scd1+/+ and Scd1−/− mice. Here, we report that TCDD induced hepatic TRG and FA accumulation, elicited differential gene expression of lipid metabolism and transport, and increased Scd1 mRNA, protein, and activity, as a result of AhR recruitment to Scd1 genic regions. Collectively, these studies suggest that AhR regulation of Scd1 alters hepatic FA composition, which influences TCDD-induced hepatotoxicity.
TCDD-altered hepatic gene expression associated with lipid transport, partitioning, and metabolism in mice (Boverhof et al., 2006; Kopec et al., 2010b, Kopec et al., 2011). For example, TCDD induced the lipolytic genes lipoprotein lipase (Lpl), phospholipase A2, group XIIA (Pla2g12a), monoglyceride lipase (Mgll), and pancreatic lipase-related protein (Pnliprp1) that hydrolyze hepatocellular TRG stores into FFAs and monoglycerides (Giller et al., 1992). It also induced low-density lipoprotein receptor (Ldlr), very low-density lipoprotein receptor (Vldlr), Cd36 antigen (Cd36), and FA binding protein (Fabp4) in Scd1+/+ and Scd1−/− mice. Ldlr and Cd36 are membrane-associated proteins that facilitate the uptake of chylomicron and very low-density lipoprotein (VLDL) remnants as well as long-chain FA (Ibrahimi and Abumrad, 2002; Iqbal and Hussain, 2009). Cd36 has been implicated in the etiology of obesity and diabetes, and Cd36 null mice exhibit minimal TCDD-elicited steatosis (Drover et al., 2005; Hajri et al., 2002; Lee et al., 2010; Rankinen et al., 2006). Fabps are cytosolic, high-affinity long-chain FA-binding proteins that target lipids to intracellular compartments (Storch and Thumser, 2010). Fabp4, which mediates lipid trafficking to the nucleus and may provide ligands for peroxisome proliferator–activated receptors (PPARs) (Gillilan et al., 2007; Tan et al., 2002), was also induced. Furthermore, inhibition of VLDL secretion has been reported in mice following TCDD treatment (Lee et al., 2010). These changes likely underlie effects that contribute to TCDD-elicited hepatic FA accumulation.
ChIP-chip analysis identified six Scd1 genic regions with AhR enrichment. Two proximal AhR-enriched regions were within 2 kb of the TSS (Dere et al., 2011b). The 3′ regions also exhibited AhR enrichment and high-scoring DREs, suggesting AhR regulation by distal enhancer sites. Although distal regulation remains largely uninvestigated, studies suggest transcription factor binding at these sites promotes chromatin looping and structural modifications that facilitate gene expression (Farnham, 2009; Li et al., 2006; Long and Miano, 2007). Two 5′ AhR-enriched regions at distal sites lacked DRE cores, providing further evidence of DRE-independent AhR-DNA interactions, which may involve tethering to other DNA interacting transcription factors (Murray et al., 2010). However, the arrays do not have uniform tiling across the genome, and several genomic regions lack probe coverage in areas containing high-scoring DREs that may confound mapping AhR enrichment to regions containing DREs.
AhR enrichment at Scd1 genic regions and the induction of Scd1 mRNA, protein, and activity by TCDD provided compelling evidence for AhR regulation of Scd1. The induction of Scd1 activity increased MUFA levels, decreased SFA levels, and increased MUFA:SFA and PUFA:SFA ratios. Scd1 deficiency did not affect TCDD-induced hepatic lipid accumulation (as measured by GC-MS analysis of FAMES). However, null mice had fewer TRGs, reduced MUFA levels, and exhibited less hepatic injury relative to treated wild-type mice. More specifically, Scd1−/− mice exhibited no increase in serum ALT, less hepatic vacuolization, and reduced immune cell infiltration compared with Scd1+/+ mice, suggesting lower overall TCDD-elicited hepatotoxicity.
