Abstract
Hair is a unique mammalian trait that is absent in all other animal forms. Hairlessness is rare in mammals and humans are exceptional among primates in lacking dense layer of hair covering. HR was the first gene identified to be implicated in hair-cycle regulation. Point mutations in HR lead to congenital human hair loss, which results in the complete loss of body and scalp hairs. HR functions are indispensable for initiation of postnatal hair follicular cycling. This study investigates the phylogenetic history and analyzes the protein evolutionary rate to provide useful insight into the molecular evolution of HR. The data demonstrates an acceleration of HR sequence evolution in human branch and suggests that the ability of HR protein to mediate postnatal hair-cycling has been altered in the course of human evolution. In particular those residues were pinpointed which should be regarded as target of positive Darwinian selection during human evolution.
Hair is a defining characteristic of mammals and their evolutionary origin is presumably one of the key steps that contributed significantly to the rapid radiation of mammals and their rise to become the dominant terrestrial vertebrate during late Triassic1. All mammals have hairs, with the exception of some, including whales, dolphins, armadillos and few others only partly covered with hairs. Being soft and decomposable, hairs are unavailable to paleontologist in fossil record and therefore their phylogenetic origin remains highly speculative. As hairs are unique to mammals and does not occur in other amniotes, they might arise specifically within late Triassic therapsid lineage (ancestor of modern mammals/mammaliaforms) approximately 200 million years ago2. The selective forces behind the origin of hairs also remain elusive. The potential selective advantages that may be responsible for the origin of thick coat of hair, the pelage, include the heat-insulating function in primitive homeothermic mammals3. Other functions of hairs include the sensory function, sexual dimorphism, attraction of mates, and skin protection.
Hairs morphology differs considerably among closely related mammalian taxa and they are highly plastic in terms of adaptation to habitat condition4. Despite of diverse macromorphology the hairs present same structural patterns throughout the class. The hair shaft is a keratinized cylindrical filament of different configuration. The outer surface of the shaft is often covered with single or multilayer cuticle. Beneath the cuticle is the cortex, whereas medullary layer constituting the core of the hair. An important aspect of hair evolution is the considerable reduction in hair cover in adult humans during their recent history (after humans-African apes split)5. Naked skin might worked as body cooling system to facilitate efficient heat emission (prevent thermal damage) in response to establishment of bipedalism and large relative brain size in hominids6.
In most mammals the hair cover need constant supply of new hairs to perform functions like, heat retention, attraction of mates and protection of skin. To produce new hairs primary hair follicles (established during early development) goes through a cycle of activity divided into three phases, i.e. growth phase (anagen), destructive phase (catagen) and resting phase (telogen)7. During anagen the hair shaft emerges from the skin surface due to the continued proliferation and differentiation of cells in the hair papilla at the base of the hair. During catagen the hair generating cells undergoes apoptosis and thus entering the degeneration stage. The resting phase follows the destructive phase, during which the hair shaft does not grow but stays attached to the follicle. At the end of telogen the follicle stem cells starts proliferating and the growth stage begins again. A number of signaling pathways/molecules have been implicated in regulating different steps of hair follicle cycling7,8. For instance, Wnt/β-catenin, BMP and Shh pathways act as anagen-stimulating signals, whereas the catagen is induced by TGFβ family pathway and growth factors such as FGF5 and EGF. Key molecular players for anagen maintenance include, IGF1, HGF and VEGF.
Alopecia universalis congenita (AUC) is characterized by the absence of scalp and body hairs causing complete baldness9. Initial hair growth is normal, but after birth once the hair is shed the follicles fails to regenerate and hair loss becomes permanent10. This led to the conclusion that gene underlying AUC is highly specific mediator of hair follicle cycling. Mutations in the human hairless gene (HR) on chromosome 8p12 have been associated with this disease phenotype through genetic linkage analysis11,12. Genetic studies with rodents and human hairless gene have revealed molecular mechanisms by which HR functions in hair development and growth. HR protein has been shown to interact with multiple nuclear receptors, including thyroid hormone receptor (TR), the retinoic acid receptor-related orphan receptors (ROR) and the vitamin D receptors (VDR)13,14,15,16. HR also interacts with histone deacetylases (HDACs), modifies chromatin structure and resulting in transcriptional repression17. During hair cycling in mammals the HR protein regulates hair follicle regeneration (telogen to anagen transition) by promoting Wnt signaling. In HR mutants overexpression of Wnt signaling inhibitors occurs, preventing the Wnt pathway and resulting in failure of hair follicles to regrow18. Thus initial hair growth is normal (during early development) but once the hair is shed it does not grow back resulting in AUC phenotype. This observation implicate the mammalian HR as one of the master regulator of hair cycle which is indispensable for telogen to anagen transition and thus to reinitiate postnatal hair growth19,20.
This study examines the molecular evolution of HR and provides a well defined phylogeny, which infer the orthologs and paralogs and reconstruct its history. The gene duplication history establishes a very distant relationship between HR and its putative paralogous counterparts KDM3A, KDM3B and JMJD1C. Phylogenetic tree confirms the presence of HR in all hairy animals (therian and prototherian), but no recognizable ortholog of mammalian HR was found in any of the non-mammalian vertebrate animal analyzed. This intriguing observation, suggested a key role of HR in hair evolution during mammalian history. In light of this interest, a comparative sequence analysis was performed to estimate the functional constraints on primates, rodents and carnivores HR. Evolutionary rate difference is coupled with structural and biochemical information to infer for potential functional changes at the sequence level among primate HR. In addition variations in domain topologies were explored by comparative analysis of known functional domains of HR protein.
