Abstract
Fibroblast growth factors (FGFs) mediate cell growth, differentiation, migration, and morphogenesis by binding to the extracellular domain of cell surface receptors, triggering receptor tyrosine phosphorylation and signal transduction [1–5]. FGF-homologous factors (FHFs) were discovered within vertebrate DNA sequence databases by virtue of their sequence similarity to FGFs [3,6,7], but the mechanism of FHF action has not been reported. We show here that FHF-1 is associated with the MAP kinase (MAPK) scaffold protein Islet Brain-2 (IB2) [8] in the brain and in specific cell lines. FHF/IB2 interaction is highly specific, as FHFs do not bind to the related scaffold protein IB1(JIP-1b) [9,10], nor can FGF-1 bind to IB2. We further show that FHFs enable IB2 to recruit a specific MAPK in transfected cells, and our data suggest that the scaffolds IB1 and IB2 have different MAPK specificities. Hence, FHFs are intracellular components of a tissue-specific protein kinase signalling module.
Results
FHFs-1,2,3,4, also designated FGFs-12, 13,11,14, respectively, bear sequence similarity to the core homology domain of FGFs, suggesting that FHFs and classical FGFs have related tertiary structures. However, FHFs share features that set them apart from other FGFs. First, unlike prior identified FGFs, FHFs bear sequence similarity to one another in both the N-terminal and C-terminal extensions flanking the core homology domain (Figure 1). Second, unlike most FGFs, FHF proteins are synthesized without secretion signal sequences and are detected within native or transfected expressing cells [6,11]. Lastly, functional interactions between FHFs and FGF receptors (FGFRs) have not been described. The intracellular distribution of FHFs and our own failures to detect FHF/FGFR interactions compelled us to search for an alternative FHF function inside cells.
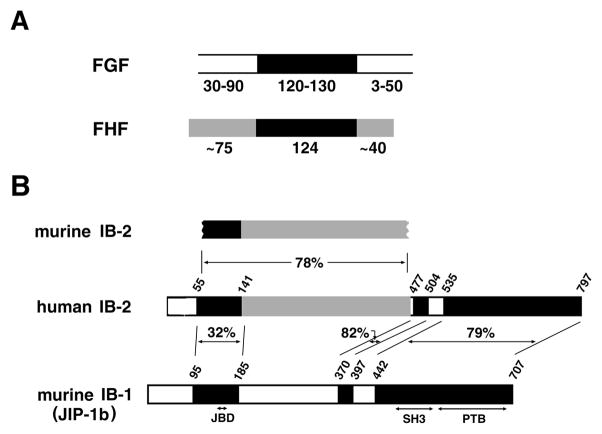
Figure 1.
FHF/FGF and IB2/IB1 sequence similarities. A, Relatedness of FHF and FGF proteins. The core homology region shared by FGFs and FHFs is denoted in black. Additional regions of sequence similarity among FHFs are in grey. The lengths (in residues) of cores and flanking regions are indicated. B, Alignment of IB2 with IB1(JIP-1b). Human IB2 is aligned with the cloned segment of murine IB2 and with IB1. Three regions of IB2/IB1 similarity are blackened, and the percent amino acid sequence identities indicated. Grey denotes additional sequence similarity between human and murine IB2. PTB, SH3, and JNK-binding (JBD) domains of IB1 are indicated.
From a yeast two-hybrid library screen for proteins which could interact with FHF-2, we isolated a partial murine cDNA encoding a protein segment homologous to the recently described human MAPK scaffold protein Islet-Brain-2 (Figure 1), which is expressed predominently in pancreatic islet cells and in the brain [8,12]. The isolated fragment of murine IB2 interacted with both FHF-1 and FHF-2, but not FGF-1, in yeast protein interaction assays (data not shown). IB2 bears sequence and functional similarities to a related scaffold protein, IB1(JIP-1b) (Figure 1)[9,10]. While FHFs interact with IB2 and FGFs interact with FGFRs, there is no detectable sequence similarity between IB2 and FGFRs.