In addition to Scd1, hepatic FA profiles indicate that AhR regulates other lipid modifying enzymes in addition to Scd1. Increases in n-6 and n-3 pathway intermediates (e.g., 20:2n6, 20:3n6, 20:4n3, and 22:5n3) suggests the induction of Elovl5 activity (Wang et al., 2008). QRT-PCR, band shift, and ChIP-chip assays (Supplementary figure 3A–C) verified AhR-mediated induction of Elovl5 by TCDD. The n-3 and n-6 PUFAs, particularly eicosapentanoic acid (EPA, 20:5n3) and arachidonic acid (AA, 20:4n6) metabolites, exhibit anti- and proinflammatory activities, respectively (Di Marzo, 1995; Marszalek and Lodish, 2005; Simopoulos, 2002; Yashodhara et al., 2009). EPA is an Elovl5 substrate, therefore excess 20:5n3 may be elongated into 22:5n3, which is increased in both genotypes. AA is not an Elovl5 substrate but is rapidly metabolized by TCDD-inducible prostaglandin-endoperoxide synthase 1 (Ptgs1), arachidonate 12-lipoxygenase (Alox12) (Kopec et al., 2010b; Dere et al., 2011b), and glutathione transferases (Boverhof et al., 2005) into proinflammatory ecosanoids. Additionally, AA is liberated from membrane phospholipids by Pla2g12a, another TCDD-inducible gene. AA levels were lower in treated Scd1+/+ mice compared with vehicles and treated nulls, and although we cannot rule out their conversion into inflammatory ecosanoids, are consistent with the increased level of inflammation in wild-type mice compared with nulls. Inflammation in Scd1+/+mice may also be due to the induction of Scd1, which would sequester cytochrome b5, uncouple P450 monooxygenases, and increase superoxide and reactive oxygen species (ROS) formation (Hardwick et al., 2009; Schenkman and Jansson, 2003). However, P450 monooxygenase and xanthine dehydrogenase induction by TCDD are the primary ROS contributors (Sugihara et al., 2001; Zimmerman and Granger, 1994).
Our results differ from studies examining the protective effects of Scd1 deficiency from steatosis and exacerbated steatohepatitis elicited using in vivo dietary-induced models of liver injury. For example, high-fat diets (HFD) induce steatosis in mice. Scd1−/− mice fed HFD exhibit no evidence of hepatomegaly or histological changes (Li et al., 2009), yet hepatomegaly is observed in all animals exposed to TCDD (Poland and Knutson, 1982). Methionine choline deficient (MCD) diets induce steatohepatitis, but in contrast to TCDD, decrease Scd1 expression, and induce Cyp4A expression, an enzyme involved in lipid peroxidation (Anstee and Goldin, 2006; Li et al., 2009; Yamaguchi et al., 2007). Cyp4a is a PPAR target, and administration of a PPAR agonist to MCD fed mice decreases liver damage (Nagasawa et al., 2006), suggesting a role for PPAR rather than AhR in MCD-elicited liver injury. Furthermore, our results are consistent with increased MUFA:SFA ratios in lipotoxic mechanisms of liver injury (Larter et al., 2008).
TCDD-induced steatosis is a significant hepatotoxic effect, and AhR-mediated induction of Scd1 exacerbates TCDD hepatotoxicity by altering the composition of accumulated lipids. Scd1-mediated increases in unsaturated FA levels may alter membrane fluidity, increase lipid peroxidation and ROS formation, as well as propagate inflammatory responses through TRAIL-mediated cytotoxicity (Malhi et al., 2007; Trauner et al., 2010). Steatosis followed by a progressive inflammatory response poses a significant risk of progression to cirrhosis and may contribute to hepatocellular carcinoma development in rodents.
The induction of hepatic lipid accumulation in mice is consistent with the occurrence of dyslipidemia in humans following TCDD exposure at high doses (Pelclova et al., 2002). Interestingly, the hepatic FA composition induced by TCDD is similar to serum and lipid profiles (e.g., increase in TRGs and 16:1n7 and 18:1n9 levels) described for human non-alcoholic fatty liver disease (NAFLD) patients (Puri et al., 2007, 2009). NAFLD is considered the hepatic manifestation of metabolic syndrome and precedes nonalcoholic steatosis and cirrhosis. This report suggests that AhR regulation of lipid transport, metabolism, and modifying enzymes, including Scd1, alters lipid composition that contributes to the hepatotoxicity of TCDD.