Results
Phylogenetic analysis
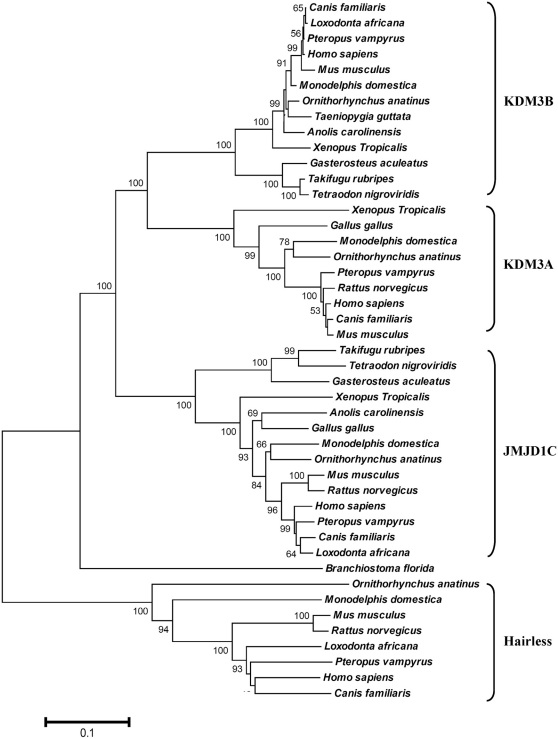
Evolutionary relationship among human HR and JmjC domain containing its putative paralogues, KDM3A, KDM3B and JMJD1C, was estimated through ML and NJ methods (Figure 1 and see Supplementary Figure). Protein sequences from representative members of teleost and tetrapod lineages were subjected to phylogenetic analysis. Amphioxus sequence was used as closest invertebrate relative of vertebrate JmjC-containing proteins. ML and NJ topologies are identical (Figure 1 and see Supplementary Figure) with branching pattern of the type (KDM3A, KDM3B) JMJD1C) invertebrate) HR))). Vertebrate, KDM3A, KDM3B and JMJD1C proteins showing the topology of the form (AB)(C) and clustered with amphioxus sequence. Cluster of HR proteins falling outside the subgroup formed by KDM3A/KDM3B/JMJD1C and amphioxus protein sequences. This pattern received the highly significant bootstrap support (100%). The tree branching pattern suggests that first duplication might predate the vertebrate-cephalochordate split producing ancestral gene of KDM3A/KDM3B/JMJD1C subgroup and HR lineage, whereas the subsequent two duplications events producing KDM3A, KDM3B and JMJD1C might have occurred within the time window of vertebrate-cephalochordate and tetrapod-teleost divergence.
Figure 1. The evolutionary history was inferred using the Neighbor-Joining method.
Uncorrected p-distance was used. Numbers on branches represent bootstrap values (based on 1000 replications) supporting that branch; only the values ≥ 50% are presented here. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 399 positions in the final dataset. Scale bar shows amino acid substitution per site.
The gene phylogeny clearly suggests that KDM3A/ KDM3B /JMJD1C are closely related duplicate genes whereas despite having a shared JmjC domain, HR lineage is very distantly related to this subgroup. Furthermore, the ML and NJ topologies indicates that HR is present in all the three main infraclass taxa of mammals, i.e. monotremes (platypus), metatherians (opossum) and eutherians (placental mammals) but missing from all the non-mammalian vertebrates analyzed (bird, reptile, amphibian and teleost fish) (Figure 1). Thus, this phylogeny reinforces the initial BLAST searches of the NCBI and Ensembl, databases with bird, reptile, amphibian and teleost fish, which found no non-mammalian HR. Absence of HR might suggest evolutionary loss or alternatively owing to relived selective constraints, orthologs of this gene in non-mammalian vertebrates might have diverged to such an extent that they are no longer identifiable through BLAST based similarity searches.
The phylogeny also indicates the absence of KDM3A ortholog form teleost fish lineage (Figure 1). However the tree branching order suggests that KDM3A along with its closest homolog KDM3B might have been originated by a duplication event prior to tetrapod-teleost split. Subsequently KDM3B was retained in both teleosts and tetrapods, whereas evolutionary loss of KDM3A might had occurred in the lineage leading to teleosts.
Comparing evolutionary rate of HR gene among various orders of the class Mammalia
In order to estimate the evolutionary rate differences among various groups of placental mammals the orthologous coding sequences of HR from representative members of primates (human, gorilla and marmoset) rodents (mouse, rat and kangaroo rat) and carnivores (cat, dog, panda) were obtained. Nonsynonymous (Ka) and synonymous (Ks) rates were estimated for primates based on human-gorilla-marmoset comparison, for rodents based on mouse-rat-kangaroo rat comparison, and for carnivores based on cat-dog-panda comparison. The t-value of difference between average Ka and Ks has then been used to estimate the significance to which they differ within each group of placental animals.