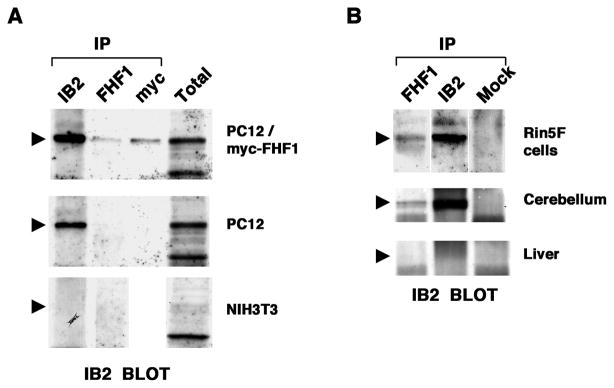
FHFs and IB2 are most prominently expressed in specific developing and mature neurons of the central and peripheral nervous systems [6–8,11,12]. Their coexpression raised the possibility that FHFs and IB2 form native complexes. In order to assay for endogenous FHF/IB2 complexes, we first assayed for FHF1 and IB2 gene expression in selected tissues and cell lines by RT-PCR and RNA blot hybridization analyses (data not shown). These data formed the basis for control immunoprecipitation/immunoblot tests on lysates of NIH3T3 cells (IB2 RNA−, FHF-1 RNA−), PC12 cells (IB2 RNA+, FHF-1 RNA−) and derivative myc-tagged FHF-1-transfected PC12 cells. Immunoprecipitation from FHF-1-transfected PC12 cells with antibodies to either FHF-1 or myc-tag coprecipitated endogenous IB2 as detected by immunoblotting with anti-IB2 (Figure 2A). The lack of IB2 or comigrating background bands in anti-FHF1 precipitates from the parental PC12 cells and 3T3 cells (Fig. 2A) demonstrated the specificity of these antibodies for FHF/IB2 complex detection. Subsequent immunoprecipitation and immunoblot analysis of lysates from insulinoma RIN5F cells (IB2 RNA+/FHF-1 RNA+), whole cerebellum (IB2 RNA+/FHF-1 RNA+) and liver (IB2 RNA−/FHF-1 RNA−) detected a portion of the total IB2 protein in anti-FHF-1 precipitates from both RIN5F cells and cerebellum (Figure 2B). These data demonstrate the existence of endogenous FHF/IB2 complexes.
Figure 2.
FHF-1/IB2 complexes in cell lines and brain. A, Complexes in FHF-transfected PC12 cells. Lysates from NIH3T3 cells (IB2 RNA−FHF-1 RNA−), PC12 cells (IB2 RNA+FHF-1 RNA−) and a stably transfected clonal derivative expressing myc-tagged FHF-1 were immunoprecipitated with antibodies against IB2, FHF-1, or myc-tag, and immunoprecipitates or total lysates were electrophoresed and immunoblot probed with anti-IB2. Anti-FHF-1 or anti-myc coprecipitated IB2 only from FHF1-transfected PC12 cells. Arrowheads mark position of IB2. B, Native FHF-1/IB2 complexes. Lysates from RIN5F insulinoma cells (IB2 RNA+FHF-1 RNA+), rat cerebellum (IB2 RNA+FHF-1 RNA+), and liver (IB2 RNA−FHF-1 RNA−) were immunoprecipitated with antibodies to IB2, FHF-1, or an unrelated antigen (SNT-1, designated Mock) and immunoblotted for IB2, as in A. Anti-FHF-1 coprecipitated IB2 from RIN5F cells and cerebellum.