SUPPLEMENTARY MATERIAL
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences Superfund Basic Research Program (P42ES04911).
Supplementary Material
Acknowledgments
We would like to thank Dr Bev Chamberlain at Michigan State University Mass Spectrometry Facility, and Bryan Mets, Michelle D’Souza, Daniel Wright, Minghui Zhao, and Dr James Ntambi for technical assistance. We would also like to thank Dr Anna Kopec for critical review.
References
- Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attie AD, Flowers MT, Flowers JB, Groen AK, Kuipers F, Ntambi JM. Stearoyl-CoA desaturase deficiency, hypercholesterolemia, cholestasis, and diabetes. Nutr. Rev. 2007;65:S35–S38. doi: 10.1111/j.1753-4887.2007.tb00326.x. [DOI] [PubMed] [Google Scholar]
- Attie AD, Krauss RM, Gray-Keller MP, Brownlie A, Miyazaki M, Kastelein JJ, Lusis AJ, Stalenhoef AF, Stoehr JP, Hayden MR, et al. Relationship between stearoyl-CoA desaturase activity and plasma triglycerides in human and mouse hypertriglyceridemia. J. Lipid Res. 2002;43:1899–1907. doi: 10.1194/jlr.m200189-jlr200. [DOI] [PubMed] [Google Scholar]
- Bank PA, Yao EF, Phelps CL, Harper PA, Denison MS. Species-specific binding of transformed Ah receptor to a dioxin responsive transcriptional enhancer. Eur. J. Pharmacol. 1992;228:85–94. doi: 10.1016/0926-6917(92)90016-6. [DOI] [PubMed] [Google Scholar]
- Bank PA, Yao EF, Swanson HI, Tullis K, Denison MS. DNA binding of the transformed guinea pig hepatic Ah receptor complex: Identification and partial characterization of two high-affinity DNA-binding forms. Arch. Biochem. Biophys. 1995;317:439–448. doi: 10.1006/abbi.1995.1186. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell Biol. 2002;22:4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-mediated hepatotoxicity. Toxicol. Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, Bradfield CA. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- Croutch CR, Lebofsky M, Schramm KW, Terranova PF, Rozman KK. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and 1,2,3,4,7,8-hexachlorodibenzo-p-dioxin (HxCDD) alter body weight by decreasing insulin-like growth factor I (IGF-I) signaling. Toxicol. Sci. 2005;85:560–571. doi: 10.1093/toxsci/kfi106. [DOI] [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 2011a;24:494–504. doi: 10.1021/tx100328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011b;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V. Arachidonic acid and eicosanoids as targets and effectors in second messenger interactions. Prostaglandins Leukot. Essent. Fatty Acids. 1995;53:239–254. doi: 10.1016/0952-3278(95)90123-x. [DOI] [PubMed] [Google Scholar]
- Drover VA, Ajmal M, Nassir F, Davidson NO, Nauli AM, Sahoo D, Tso P, Abumrad NA. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J. Clin. Invest. 2005;115:1290–1297. doi: 10.1172/JCI21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr. Opin. Lipidol. 2008;19:248–256. doi: 10.1097/MOL.0b013e3282f9b54d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller T, Buchwald P, Blum-Kaelin D, Hunziker W. Two novel human pancreatic lipase related proteins, hPLRP1 and hPLRP2. Differences in colipase dependence and in lipase activity. J. Biol. Chem. 1992;267:16509–16516. [PubMed] [Google Scholar]
- Gillesby BE, Stanostefano M, Porter W, Safe S, Wu ZF, Zacharewski TR. Identification of a motif within the 5' regulatory region of pS2 which is responsible for AP-1 binding and TCDD-mediated suppression. Biochemistry. 1997;36:6080–6089. doi: 10.1021/bi962131b. [DOI] [PubMed] [Google Scholar]