The primates Ka-Ks difference is 0.0146 with higher frequency of non-silent (0.0707) than silent (0.0561) substitutions, whereas the rodent Ka-Ks difference is −0.3789 with higher frequency of silent (0.4608) than non-silent (0.0818) substitutions. In carnivores Ka-Ks difference is − 0.165 with higher frequency of silent (0.2147) than non-silent (0.0499) substitutions. In general, Ka value lower than Ks (Ka<Ks) suggests negative selection, i.e. non-silent substitutions have been purged by natural selection, whereas the converse scenario (Ka>Ks) implies positive selection, i.e. advantageous mutations have accumulated during the course of evolution21. However the evidence for positive or negative selection requires the values to be significantly different from each other. Estimation of t-value of difference between average Ka and Ks within each group of placental mammals analyzed indicates that in primates the HR gene experienced replacement substitutions at higher rate than expected by chance (T = 3.175, P < 0.05) and thus under positive selection. In contrast to primates, rodents rate (T = 25.167, P < 0.0001) and the carnivore rate (T = 16.556, P < 0.0001) suggest that in these two lineages the HR gene is under strong selective constraints.
Evolutionary rate of HR within primates
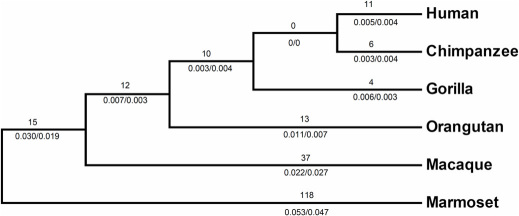
To further explore the molecular evolution in primates, the phylogenetic tree was constructed by using the orthologous coding sequences of HR from human, chimpanzee, gorilla, orangutan, macaque and marmoset. Ka/Ks values were then calculated for each branch of the tree (Figure 2). This analysis revealed that the replacement substitutions outnumber the silent ones for all terminal branches analyzed with the exception of chimpanzee and macaque branches (Figure 2). Estimation of Ka and Ks values for reconstructed ancestral DNA sequences representing all internal nodes on the tree pinpointed three episodes of HR sequence evolution in ancestral lineages of extant primate animals analyzed (Figure 2). In the ancestral lineage leading to hominoids and Old World monkey (macaque), the replacement substitutions outnumber the silent ones (Ka/Ks = 1.58) and is indicative of adoptive selection. Another episode of positive selection, which is the highest found in this analysis (Ka/Ks = 2.33), was identified on the ancestral hominoid lineage. In ancestral African ape (chimpanzee, gorilla and human) lineage (after its divergence from Asian ape/orangutan) Ka/Ks ratio was less than one (Ka/Ks = 0.75) (Figure 2).
Figure 2. Molecular evolution of HR in primates.
Ka and Ks values were estimated for each branch of the HR tree with the reconstructed sequences at ancestral nodes. Number above the lineage indicates the minimum number of amino acid replacements to explain differences among reconstructed sequences. Ka/Ks ratios are shown below branches. Branch lengths are drawn arbitrarily and do not reflect evolutionary time.
Human polymorphisms and tests of departure from neutrality
Ks/Ka values of terminal branches revealed different evolutionary rate of HR among very recently diverged human-chimpanzee lineages (∼6 Mya), with human gene evolving faster (Ka/Ks>1) than its orthologous copy in chimpanzee (Ka/Ks<1). This evidence might suggest that increased rate of amino acid substitutions of human HR (after its divergence from chimpanzee lineage) was driven by positive selection towards functional diversification. To confirm this assumption diversity among human HR is examined by exploiting the large data sets of publicly available human polymorphisms. Information about the dbSNPs (dbSNP build 131) across human HR was obtained from UCSC Genome Browser22. In total 114 SNPs were identified covering the entire human HR interval, with 75 SNPs in intronic regions, 30 SNPs in coding exons, and 7 SNPs were located in untranslated exonic regions. To investigate if the observed patterns of variability in human HR is consistent with the neutral model, the tests of Tajima's D23, Fu and Li's D and Fu and Li's F24 (with or without outgroup) were performed on the panel of 24 validated polymorphisms within the coding intervals of HR (6 non-validated coding SNPs were not included in final analyses) (Supplementary Table). Of these 13 are non-synonymous and 11 are synonymous polymorphisms. Nucleotide diversity (π) is 0.00056 per site and Watterson's θ is 0.00181 per site. Both Tajima's test (D = −2.55327, P < 0.001) and Fu and Li's test without using outgroup (D* = −4.248, P < 0.02; F* = −4.35, P < 0.02) give significant negative values. Similarly, Fu and Li's D and F values using chimpanzee sequence as an outgroup were also significantly negative (D = −3.87, P < 0.02; F = −4.15, P < 0.02). Thus Tajima's D and Fu and Li's D and F (with or without using outgroup) statistics rejects neutrality and indicates a sharp excess of rare polymorphisms. This is expected under positive selection in the human lineage and could explain the observed pattern for the human HR gene variation.