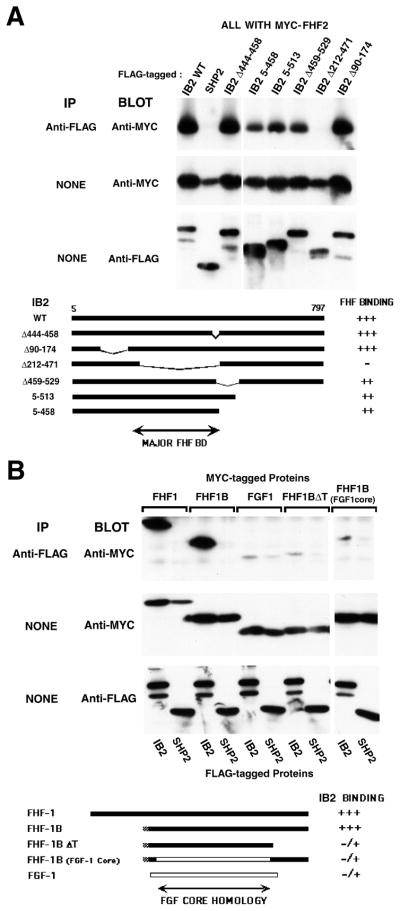
The FHF-binding domain in IB2 was mapped by communoprecipitation/immunoblot analysis from 293T cells cotransfected with FHF-2 and deletion mutants of IB2. A major FHF-binding domain was identified within the region spanning IB2 residues 212–471, as deletion of this region almost completely abolished FHF-2 interaction, while deletions of downstream or upstream regions had far lesser or undetectable impact upon FHF-2 binding (Figure 3A). IB2 residues 212–471 were also sufficient for strong interaction with FHF in cotransfected 293T cells and between bacterially expressed proteins incubated in vitro (data not shown). The FHF-binding domain on IB2 bears no sequence similarity to the related scaffold IB1 (Figure 1). Not surprisingly, IB1 could not interact with FHFs in transfected 293T cells (Figure 4A) nor in yeast protein interaction assays (data not shown).
Figure 3.
Sequence requirements for FHF/IB2 interaction. A, Major FHF-binding domain on IB2 is unrelated to IB1. Total lysates or M2 anti-flag immunoprecipitates from transfected 293T cells expressing flag-tagged wild-type IB2, mutant IB2 derivatives, or SHP-2 together with myc-FHF-2 were electrophoresed, and blots probed with 9E10 anti-myc and rabbit anti-flag antibodies. Schematic depicts alignment of mutant IB2 proteins and relative magnitude of interactions with FHF-2. B, FGF core homology region and C-terminus of FHF-1 are required for IB2 interaction. Total lysates or M2 anti-flag immunoprecipitates from transfected 293T cells expressing flag-tagged IB2 or SHP-2 and myc-tagged FGF-1, FHF-1, FHF-1B, FHF-1BFGF1core, or FHF-1BΔT were electrophoresed, and blots probed with anti-myc and anti-flag antibodies. Schematic shows different FHF and FGF proteins employed and relative magnitude of interaction with IB2. White bar, FGF-1 derived sequence; black bar, FHF-1 derived sequence.
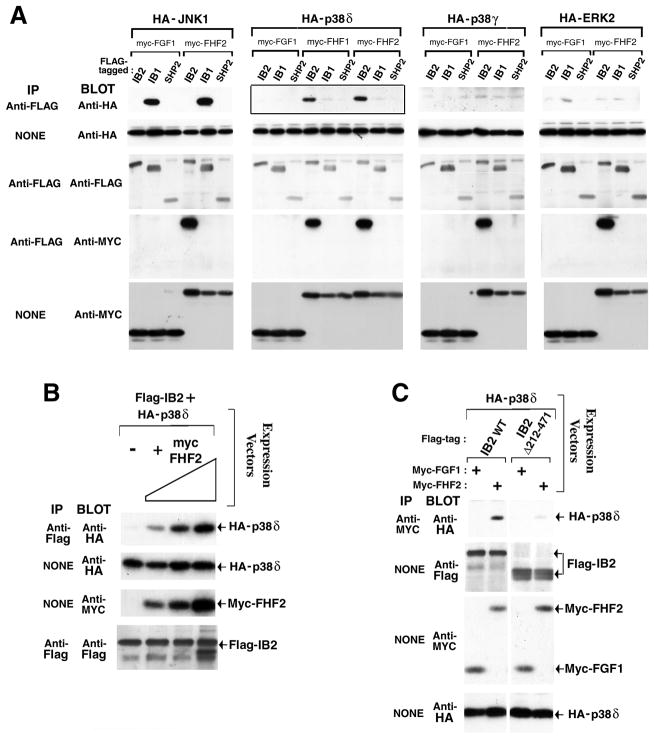
Figure 4.