- Gillilan RE, Ayers SD, Noy N. Structural basis for activation of fatty acid-binding protein 4. J. Mol. Biol. 2007;372:1246–1260. doi: 10.1016/j.jmb.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J. Clin. Invest. 2002;109:1381–1389. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hardwick JP, Osei-Hyiaman D, Wiland H, Abdelmegeed MA, Song BJ. PPAR/RXR regulation of fatty acid metabolism and fatty acid omega-hydroxylase (CYP4) isozymes: Implications for prevention of lipotoxicity in fatty liver disease. PPAR Res. 2009;2009:952734. doi: 10.1155/2009/952734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J, Cirvello J, Haseman J, Bucher J. Chemicals associated with site-specific neoplasia in 1394 long-term carcinogenesis experiments in laboratory rodents. Environ. Health Perspect. 1991;93:247–270. doi: 10.1289/ehp.9193247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport of long-chain fatty acids. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:139–145. doi: 10.1097/00075197-200203000-00004. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Hussain MM. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issandou, M., Bouillot, A., Brusq, J. M., Forest, M. C., Grillot, D., Guillard, R., Martin, S., Michiels, C., Sulpice, T., and Daugan, A. (2009). Pharmacological inhibition of stearoyl-CoA desaturase 1 improves insulin sensitivity in insulin-resistant rat models. Eur. J. Pharmacol.618, 28–36. [DOI] [PubMed]
- Jiang G, Li Z, Liu F, Ellsworth K, Dallas-Yang Q, Wu M, Ronan J, Esau C, Murphy C, Szalkowski D, et al. Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J. Clin. Invest. 2005;115:1030–1038. doi: 10.1172/JCI23962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM, Kaur K, Swanson HI. The aryl hydrocarbon receptor interacts with estrogen receptor alpha and orphan receptors COUP-TFI and ERRalpha1. Arch. Biochem. Biophys. 2000;373:163–174. doi: 10.1006/abbi.1999.1552. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Burgoon LD, Ibrahim-Aibo D, Harkema JR, Tashiro C, Chittim B, Zacharewski TR. Comparative toxicogenomic examination of the hepatic effects of PCB126 and TCDD in immature, ovariectomized C57BL/6 mice. Toxicol. Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Burg AR, Lee AW, Tashiro C, Potter D, Sharratt B, Harkema JR, et al. Automated dose-response analysis and comparative toxicogenomic evaluation of the hepatic effects elicited by TCDD, TCDF, and PCB126 in C57BL/6 mice. Toxicol. Sci. 2010a;118:286–297. doi: 10.1093/toxsci/kfq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Mets BD, Tashiro C, Potter D, Sharratt B, Harkema JR, Zacharewski TR, et al. PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: Comparative toxicogenomic effects of dioxin and non-dioxin-like ligands. Toxicol. Appl. Pharmacol. 2010b;243:359–371. doi: 10.1016/j.taap.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec, A. K., D'Souza, M. L., Mets, B. D., Burgoon, L. D., Reese, S. E., Archer, K. J., Potter, D., Tashiro, C., Sharratt, B., Harkema, J. R., (2011). Non-additive hepatic gene expression elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) co-treatment in C57BL/6 mice. Toxicol. Appl. Pharmacol.256, 154–167. [DOI] [PMC free article] [PubMed]
- Larter, C. Z., Yeh, M. M., Haigh, W. G., Williams, J., Brown, S., Bell-Anderson, K. S., Lee, S. P., and Farrell, G. C. (2008). Hepatic free fatty acids accumulate in experimental steatohepatitis: Role of adaptive pathways. J. Hepatol.48, 638–647. [DOI] [PubMed]
- Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, Gonzalez FJ, Xie W. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139:653–663. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Li ZZ, Berk M, McIntyre TM, Feldstein AE. Hepatic lipid partitioning and liver damage in nonalcoholic fatty liver disease: Role of stearoyl-CoA desaturase. J. Biol. Chem. 2009;284:5637–5644. doi: 10.1074/jbc.M807616200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Miano JM. Remote control of gene expression. J. Biol. Chem. 2007;282:15941–15945. doi: 10.1074/jbc.R700010200. [DOI] [PubMed] [Google Scholar]