Sliding window analysis of HR
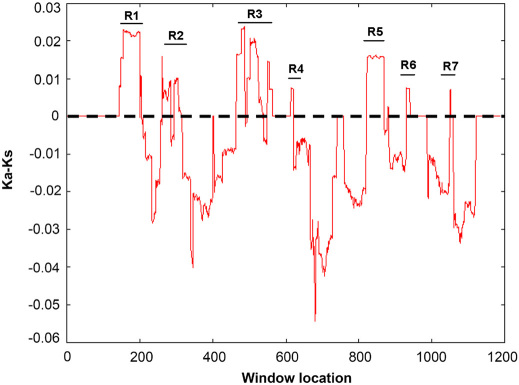
To pinpoint protein segments that might have contributed in functionally diversifying the human HR during its recent history, the sliding window analysis of Ka/Ks was performed along the coding sequence of HR for the human-chimpanzee pairwise comparison (Figure 3).
Figure 3. Sliding window analysis of human-chimpanzee Ka-Ks along the Hairless coding region.
Ka-Ks was calculated at the sliding increment of 10 codons (30 nucleotides). Peaks (R1–R7) above the dotted line indicates an excess of non-synonymous substitutions over the neutral expectations (Ka-Ks > 0).
Sliding window profile revealed seven regions (Figure 3, R1–R7) of high peaks consistent with positive selection, and many regions with very low Ka/Ks values that are consistent with purifying selection (Figure 3). The non-synonymous changes within positively selected (Ka/Ks >1) segments are classified according to their location within human HR protein and their putative physicochemical impact on protein structure/function. It appeared that, after the divergence from last common ancestor, eleven and six amino acid replacements fixed independently in human and chimpanzee HR proteins respectively (Figure 2). Careful comparison of these replacements with inferred human-chimpanzee ancestral residues at corresponding positions revealed that 7/11 (∼64%) replacements in human and 4/6 (∼67%) in chimpanzee might have profound effect on protein structure/function (Table 1). Among protein segments with Ka/Ks >1, region-1 fixed two radical replacements in chimpanzee and one neutral replacement in human lineage within putative nuclear matrix targeting signal, region-2 involves two radical amino acid changes within repression domain-1 (RD1) of human and one radical and one neutral replacement within corresponding segment of chimpanzee protein, region-3 underwent one radical and one neutral replacement in both human and chimpanzee lineage within an uncharacterized medial portion of protein (Table 1). Intriguingly, after divergence from last common ancestor the C-terminal portion of HR protein (residue 645 to carboxy-terminus end) appeared to be unaltered in chimpanzee branch but showed signatures of accelerated evolution in human branch with region-4 fixed one radical change within an uncharacterized segment, region-5 and 6 together involves three radical changes within repression domain-3 (RD3), region-7 experienced no radical amino acid replacement but underwent one physicochemically neutral amino acid change within JmjC domain (Table 1). Thus, this analysis not only pinpointed the amino acid changes that fixed independently in human and chimpanzee HR proteins, but also discriminated the replacements that might have little or no impact on protein structure /function and the ones that are likely to be involved in positive selection and altering the HR protein structure/function in the course of human and chimpanzee evolution.
Table 1. After the divergence of human and chimpanzee lineages, eleven fixed amino-acid changes occurred on the human lineage, whereas six occurred on the chimpanzee lineage.
| Ka/Ks > 1 | Position | Ancestral residue | Replacement in Chimpanzee | Replacement in Human | Neutral/Radical | Region |
|---|---|---|---|---|---|---|
| Region-1 | ||||||
| 175 | R | H | Neutral (0) | Nuclear matrix targeting signal | ||
| 180 | P | A | Radical (-1) | |||
| 186 | W | C | Radical (-2) | |||
| Region-2 | ||||||
| 233 | P | S | Radical (-1) | RD1 | ||
| 276 | R | W | Radical (-3) | |||
| 289 | T | S | Radical (1) | |||
| 287 | V | A | Neutral (0) | |||
| Region-3 | ||||||
| 458 | E | D | Radical (2) | Uncharacterized | ||
| 496 | Q | E | Radical (2) | |||
| 511 | A | T | Neutral (0) | |||
| 534 | A | T | Neutral (0) | |||
| Region-4 | ||||||
| 645 | T | A | Neutral (0) | Uncharacterized | ||
| 660 | T | M | Radical (-1) | |||
| Region-5 | ||||||
| 852 | R | Q | Radical (1) | RD3 | ||
| 854 | W | C | Radical (-2) | |||
| Region-6 | ||||||
| 910 | T | S | Radical (1) | RD3 | ||
| Region-7 | ||||||
| 1022 | A | T | Neutral (0) | JmjC | ||
The table shows the putative ancestral amino acid residues and the type of replacement occurred in each lineage since their divergence. The 6th column depicts the putative physicochemical impact of each replacement on protein structure/function. The numbers within bracket are the log odds scores associated with changing the amino acids. Positive numbers imply a preferred change, zero implies a neutral change, and negative numbers imply an un-preferred change.