FHFs promote IB2 interaction with p38δ MAPK. A, FHF-dependent IB2/p38δ interaction. Lysates from transfected 293T cells expressing flag-tagged IB2, IB1, or SHP-2, myc-tagged FGF-1, FHF-1, or FHF-2, and HA-tagged MAPKs (p38δ, p38γ, ERK-2, or JNK-1) were assayed for scaffold-associated kinases and scaffold-associated FHFs by anti-flag immunoprecipitation and anti-HA or anti-myc immunoblot, respectively. Protein expression was assayed by direct immunoblots with appropriate antibodies. FHF-dependent IB2/p38δ interaction (solid-boxed panel) is highlighted. B, IB2/p38δ interaction is FHF-2 dose dependent. Expression plasmids for flag-IB2 (0.12 μg), HA-p38δ (0.02 μg), and myc-FHF-2 (0, 0.12, 0.35, or 1.2 μg) were cotransfected into 293T cells, and IB2/p38δ complexes were detected by anti-flag immunoprecipitation and anti-HA immuoblot. IB2, p38δ, and FHF expression levels were also assayed on immunoblots. C, Heterotrimeric FHF/IB2/p38δ complexes. Anti-myc immunoprecipitates from transfected 293T cells expressing HA-p38δ, myc-tagged FGF-1 or FHF-2, and flag-tagged IB2 or IB2Δ212–471 were immunoblot probed with anti-HA to detect wild-type IB2-dependent FHF/p38δ interaction. Protein expression levels were assayed on anti-flag, anti-myc, and anti-HA immunoblots.
Variant and mutant derivatives of FHF-1 were tested for their ability to interact with IB2. FHF-1 and FHF-1B are alternative products of the same gene resulting from different first coding exon usage [7]; FHF-1B is 62 residues shorter in length than FHF-1 and bears only 14 residues N-terminal to the FGF core homology domain. FHF-1B complexed with IB2 as effectively as did FHF-1 (Figure 3B), demonstrating that the long N-terminal extension of FHF-1 is not required for IB2 interaction. By contrast, FHF-1BΔT bearing an engineered deletion of the C-terminal 39 residues downstream of the core failed to bind IB2 appreciably. However, sequences C-terminal to the core were not sufficient for IB2 interaction (data not shown). To test the importance of the core region, FHF-1B was mutated by replacing its core homology region with that of FGF-1. The resultant chimera, FHF-1BFGF1core, also failed to appreciably interact with IB2 (Figure 3B). These data demonstrate the participation of both the core and C-terminal extension in mediating FHF interaction with IB2. Hence, through gene duplication and sequence divergence, the FGF core homology region has been exploited for interactions with unrelated FGF extracellular or FHF intracellular binding partners.
We have analyzed the biochemical consequence of FHF/IB2 interaction within the context of IB2’s function as a MAPK scaffold protein. MAPK scaffold proteins are thought to enhance signalling efficiency and specificity by bringing together specific MAPKs and upstream kinases [13,14], as best exemplified by the yeast STE5 scaffold protein. STE5 interacts with pheromone receptor-coupled Gβγ as well as a specific set of three protein kinases which establish a MAP kinase cascade MAPKKK(MAPK kinase kinase) --> MAPKK (MAPK kinase) --> MAPK [15–17]. STE5 thereby coordinates the activation of a specific MAPK, FUS3, and as a consequence, the regulation of specific target genes in response to pheromone stimulation. Among vertebrate MAPK scaffolds, IB1(JIP-1b) binds the MAPKKKs MLK3 or DLK, the MAPKK MKK7, and Jun N-terminal kinase (JNK) MAPKs, and IB1 facilitates JNK activation in cells transfected with MLK3 or MKK7 [9].
We tested the influence of FHFs upon the binding of specific MAPKs to IB2, and compared the MAPK binding specificity of IB2 to that of IB1. As negative controls to FHF-1 transfections, cells were alternatively transfected with FGF-1, which is also localized intracellularly [1], but cannot interact with IB2. Confirming and extending previous findings [9], IB1 efficiently bound JNK1, but not ERK-2, p38γ nor p38δ (Figure 4A). By contrast, IB2 expressed at levels comparable to IB1 failed to efficiently bind any MAPK tested when FHFs were absent (Figure 3A). In the presence of FHF-2, IB2 acquired the ability to complex with p38δ [18–20], but not with other MAPKs tested, including JNK1 (Figure 4A). FHF-1 could also promote IB2/p38δ interaction (Figure 4A). Additionally, FHF-2 promoted IB2/p38δ interaction in a dose-dependent manner (Figure 4B).