- Lusska A, Shen E, Whitlock JP., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor. J. Biol. Chem. 1993;268:6575–6580. [PubMed] [Google Scholar]
- Maines MD Wiley online library. Current Protocols in Toxicology. New York, NY: John Wiley; 2002. [Google Scholar]
- Malhi H, Barreyro FJ, Isomoto H, Bronk SF, Gores GJ. Free fatty acids sensitise hepatocytes to TRAIL mediated cytotoxicity. Gut. 2007;56:1124–1131. doi: 10.1136/gut.2006.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe JL, Fan Y, Chang X, Peng L, Knudsen ES, Xia Y, Puga A. The aryl hydrocarbon receptor binds to E2F1 and inhibits E2F1-induced apoptosis. Mol. Biol. Cell. 2008;19:3263–3271. doi: 10.1091/mbc.E08-04-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Lodish HF. Docosahexaenoic acid, fatty acid-interacting proteins, and neuronal function: Breastmilk and fish are good for you. Annu. Rev. Cell Dev. Biol. 2005;21:633–657. doi: 10.1146/annurev.cellbio.21.122303.120624. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Bruggink SM, Ntambi JM. Identification of mouse palmitoyl-coenzyme A Delta9-desaturase. J. Lipid Res. 2006;47:700–704. doi: 10.1194/jlr.C500025-JLR200. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Flowers MT, Sampath H, Chu K, Otzelberger C, Liu X, Ntambi JM. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab. 2007;6:484–496. doi: 10.1016/j.cmet.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Jacobson MJ, Man WC, Cohen P, Asilmaz E, Friedman JM, Ntambi JM. Identification and characterization of murine SCD4, a novel heart-specific stearoyl-CoA desaturase isoform regulated by leptin and dietary factors. J. Biol. Chem. 2003;278:33904–33911. doi: 10.1074/jbc.M304724200. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kim YC, Gray-Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J. Biol. Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kim YC, Ntambi JM. A lipogenic diet in mice with a disruption of the stearoyl-CoA desaturase 1 gene reveals a stringent requirement of endogenous monounsaturated fatty acids for triglyceride synthesis. J. Lipid Res. 2001;42:1018–1024. [PubMed] [Google Scholar]
- Murray IA, Morales JL, Flaveny CA, Dinatale BC, Chiaro C, Gowdahalli K, Amin S, Perdew GH. Evidence for ligand-mediated selective modulation of aryl hydrocarbon receptor activity. Mol. Pharmacol. 2010;77:247–254. doi: 10.1124/mol.109.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, T., Inada, Y., Nakano, S., Tamura, T., Takahashi, T., Maruyama, K., Yamazaki, Y., Kuroda, J., and Shibata, N. (2006). Effects of bezafibrate, PPAR pan-agonist, and GW501516, PPARdelta agonist, on development of steatohepatitis in mice fed a methionine- and choline-deficient diet. Eur. J. Pharmacol. 536, 182–191. [DOI] [PubMed]
- Ntambi JM, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog. Lipid Res. 2004;43:91–104. doi: 10.1016/s0163-7827(03)00039-0. [DOI] [PubMed] [Google Scholar]
- Pelclova D, Fenclova Z, Preiss J, Prochazka B, Spacil J, Dubska Z, Okrouhlik B, Lukas E, Urban P. Lipid metabolism and neuropsychological follow-up study of workers exposed to 2,3,7,8-tetrachlordibenzo-p-dioxin. Int. Arch. Occup. Environ. Health. 2002;75(Suppl.):S60–S66. doi: 10.1007/s00420-002-0350-4. [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Pollenz RS, Sattler CA, Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct subcellular localizations in Hepa 1c1c7 cells by immunofluorescence microscopy. Mol. Pharmacol. 1994;45:428–438. [PubMed] [Google Scholar]
- Popeijus HE, Saris WH, Mensink RP. Role of stearoyl-CoA desaturases in obesity and the metabolic syndrome. Int. J. Obes. (Lond.) 2008;32:1076–1082. doi: 10.1038/ijo.2008.55. [DOI] [PubMed] [Google Scholar]
- Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, Cheung O, Sargeant C, Contos MJ, Sanyal AJ. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, Contos MJ, Sterling RK, Fuchs M, Zhou H, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: The 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath H, Miyazaki M, Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of dietary saturated fat. J. Biol. Chem. 2007;282:2483–2493. doi: 10.1074/jbc.M610158200. [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol. Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. St Louis, MO: Mosby; 1980. Theory and Practice of Histotechnology. [Google Scholar]
- Shen ES, Whitlock JP., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. J. Biol. Chem. 1992;267:6815–6819. [PubMed] [Google Scholar]
- Simopoulos AP. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002;56:365–379. doi: 10.1016/s0753-3322(02)00253-6. [DOI] [PubMed] [Google Scholar]
- Stefan N, Peter A, Cegan A, Staiger H, Machann J, Schick F, Claussen CD, Fritsche A, Haring HU, Schleicher E. Low hepatic stearoyl-CoA desaturase 1 activity is associated with fatty liver and insulin resistance in obese humans. Diabetologia. 2008;51:648–656. doi: 10.1007/s00125-008-0938-7. [DOI] [PubMed] [Google Scholar]
- Storch J, Thumser AE. Tissue-specific functions in the fatty acid-binding protein family. J. Biol. Chem. 2010;285:32679–32683. doi: 10.1074/jbc.R110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Kitamura S, Yamada T, Ohta S, Yamashita K, Yasuda M, Fujii-Kuriyama Y. Aryl hydrocarbon receptor (AhR)-mediated induction of xanthine oxidase/xanthine dehydrogenase activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001;281:1093–1099. doi: 10.1006/bbrc.2001.4464. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauner M, Arrese M, Wagner M. Fatty liver and lipotoxicity. Biochim. Biophys. Acta. 2010;1801:299–310. doi: 10.1016/j.bbalip.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Torres-Gonzalez M, Tripathy S, Botolin D, Christian B, Jump DB. Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition. J. Lipid Res. 2008;49:1538–1552. doi: 10.1194/jlr.M800123-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warensjo E, Ingelsson E, Lundmark P, Lannfelt L, Syvanen AC, Vessby B, Riserus U. Polymorphisms in the SCD1 gene: Associations with body fat distribution and insulin sensitivity. Obesity (Silver Spring) 2007;15:1732–1740. doi: 10.1038/oby.2007.206. [DOI] [PubMed] [Google Scholar]
- Warensjo E, Ohrvall M, Vessby B. Fatty acid composition and estimated desaturase activities are associated with obesity and lifestyle variables in men and women. Nutr. Metab. Cardiovasc. Dis. 2006;16:128–136. doi: 10.1016/j.numecd.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Warensjo E, Rosell M, Hellenius ML, Vessby B, De Faire U, Riserus U. Associations between estimated fatty acid desaturase activities in serum lipids and adipose tissue in humans: Links to obesity and insulin resistance. Lipids Health Dis. 2009;8:37. doi: 10.1186/1476-511X-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Yang, L., McCall, S., Huang, J., Yu, X. X., Pandey, S. K., Bhanot, S., Monia, B. P., Li, Y. X. and Diehl, A. M. (2007). Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology45, 1366–1374. [DOI] [PubMed]
- Yashodhara BM, Umakanth S, Pappachan JM, Bhat SK, Kamath R, Choo BH. Omega-3 fatty acids: A comprehensive review of their role in health and disease. Postgrad. Med. J. 2009;85:84–90. doi: 10.1136/pgmj.2008.073338. [DOI] [PubMed] [Google Scholar]
- Zimmerman BJ, Granger DN. Mechanisms of reperfusion injury. Am. J. Med. Sci. 1994;307:284–292. doi: 10.1097/00000441-199404000-00009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.