HR domain topologies
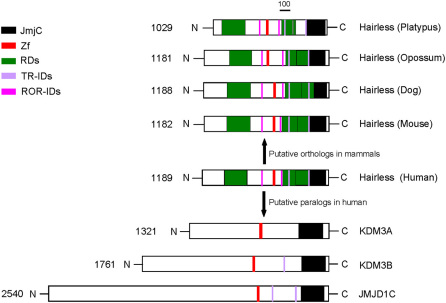
In order to have an insight into comparative domain organization, the key functional domains of the human HR protein were mapped on its paralogous copies in human and orthologous copies in various mammalian lineages. Figure 4 highlights the organization of key functional domains along the human HR protein (JmjC, zinc finger, RDs, TR-IDs, ROR-IDs, LXXL-motif) and their relative topology in human paralogs JMJD1C, KDM3A and KDM3B, and orthologs in mouse, dog, opossum and platypus.
Figure 4. Domain organization of HR protein.
Schematic view of comparative organization of key functional domains of HR across human paralogous proteins and orthologous proteins from phylogenetically distant mammalian species. Protein lengths are drawn approximately to scale and domains are color coded. JmjC; Jumonji C, Zf; Zinc finger, TR-IDs; TR-interacting domains, ROR-IDs; ROR-interacting domains.
The JmjC domain is responsible for histone demethylase activity and is present at the carboxyl terminus of human HR (946–1157 amino acids). This analysis has detected the occurrence of JmjC domain at conserved position in all orthologous and paralogous copies analyzed (Figure 4).
Three major repression domains of human HR protein, including one at the amino terminal end (RD1: 210–426 aa) and two juxtaposed domains at the carboxyl portion (RD2: 730–845 aa; RD3: 845–967 aa) showed the conserved location and span among all orthologous copies analyzed with the exception of platypus where RD3 domain was considerably reduced in length, i.e. human-platypus conservation of RD3 was confined to a protein fragment of 24 amino acids (platypus 725–749 aa) (Figure 4). In contrast the paralogous comparison, suggests the absence of HR repression domains (RD1, RD2 and RD3) counterparts from KDM3A, KDM3B and JMJD1C proteins.
HR binds with ROR (Retinoic acid receptor-related Orphan Receptor) through two motifs containing LxxLL consensus sequence (human; RORID-1: 566–570, RORID-2: 758–762). This interaction leads to transcriptional inhibition by all ROR isoforms (α, β and γ). Multiple sequence alignments, although suggests the presence of these two motifs at conserved location (one on either side of zinc finger domain) across mammalian HR proteins, but fail to identify RORID-1 and RORID-2 in putative paralogous copies of HR protein in human (Figure 4).
HR is known to be an important mediator of thyroid hormone (TH) action in the brain. As corepressor protein, HR interacts with unliganded TH receptors (TRs) and thus triggers transcriptional repression in the absence of TH. HR interacts with TR via two independent domains, i.e. TR-ID1 (human; 786–810 aa) and TR-ID2 (human 1008–1020 aa). Multiple sequence alignments predicted the presence of these two domains at conserved location within carboxyl portion of all mammalian HR proteins analyzed (Figure 4). In addition comparisons of human HR with its paralogous counterparts identify two conserved TR-ID like sequence blocks at the C-terminus portion of JMJD1C protein and one within C-terminus portion of KDM3B protein (Figure 4). However this homology searching fails to identify TR-ID like segments in human KDM3A.
Homology searching demonstrates the conservation of cysteine rich putative C6-type zinc finger domain across mammalian HR proteins and their putative paralogs (JMJD1C, KDM3A and KDM3B) (Figure 4).
Discussion
The increasing availability of genomic sequence data and high throughput annotation of genes from a wide range of animal taxa enables bioinformatics analysis of genes of interest and to provide important insight into their evolutionary link with particular phenotypic trait and association with human disease25,26. Mutations in human hairless gene (HR) have been reported to cause severe type of hair loss phenotype resulting in complete absence of scalp and body hairs11,12. Biochemical and genetic studies have confirmed the pivotal role of HR protein in mammalian hair cycle19,20. This study presents the phylogenetic history of HR based on representative vertebrate genomes and shed insight into the comparative evolutionary rates of HR coding sequence across various mammalian lineages.
The ML and NJ gene phylogenies (see Figure 1 and Supplementary Figure) well defined by bootstrap scores, establishes a distant evolutionary relationship between KDM3A/KDM3B/JMJD1C subfamily and HR. The branching pattern indicates the diversification of KDM3A, KDM3B and JMJD1C during chordate history prior to fish-tetrapod split, whereas the HR clade separated earlier in evolution forming the most basal branch (Figure 1). The close historical/sequence relationship among KDM3A, KDM3B and JMJD1C might indicate biological similarity. This is reflected in their functional resemblance; as these vertebrate proteins are know to share the H3K9 histone demethylase activity and contributes to nuclear receptor mediated gene activation27. The most divergent phylogenetic positioning of HR might account for large differences in the functional aspects of this protein and its putative paralogous counter parts in vertebrates27. In fact, surveying domain topologies revealed highly preserved domain features among orthologous HR proteins; a single C-terminal JmjC domain, a highly conserved C6-type zinc finger (ZF) domain, three repression domains (RD1, RD2 & RD3), two TR-interacting domains and two ROR-interacting domains, whereas comparing HR domain features with JMJD1C, KDM3A and KDM3B identified limited homology (restricted only to JmjC and ZF domains) and thus further confirming considerable functional divergence among HR and its putative paralogs (Figure 4).