FHF/IB2 interaction and FHF-dependent IB2/p38δ interaction suggested the formation of a FHF/IB2/p38δ trimeric complex. Evidence for such trimeric complexes was obtained using lysates from cells cotransfected with FHF-2, p38δ, and either wild-type IB2 or a mutant derivative lacking the major FHF-binding domain, IB2Δ212–471. Immunoprecipitation of FHF-2 coprecipitated p38δ more efficiently in the presence of wild-type IB2 as opposed to IB2Δ212–471 (Figure 4C), supporting the existence of the trimeric complex. These data show that FHFs have the potential to serve as cofactors for MAPK recruitment to the IB2 scaffold.
Discussion
The core homology region of FGFs assumes a β-trefoil fold which possesses the ability to cluster and activate FGF receptors [21–23]. While FHF structure has not been described, FHF amino acid similarity to FGFs, most notably at all nine residues which define the hydrophobic core of the fold [21], suggests that FHFs and FGFs assume similar tertiary structures. Nevertheless, the core-homology segment of FHF is required for interaction with IB2, and the homologous segment of FGF-1 cannot substitute (Figures 3B, 4A). Reciprocally, FHFs-1 and -2 cannot activate any of the known FGF receptors which we have tested, including FGFR1(IIIc), R2(IIIb), R2(IIIc), R3(IIIb), and R4 (M.G., unpublished data). Our data demonstrate that FGF gene duplication and divergence in vertebrates has generated sequence-related proteins which can interact with unrelated intracellular or extracellular protein targets. With the ongoing rapid identification of new genes through whole genome sequence initiatives, our findings should sound a cautionary note regarding inference of gene function based upon sequence homology.
We have shown that FHFs enable IB2 to recruit a specific MAPK, p38δ, in transfected cells. FHFs failed to interact with IB1 or modulate MAPK recruitment to this related scaffold. The IB2 scaffold also interacts with MLK3, DLK and MKK7 [8,12](and J.S., data not shown). As MKK7 is a substrate for MLK3 or DLK [24] and p38δ is a substrate for MKK7 [25] (and J.S., data not shown), our data suggest that FHFs support the assembly of a signalling module for p38δ activation. While these findings do not exclude potential interaction of IB2 with other MAPKs, they support the conclusion that IB1 and IB2 have different MAPK binding specificities (Figure 4A). By contrast, the stimuli which engage each of these scaffolds may be similar. Both scaffolds can interact with the same upstream kinases. Furthermore, both scaffolds interact with the Rho GTPase p190 guanine nucleotide release factor [26] and share affinity for the cytoplasmic tails of several lipoprotein receptors [27]. We speculate that neurons and pancreatic endocrine cells expressing FHFs and IB2 along with IB1 [6–12] respond to specific stimuli which coordinately engage both scaffolds to activate different MAP kinases.
Materials and Methods
Yeast strain CG-1945 was transformed with bait plasmid pAS2-1/FHF-2-HA which expresses HA-tagged FHF-2 fused to GAL4 DNA binding domain. A GAL4 activation domain-tagged cDNA library from day E11.5 whole mouse embryo RNA (Clontech Labs) was transformed into CG1945:pAS2-1/FHF2-HA and plated for histidine prototrophs (Trp−Leu−His−, 2 mM 3-aminotriazole), with some cells used to score plasmid transformation efficiency (Trp−Leu−). One HIS+ clone among 6 × 106 primary transformants also tested positive for βgalactosidase activity and harbored a partial murine IB2 cDNA clone. Rabbits were immunized with either bacterially expressed human IB2 protein fragment (residues 77–458) or with carrier-conjugated synthetic peptide corresponding to the C-terminus of murine FHF-1 (residues 234–243). Antibodies were affinity purified with corresponding protein- or peptide-agarose resins. Antibody specificities were assayed by immunoprecipitation of epitope tagged proteins expressed in transfected 293T cells. IB2 antibodies recognize IB2, but not IB1. FHF-1 antibodies recognize FHF-1, weakly crossreact with FHF-2, and do not recognize FGF-1, reflecting the C-terminal homology between FHFs not shared with FGFs.