BLAST searches complemented by phylogenetic data confirm the absence of HR orthologs from very well sequenced non-mammalian vertebrate genomes (e.g. chicken, zebrafinch, lizard, frog, teleost-fish). This intriguing observation might have two alternative explanations, one is that in the ancestral mammalian lineage HR was subjected to relaxed functional constraints and accelerated sequence evolution which might have allowed the recruitment of this ancient gene for new mammalian-specific biological mechanisms. If this was the case, then HR orthologs are likely to be maintained under different functional constraints in mammalian and non-mammalian vertebrates and thus have diverged to such an extent that they are no longer identifiable through BLAST based similarity searches. Another parsimonious explanation of HR absence in all non-mammalian vertebrate genomes is based on the assumption that the birth of this gene coincides with the origin of mammals. This suggests that the HR gene might have been originated via duplication of the JmjC-domain-containing histone demethylase gene in the ancestor of mammalian vertebrates. In this case, instead of distant evolutionary separation, the remarkable phylogenetic divergence among mammalian HR and its putative ancestral clades (KDM3A/KDM3B/JMJD1C) might be the effect of selective forces which have acted during their independent evolution.
Hairs are typical to mammals and it seems HR as well. Therefore it is conceivable to argue that both explanations, i.e. recruitment of ancient gene for new functions or mammalian specific post-duplication neofunctionalization of one gene copy, reconcile with the indispensable role of HR not only in mammalian hair growth but also in origin of this novel trait (hair cover) in Mesozoic mammalian ancestors. It is of note that, HR might be dispensable for hair follicle development because in mammals (human/mouse) null and hypomorphic HR alleles leads to AUC after a single cycle of normal hair growth28. The hair loss usually begins soon after birth and within first few weeks of postnatal life the animals are completely hairless11. Biochemical and genetic data suggests that HR protein corepressor functions induce hair follicle rest to regrowth (telogen-anagen) transition by promoting Wnt signaling in hair follicles18. In this respect, HR functions are considered indispensable for hair regrowth once they shed after birth (first hair cycle). Therefore it is advocated here that the HR mediated deployment of Wnt signaling in hair cycle was one of the key evolutionary steps that lead to the establishment of postnatal hair cover in ancestral mammalian forms.
The study also examined the molecular evolution of hairless gene specifically in mammalian lineage. For this purpose the average Ka and Ks values were calculated within different phylogenetic groups of mammals. Estimation of statistical significance of difference between average Ka and Ks within each group, show a higher rate of protein evolution in primates than rodents and carnivores. This analysis suggests that positive selection for amino acid replacements occurred during the evolution of primate HR. To test this hypothesis, the Ka and Ks values were estimated for each branch with the reconstructed DNA sequences representing key primate ancestors (Figure 2). This ancestral analysis revealed a period, extending from catarrhine ancestor to hominoid ancestor, when primate HR experienced the increased rate of non-silent substitutions. This episode driven by positive selection is followed by a period when primate HR evolutionary rate was slowed down (purifying selection) considerably in chimpanzee/gorilla/human ancestry. Terminal branches showed an overall trend of inflated evolutionary rate leading to diversification of HR in extant group of primates (Figure 2). Therefore, partitioning of molecular variation along the primate HR tree not only confirm that adoptive evolution of HR occurred during primate evolution but allowed the detection of specific episodes of positive and negative selection and localization of these episodes to distinct branches of tree.
Maximum-likelihood analysis assign eleven and six amino acid replacements to terminal human and chimpanzee branches, suggesting that positive selection continued to alter amino-acid composition of HR after the divergence of these two lineages (Figure 2). To test further the hypothesis of positive selection in humans, neutrality statistics based on the variation within humans was employed. Neutral models of sequence evolution provide guess of expected allele-frequency distinctiveness, and observed patterns can be compared with these. Tajima's D and Fu and Li's D and F (with or without using chimpanzee sequence as an outgroup) values were significantly lower than zero and thus rejects neutrality for HR coding sequence. The results obtained with neutrality statistics can readily be understood in terms of a recent phase of positive selection on human HR.
The sliding window analysis of Ka/Ks coupled with discrimination among fixed radical and conservative substitutions on human and chimpanzee branches not only suggests remarkable heterogeneity in amino acid replacement among positions but also pinpointed seven amino acid sites that are likely to be involved in altering HR protein structure/function in the course of human evolution (Table 1). Given the fact that, functional shifts has been assigned to even single amino acid replacement during human evolution29,30, it is conceivable to argue that seven recovered positively selected positions provide a set of specific candidates for future functional experiments to elucidate biological differences between human versus chimpanzee HR.
With keeping in view the indispensable role of HR in the onset of anagen of the postnatal hair follicle cycle, severe phenotypic effects of mutation in this gene, and strong evidence of positive selection, it seems logical to speculate that there are selective forces at work in primates on the molecular mechanisms regulating postnatal patterns of hair follicle activity. If this is the case, then fine tuning of these mechanisms through subtle changes in protein activity might be one of the contributing factors in brining vital evolutionary changes in postnatal hair follicle morphogenesis over short time scale to match the different environmental and ecological needs.
Hair is a defining feature of mammals performing wide verity of pivotal functions including protection of skin, retention of heat and social interaction. As mentioned earlier, despite of sharing the same basic structural pattern hair macromorphology and distribution pattern differ considerably among taxa31. They show wide adoptive radiation to match the different environmental and ecological requirements. For instance, among traits that distinguish human from all other apes is the reduced hair cover32. Nearly all nonhuman primates are covered with thick furry hair that often differs among phylogenetically closely related species, i.e. it can be thick or thin, short or long, woolly or shaggy, dense or sparse33. Genetic underpinning of hair polymorphism remains elusive and might be quite complex and diverse set of genes are likely to be involved in the process. This study revealed the complex history of important hair cycling mediator HR and suggests that like hairs this gene is also specific to mammals. The data presented here demonstrate that HR is mainly under negative selection in mammals with the exception of primates, where it is driven by bursts of positive selection towards functional diversification. In particular, an accelerated rate of HR sequence evolution was observed in human branch and those amino acid sites were pinpointed that should be regarded as target of positive Darwinian selection during human evolution. This study, therefore, set the stage for future functional and evolutionary studies to elucidate the genetic basis of hair evolution and polymorphism and to explore further the HR role in hair morphogenesis and inherited human disease.
Methods
Sequence acquisition
Putative paralogues of human HR gene are determined by using Ensembl paralogy prediction where maximum likelihood phylogenetic gene trees (generated by TreeBeST) play a central role34. The closest putative orthologous protein sequences of human HR and its paralogs (KDM3A, KDM3B and JMJD1C) in other species were obtained through BLASTP35 searches against the protein database available at Ensembl (http://www.ensembl.org), National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the Joint Genome Institute (http://genome.jgi-psf.org). Confirmation about ancestral-descendents relationship among putative orthologs was done through clustering of homologous proteins within phylogenetic trees. Sequences whose position within a tree was sharply in conflict with the uncontested animal phylogeny were excluded. The list of all used sequences (protein and transcript sequence data) is given as Supplementary data file.
The species that were chosen are Homo sapiens (human), Mus musculus (mouse), Rattus norvegicus (rat), Gallus gallus (chicken), Canis familiaris (dog), Monodelphis domestica (opossum), Xenopus tropicalis (Frog), Erinaceus europaeus (hedgehog), Loxodonta Africana (Elephant), Pteropus vampyrus (Megabat), Ornithorhynchus anatinus (Platypus), Taeniopygia guttata (Zebra Finch), Anolis carolinensis (Anole Lizard), Takifugu rubripes (Fugu), Tetraodon nigroviridis, Gasterosteus aculeatus (Stickleback), Branchiostoma floridae (Amphioxus).
Sequence analysis
The phylogenetic tree of HR family was reconstructed by using the neighbor-joining (NJ) method36,37, the complete deletion option was used to exclude any site which postulated a gap in the sequences. Poisson corrected (PC) amino acid distance and uncorrected proportion (p) of amino acid difference were used as amino acid substitution models. Because both methods produced similar results, only the results from NJ tree based on uncorrected p-distance are presented here. Reliability of the resulting tree topology was tested by the bootstrap method38 (at 1000 pseudoreplicates) which generated the bootstrap probability for each interior branch in the tree. Maximum Likelihood tree was also constructed by using the Whelan And Goldman (WAG) model of amino acid replacement39 (Supplementary Figure). In case of both NJ and ML trees the mammalian HR sequences served as an outgroup to root the remainder of the tree, while the remaining sequences served to root Mammalian HR sequences.
To estimate the evolutionary rates of primate HR the primate phylogenetic tree was constructed using human, chimpanzee, gorilla, orangutan, macaque and marmoset orthologs. Ancestral sequences were inferred for each node of the primate tree by using ML method and WAG model of amino acid evolution, and the amino acid replacements for each branch of the tree were calculated40.
To investigate if the observed patterns of variability in HR sequence in human population is consistent with the neutral model, the tests of Tajima's D23, Fu and Li's D and Fu and Li 's F24 were performed on the panel of 24 coding SNPs downloaded from SNP data (dbSNP build 131) available at UCSC genome browser22 (Supplementary Table). Tests of neutrality were performed using the program DNAsp Version 541.
To detect the regions under positive selection sliding–window analysis of the Ka /Ks ratio was performed on human and chimpanzee HR coding sequences in pairwise comparison42. Ka-Ks was calculated at the sliding increment of 10 codons (30 nucleotides) and the results are obtained in the graph drawn by the GNUPLOT software implemented in SWAKK42. The non-synonymous changes within segments having Ka/Ks >1 are classified according to their physicochemical properties such as charge, polarity, and volume into neutral and radical43,44.
Domains were assigned to the human HR protein as described previously (Thompson CC et al 2009). Clustal W based multiple sequence alignments were used to map the putative positioning of these domains to paralogs of HR protein in human and its orthologs in various mammalian species45.
Supplementary Material
Supplementary Figure
Supplementary Table
Supplementary Data
Acknowledgments
This study was supported by Higher Education Commission (HEC) of Pakistan and the National Center for Bioinformatics, Quaid-i-Azam University, Islamabad.