Supplementary Material
Acknowledgments
We thank Xiuyan Wang and Shahryar Saba for valuable technical assistance. We are grateful to Silvio Gutkind for generously providing kinase cDNAs and to Ravi Iyengar and Stuart Aaronson for comments on the manuscript. We thank Robert Lazzarini and the Brookdale Center for Molecular Biology for institutional funds and moral support. This work was supported by a March of Dimes Birth Defects Foundation grant and PHS grants R01-GM59432 and R01-NS39906 awarded to M.G.
Footnotes
A more comprehensive description of all Materials and Methods is available with the electronic version of this article at http://current-biology.com/supmatin.htm.
References
- 1.Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–65. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 2.Goldfarb M. Functions of FGFs in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 3.Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D. Of worms and men: an evolutionary perspective on the fibroblast growth factor (FGF) and FGF receptor families. J Mol Evol. 1997;44:43–56. doi: 10.1007/pl00006120. [DOI] [PubMed] [Google Scholar]
- 4.Bellot F, Crumley G, Kaplow JM, Schlessinger J, Jaye M, Dionne CA. Ligand-induced transphosphorylation between different FGF receptors. Embo J. 1991;10:2849–54. doi: 10.1002/j.1460-2075.1991.tb07834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–7. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 6.Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, et al. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci U S A. 1996;93:9850–7. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartung H, Feldman B, Lovec H, Coulier F, Birnbaum D, Goldfarb M. Murine FGF-12 and FGF-13: expression in embryonic nervous system, connective tissue and heart. Mech Dev. 1997;64:31–9. doi: 10.1016/s0925-4773(97)00042-7. [DOI] [PubMed] [Google Scholar]
- 8.Negri S, Oberson A, Steinmann M, Sauser C, Nicod P, Waeber G, et al. cDNA cloning and mapping of a novel islet-brain/JNK-interacting protein. Genomics. 2000;64:324–30. doi: 10.1006/geno.2000.6129. [DOI] [PubMed] [Google Scholar]
- 9.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–4. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 10.Bonny C, Nicod P, Waeber G. IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. J Biol Chem. 1998;273:1843–6. doi: 10.1074/jbc.273.4.1843. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, McEwen DG, Ornitz DM. Subcellular and developmental expression of alternatively spliced forms of fibroblast growth factor 14. Mech Dev. 2000;90:283–7. doi: 10.1016/s0925-4773(99)00241-5. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda J, Whitmarsh AJ, Cavanagh J, Sharma M, Davis RJ. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–54. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481–5. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 14.Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–6. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 15.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 16.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci U S A. 1994;91:7762–6. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–97. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XS, Diener K, Manthey CL, Wang S, Rosenzweig B, Bray J, et al. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–74. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, et al. Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p38delta. J Biol Chem. 1997;272:30122–8. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- 20.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases. EMBO J. 1997;16:3563–71. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, Komiya H, Chirino A, Faham S, Fox GM, Arakawa T, et al. Three-dimensional structures of acidic and basic fibroblast growth factors. Science. 1991;251:90–3. doi: 10.1126/science.1702556. [DOI] [PubMed] [Google Scholar]
- 22.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–50. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 23.Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci U S A. 2000;97:49–54. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merritt SE, Mata M, Nihalani D, Zhu C, Hu X, Holzman LB. The mixed lineage kinase DLK utilizes MKK7 and not MKK4 substrate. J Biol Chem. 1999;274:10195–202. doi: 10.1074/jbc.274.15.10195. [DOI] [PubMed] [Google Scholar]
- 25.Hu MC, Wang YP, Mikhail A, Qiu WR, Tan TH. Murine p38-delta mitogen-activated protein kinase, a developmentally regulated protein kinase that is activated by stress and proinflammatory cytokines. J Biol Chem. 1999;274:7095–102. doi: 10.1074/jbc.274.11.7095. [DOI] [PubMed] [Google Scholar]
- 26.Meyer D, Liu A, Margolis B. Interaction of c-Jun amino-terminal kinase interacting protein-1 with p190 rhoGEF and its localization in differentiated neurons. J Biol Chem. 1999;274:35113–8. doi: 10.1074/jbc.274.49.35113. [DOI] [PubMed] [Google Scholar]
- 27.Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, et al. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem. 2000;275:25616–24. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.