References
- Maderson P. F. Mammalian skin evolution: a reevaluation. Exp Dermatol 12, 233–236 (2003). [DOI] [PubMed] [Google Scholar]
- Meng J. & Wyss A. R. Multituberculate and other mammal hair recovered from Palaeogene excreta. Nature 385, 712–714 (1997). [DOI] [PubMed] [Google Scholar]
- Brink A. S. Speculations on some advanced mammalian characteristics in the higher mammallike reptiles. Palaeontol. Afr. 4, 77–96 (1956). [Google Scholar]
- Noback C. R. Morphology and phylogeny of hair. Ann N Y Acad Sci 53, 476–492 (1951). [DOI] [PubMed] [Google Scholar]
- Schwartz G. G. & Rosenblum L. A. Allometry of primate hair density and the evolution of human hairlessness. Am J Phys Anthropol 55, 9–12 (1981). [DOI] [PubMed] [Google Scholar]
- Wheeler P. E. The evolution of bipedality and loss of functional body hair in hominids. J Hum Evol 13, 91–98 (1984). [Google Scholar]
- Alonso L. & Fuchs E. The hair cycle. J Cell Sci 119, 391–393 (2006). [DOI] [PubMed] [Google Scholar]
- Schneider M. R., Schmidt-Ullrich R. & Paus R. The hair follicle as a dynamic miniorgan. Curr Biol 19, R132–142 (2009). [DOI] [PubMed] [Google Scholar]
- Cantu J. M. et al. Autosomal recessive inheritance of atrichia congenita. Clin. Genet. 17, 209–212 (1980). [DOI] [PubMed] [Google Scholar]
- Mann S. J. Hair loss and cyst formation in hairless and rhino mutant mice. Anat Rec 170, 485–499 (1971). [DOI] [PubMed] [Google Scholar]
- Ahmad W. et al. Alopecia universalis associated with a mutation in the human hairless gene. Science 279, 720–724 (1998). [DOI] [PubMed] [Google Scholar]
- Nothen M. M. et al. A gene for universal congenital alopecia maps to chromosome 8p21-22. Am J Hum Genet 62, 386–390 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. C. et al. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J Biol Chem 278, 38665–38674 (2003). [DOI] [PubMed] [Google Scholar]
- Moraitis A. N. & Giguere V. The co-repressor hairless protects RORalpha orphan nuclear receptor from proteasome-mediated degradation. J Biol Chem 278, 52511–52518 (2003). [DOI] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V. & Thompson C. C. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol 22, 6831–6841 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G. B. et al. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev 15, 2687–2701 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G. B., Zarach J. M., Sisk J. M. & Thompson C. C. The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain. Mol Endocrinol 16, 2547–2560 (2002). [DOI] [PubMed] [Google Scholar]
- Beaudoin G. M. 3rd, Sisk J. M., Coulombe P. A. & Thompson C. C. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc Natl Acad Sci U S A 102, 14653–14658 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. C. Hairless is a nuclear receptor corepressor essential for skin function. Nucl Recept Signal 7, e010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. C., Sisk J. M. & Beaudoin G. M. 3rd Hairless and Wnt signaling: allies in epithelial stem cell differentiation. Cell Cycle 5, 1913–1917 (2006). [DOI] [PubMed] [Google Scholar]
- Wagner A. Selection and gene duplication: a view from the genome. Genome Biol 3, reviews1012 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita P. A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39, D876–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X. & Li W. H. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi A. A., Goode D. K., Amir S. & Grzeschik K. H. Evolution and functional diversification of the GLI family of transcription factors in vertebrates. Evol Bioinform Online 5, 5–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S. & Kaestner K. H. The evolution of Fox genes and their role in development and disease. Nat Rev Genet 10, 233–240 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Kallin E. M. & Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- Cachon-Gonzalez M. B. et al. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A 91, 7717–7721 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding G. B. & Dean A. M. The structural basis of molecular adaptation. Mol Biol Evol 15, 355–369 (1998). [DOI] [PubMed] [Google Scholar]
- Enard W. et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418, 869–872 (2002). [DOI] [PubMed] [Google Scholar]
- Chernova O. F. [Evolutionary aspects of hair polymorphism]. Izv Akad Nauk Ser Biol, 52–62 (2006). [PubMed] [Google Scholar]
- Carroll S. B. Genetics and the making of Homo sapiens. Nature 422, 849–857 (2003). [DOI] [PubMed] [Google Scholar]
- Montagna W. The Skin of Nonhuman Primates. Integrative and Comparative Biology 12, 109–124 (1972). [Google Scholar]
- Hubbard T. J. et al. Ensembl 2009. Nucleic Acids Res 37, D690–697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Russo C. A. M., Takezaki N. & Nei M. Efficiencies of Different Genes and Different Tree-building Methods in Recovering a Known Vertebrate Phylogeny. Mol. Biol. Evol 13, 525–536 (1996). [DOI] [PubMed] [Google Scholar]
- Felsentein J. Confidence limit on phylogenies: An approach using the bootstrap. Evolution 39, 95–105 (1985). [DOI] [PubMed] [Google Scholar]
- Whelan S. & Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18, 691–699 (2001). [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J. & Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9, 299–306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Liang H., Zhou W. & Landweber L. F. SWAKK: a web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res 34, W382–384 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974). [DOI] [PubMed] [Google Scholar]
- Betts M. J. & Russell R. B. Amino acid properties and consequences of subsitutions. Bioinformatics for Geneticists (2003). [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Table
Supplementary Data